Fur regulation of the capsular polysaccharide
biosynthesis and iron-acquisition systems in
Klebsiella pneumoniae CG43
Ching-Ting Lin,
1Chien-Chen Wu,
2Yu-Sheng Chen,
1Yi-Chyi Lai,
3Chia Chi,
1Jing-Ciao Lin,
1Yeh Chen
4and Hwei-Ling Peng
2Correspondence Ching-Ting Lin gingting@mail.cmu.edu.tw Hwei-Ling Peng hlpeng@mail.nctu.edu.tw Received 23 July 2010 Revised 9 November 2010 Accepted 11 November 2010
1School of Chinese Medicine, China Medical University, Taichung, 40402, Taiwan, ROC 2Department of Biological Science and Technology, National Chiao Tung University, Hsin Chu
30068, Taiwan, ROC
3Department of Microbiology and Immunology, Chung-Shan Medical University, Taichung 40201,
Taiwan, ROC
4Research Institute of Biotechnology, Hungkuang University, Taichung 43302, Taiwan, ROC
The ferric uptake regulator Fur has been reported to repress the expression of rmpA, a regulatory gene for the mucoid phenotype, leading to decreased capsular polysaccharide (CPS)
biosynthesis in Klebsiella pneumoniae CG43. Here, quantitative real-time PCR (qRT-PCR) analyses and electrophoretic mobility shift assays showed that Fur also repressed the expression of the CPS regulatory genes rmpA2 and rcsA. Interestingly, deletion of rmpA or rcsA but not rmpA2 from the Dfur strain was able to suppress the deletion effect of Fur. The availability of extracellular iron affected the amount of CPS, suggesting that Fur regulates CPS biosynthesis in an Fe(II)-dependent manner. Increased production of siderophores was observed in the Dfur strain, suggesting that uptake of extracellular iron in K. pneumoniae is regulated by Fur. Fur titration assays and qRT-PCR analyses demonstrated that at least six of the eight putative iron-acquisition systems, identified by aBLASTsearch in the contig database of K. pneumoniae CG43, were directly repressed by Fur. We conclude that Fur has a dual role in the regulation of CPS biosynthesis and iron acquisition in K. pneumoniae.
INTRODUCTION
Klebsiella pneumoniae is a rod-shaped Gram-negative
bacterium
that
causes
community-acquired
diseases
including pneumonia, bacteraemia, septicaemia, and
urinary and respiratory tract infections, particularly in
immunocompromised patients (Podschun & Ullmann,
1998). In Asian countries, especially in Taiwan and
Korea, K. pneumoniae is the predominant pathogen
responsible for pyogenic liver abscess in diabetic patients
(Han, 1995; Lau et al., 2000; Yang et al., 2009). Among the
virulence factors identified in K. pneumoniae, capsular
polysaccharide (CPS) is considered as the major
determi-nant for K. pneumoniae infections. Pyogenic liver abscess
isolates often carry heavy CPS which could protect the
bacteria from phagocytosis and killing by serum factors
(Lin et al., 2004; Sahly et al., 2000). Apart from the
anti-phagocytic function, Klebsiella CPS also helps bacterial
colonization and biofilm formation at the infection sites
(Boddicker et al., 2006; Favre-Bonte et al., 1999; Moranta
et al., 2010).
The Rcs system is a well-known two-component system
(2CS) that regulates the expression of cps genes in bacteria
(Stout, 1994). The transcription of cps genes is controlled
by the response regulator RcsB in complex with the
auxiliary regulatory protein RcsA (Gottesman & Stout,
1991; Majdalani & Gottesman, 2005). We have
demon-strated that cps expression in K. pneumoniae CG43 is also
affected by the coordinated action of the 2CSs KvgAS,
KvhAS and KvhR, and in this case is independent of RcsB
(Lin et al., 2006). Besides RcsA, the regulators RmpA and
RmpA2 also interact with RcsB for CPS biosynthesis
regulation. Moreover, rmpA expression was repressed by
Fur (ferric uptake regulator), the global regulator for the
expression of iron-acquisition systems (Cheng et al., 2010).
Here we demonstrate that Fur also affects rcsA and rmpA2
expression.
In Escherichia coli, under iron-replete conditions, dimeric
Fur in complex with Fe(II) binds to a 19 bp consensus
Abbreviations: 2CS, two-component system; CPS, capsularpolysac-charide; Dip, 2,2-dipyridyl; CAS, chrome azurol S; EMSA, electrophoretic mobility shift assay; FURTA, Fur titration assay; qRT-PCR, quantitative real-time PCR.
DNA sequence, the Fur box
(GATAATGATwATCATT-ATC; w5A or T), in the promoters of the genes required
for iron uptake, thereby preventing transcription from
these genes (Griggs & Konisky, 1989). The regulation helps
bacteria to avoid iron overload, which may lead to the
formation of hydroxyl radicals. Multiple iron-acquisition
systems are commonly present in bacteria for the uptake of
iron in the environment (Andrews et al., 2003). In an
anaerobic environment, Fe(II) is prevalent and is imported
into the bacterial cytoplasm via the Feo system (Hantke,
2003). However, in aerobic conditions and in mammalian
tissues (in vivo), the majority of iron is found as Fe(III),
and iron in vivo is almost entirely sequestered by
iron-binding proteins (transferrin and lactoferrin) and
haemo-proteins (haemoglobin and myoglobin) (Wandersman &
Delepelaire, 2004).
Bacteria are generally equipped with iron/haem acquisition
systems to transport iron directly from the exogenous iron/
haem sources or release siderophore and haemophore
compounds into the extracellular medium to scavenge
iron/haem
from
various
sources
(Wandersman
&
Delepelaire, 2004). In K. pneumoniae NTUH-K2044, the
expression of the ten putative iron-acquisition genes was
highly upregulated in response to human serum, and
bacterial virulence was decreased by the triple mutation of
siderophore genes (Hsieh et al., 2008). The siderophore
genes iucABCDiutA and iroNDCB have also been reported
to be the determinants of K. pneumoniae-caused liver
abscess (Hsieh et al., 2008; Koczura & Kaznowski, 2003;
Tang et al., 2010). Nevertheless, until now the regulation of
iron-acquisition gene expression in K. pneumoniae has not
been studied.
In this study, we investigated the regulatory roles of Fur on
the expression of the cps regulators RmpA, RmpA2 and
RcsA, and the expression of eight iron-acquisition systems
in K. pneumoniae CG43.
METHODS
Bacterial strains, plasmids and media. Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely cultured at 37uC in Luria–Bertani (LB) medium or M9 minimal medium supplemented with appropriate antibiotics. The antibiotics used include ampicillin (100 mg ml21), kanamycin (25 mg ml21), streptomycin (500 mg ml21) and tetracycline (12.5 mg ml21).
Construction of deletion mutants. Specific gene deletions were introduced into K. pneumoniae CG43 by using an allelic exchange strategy as described by Lai et al. (2003). The pKAS46 system was used in the selection of the mutants (Skorupski & Taylor, 1996), and the mutations were confirmed by PCR and Southern hybridization (data not shown).
Quantitative real-time PCR (qRT-PCR).Total RNA was isolated from bacterial cells grown to early exponential phase by using the RNeasy midi-column (Qiagen) according to the manufacturer’s instructions. RNA was treated with RNase-free DNase I (MoBioPlus) to eliminate DNA contamination. RNA (100 ng) was
reverse-transcribed with the transcriptor first strand cDNA synthesis kit (Roche) by using random primers. qRT-PCR was performed in a Roche LightCycler 1.5 instrument by using LightCycler TaqMan master (Roche). Primers and probes were designed for selected target sequences by using the universal ProbeLibrary assay design center (Roche–Applied science) and are listed in Table 2. Data were analysed using the real-time PCR software of the Roche LightCycler 1.5 instrument. Relative gene expression was quantified by using the comparative threshold cycle 22DDCTmethod with 23S rRNA as the endogenous reference.
Electrophoretic mobility shift assay (EMSA). Recombinant K. pneumoniae Fur protein was expressed in E. coli and purified as described previously (Cheng et al., 2010). DNA fragments of the putative promoter regions of rmpA, rmpA2 and rcsA were amplified by PCR using specific primer sets. The purified His6-Fur was
incubated with 10 ng DNA in a 15 ml solution containing 50 mM Tris/HCl (pH 7.5), 100 mM NaCl, 100 mM dithiothreitol, 200 mM MnCl2 and 1 mg BSA ml21 at room temperature for 20 min. The
samples were then loaded onto 5 % native (non-denaturing) polyacrylamide gel containing 5 % glycerol in 0.56 TB buffer (45 mM Tris/HCl, pH 8.0, 45 mM boric acid) and electrophoresed at 20 mA constant current at 4uC for 2 h. The gel was stained with SYBR Green EMSA stain (Invitrogen), and was then visualized by using a Safe Imager blue-light transilluminator.
Extraction and quantification of CPS. CPS was extracted and quantified as described by Domenico et al. (1989). The glucuronic acid content, representing the amount of K. pneumoniae K2 CPS, was determined from a standard curve of glucuronic acid (Sigma-Aldrich) and was expressed as mg (109 c.f.u.)21 (Blumenkrantz & Asboe-Hansen, 1973).
Identification of the iron-acquisition genes in K. pneumoniae CG43.The ten genes encoding different iron-acquisition systems in K. pneumoniae NTUH-K2044 (Hsieh et al., 2008) were used as query sequences to search for homologues in the K. pneumoniae CG43 contig database (unpublished results from Dr S.-F. Tsai, National Health Research Institutes, Taiwan) as assessed by theBLAST search program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., 1997).
Fur titration assay (FURTA).FURTA was performed according to the method described by Stojiljkovic et al. (1994). DNA sequences containing a putative Fur box were amplified by PCR with specific primer sets and then cloned into pT7-7. The resulting plasmids were introduced into the E. coli strain H1717, and the transformants were plated onto MacConkey-lactose plates containing 100 mg ampicillin ml21 and 30 mM Fe(NH
4)2(SO4)2. The indicator strain H1717
contained a chromosomal fhuF : : lacZ fusion, and a low-affinity Fur box has been identified in the fhuF promoter. The introduction of pT7-7-derived plasmids carrying Fur-binding sequences could thus cause the removal of Fur from the fhuF Fur box (Hantke, 1987). H1717 harbouring pT7-7 was used as a negative control. Colony phenotype was observed after incubation at 37uC for 10 h. A red colony colour (Lac+) denoted a FURTA-positive phenotype and indicated the binding of Fur to the DNA sequence cloned into the pT7-7 plasmid.
Chrome azurol S (CAS) assay. The CAS assay was performed according to the method described by Schwyn & Neilands (1987). Each of the bacterial strains was grown overnight in LB medium, and then 5 ml of culture was added onto a CAS agar plate. After 16 h incubation at 37uC, effects of the bacterial siderophore production could be observed. Siderophore production was apparent as an orange halo around the colonies; absence of a halo indicated the inability to produce siderophores.
Statistical method.An unpaired t-test was used to determine the statistical significance and values of P,0.001 were considered significant. The results of CPS quantification and qRT-PCR analysis were derived from a single experiment representative of three independent experiments. Each sample was assayed in triplicate and the mean activity and standard deviation are presented.
RESULTS
Fur regulates the expression of RmpA, RmpA2
and RcsA
To investigate whether Fur affects the expression of the cps
regulatory proteins RcsA, RcsB, RmpA2, KvgA and KvhR
(Cheng et al., 2010; Lai et al., 2003; Lin et al., 2006), in
addition to RmpA (Cheng et al., 2010), qRT-PCR analyses
were performed to compare expression levels in K.
pneumoniae CG43S3 and its isogenic Dfur strain. As shown
in Fig. 1(a), when the bacteria were grown in LB, the
deletion of fur increased the expression of not only rmpA
but also rmpA2 and rcsA. By contrast, fur deletion appeared
to have no effect on the expression of rcsB, kvgA or kvhR.
Addition of the iron chelator 2,2-dipyridyl (Dip) to the
growth medium also increased the expression of rmpA and
rcsA in the wild-type strain, suggesting that a Fur–Fe(II)
complex is involved in regulating the expression of rmpA
and rcsA. However, rmpA2 expression did not appear to
change, suggesting a novel mechanism that requires further
Table 1. Bacterial strains and plasmids used in this study
Strain or plasmid Description Reference or source
K. pneumoniae
CG43S3 CG43 Smr Lai et al. (2001)
DrmpA CG43S3DrmpA Cheng et al. (2010)
DrmpA2 CG43S3DrmpA2 Lai et al. (2001)
Dfur CG43S3Dfur Cheng et al. (2010)
DrcsA CG43S3DrcsA This study
DrmpADrcsA CG43S3DrmpADrcsA This study
DrmpADrmpA2DrcsA CG43S3DrmpADrmpA2DrcsA This study
DfurDrmpA CG43S3DfurDrmpA This study
DfurDrmpA2 CG43S3DfurDrmpA2 This study
DfurDrcsA CG43S3DfurDrcsA This study
DfurDrmpADrcsA CG43S3DfurDrmpADrcsA This study
DfurDrmpADrmpA2DrcsA CG43S3DfurDrmpADrmpA2DrcsA This study
E. coli
DH5a supE44 DlacU169 (f80 lacZDM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 Hanahan (1983)
BL21-RIL F2ompT hsdSB[rB2mB2]gal dcm [DE3] Laboratory stock
S17-1 l pir hsdR recA pro RP4-2[Tc : : Mu; Km : : Tn7] [lpir] Skorupski & Taylor (1996) H1717 araD139 DlacU169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB fhuF : : l placMu Hantke (1987)
Plasmid
pKAS46 Positive selection suicide vector, rpsL AprKmr Skorupski & Taylor (1996)
pET30a-c His-tagging protein expression vector, Kmr Novagen
yT&A TA cloning vector Yeastern
pRK415 Broad-host-range IncP cloning vector, Tcr Keen et al. (1988)
pT7-7 Cloning vector, Apr Tabor & Richardson (1985)
pfur03 1.7 kb fragment containing an internal 454 bp deletion in fur, cloned into pKAS46 Cheng et al. (2010) prcsA03 2.0 kb fragment containing an internal 620 bp deletion in rcsA, cloned into pKAS46 This study piroB_2 928 bp fragment containing the putative iroBCD promoter, cloned into pT7-7 This study pentC_2 284 bp fragment containing the putative entC promoter, cloned into pT7-7 This study piucA_2 700 bp fragment containing the putative iucABCD promoter, cloned into pT7-7 This study phmuR_2 500 bp fragment containing the putative hmuRSTUV promoter, cloned into pT7-7 This study pfeo_2 564 bp fragment containing the putative feoABC promoter, cloned into pT7-7 This study pfec_2 296 bp fragment containing the putative fecIRA promoter, cloned into pT7-7 This study pfhuA_2 313 bp fragment containing the putative fhuA promoter, cloned into pT7-7 This study psitA_2 283 bp fragment containing the putative sitABCD promoter, cloned into pT7-7 This study pFT01 0.5 kb fragment containing the putative orf1-2 promoter, cloned into pT7-7 This study pFT02 0.9 kb fragment containing the putative orf3-15 promoter, cloned into pT7-7 This study pFT03 0.3 kb fragment containing the putative orf16-17 promoter, cloned into pT7-7 This study pFT04 0.5 kb fragment containing the putative rmpA promoter, cloned into pT7-7 This study pFT05 0.5 kb fragment containing the putative rmpA2 promoter, cloned into pT7-7 This study pFT06 0.5 kb fragment containing the putative rcsA promoter, cloned into pT7-7 This study
Table 2. Primers used in this study
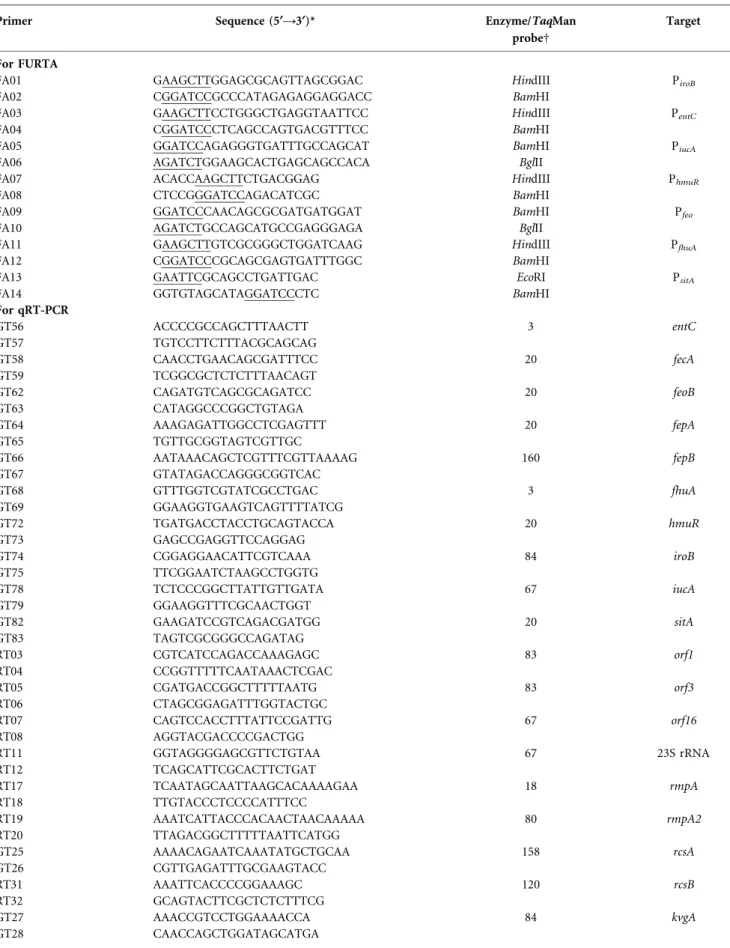
Primer Sequence (5§A3§)* Enzyme/TaqMan
probeD
Target
For FURTA
FA01 GAAGCTTGGAGCGCAGTTAGCGGAC HindIII PiroB
FA02 CGGATCCGCCCATAGAGAGGAGGACC BamHI
FA03 GAAGCTTCCTGGGCTGAGGTAATTCC HindIII PentC
FA04 CGGATCCCTCAGCCAGTGACGTTTCC BamHI
FA05 GGATCCAGAGGGTGATTTGCCAGCAT BamHI PiucA
FA06 AGATCTGGAAGCACTGAGCAGCCACA BglII
FA07 ACACCAAGCTTCTGACGGAG HindIII PhmuR
FA08 CTCCGGGATCCAGACATCGC BamHI
FA09 GGATCCCAACAGCGCGATGATGGAT BamHI Pfeo
FA10 AGATCTGCCAGCATGCCGAGGGAGA BglII
FA11 GAAGCTTGTCGCGGGCTGGATCAAG HindIII PfhuA
FA12 CGGATCCCGCAGCGAGTGATTTGGC BamHI
FA13 GAATTCGCAGCCTGATTGAC EcoRI PsitA
FA14 GGTGTAGCATAGGATCCCTC BamHI
For qRT-PCR GT56 ACCCCGCCAGCTTTAACTT 3 entC GT57 TGTCCTTCTTTACGCAGCAG GT58 CAACCTGAACAGCGATTTCC 20 fecA GT59 TCGGCGCTCTCTTTAACAGT GT62 CAGATGTCAGCGCAGATCC 20 feoB GT63 CATAGGCCCGGCTGTAGA GT64 AAAGAGATTGGCCTCGAGTTT 20 fepA GT65 TGTTGCGGTAGTCGTTGC GT66 AATAAACAGCTCGTTTCGTTAAAAG 160 fepB GT67 GTATAGACCAGGGCGGTCAC GT68 GTTTGGTCGTATCGCCTGAC 3 fhuA GT69 GGAAGGTGAAGTCAGTTTTATCG GT72 TGATGACCTACCTGCAGTACCA 20 hmuR GT73 GAGCCGAGGTTCCAGGAG GT74 CGGAGGAACATTCGTCAAA 84 iroB GT75 TTCGGAATCTAAGCCTGGTG GT78 TCTCCCGGCTTATTGTTGATA 67 iucA GT79 GGAAGGTTTCGCAACTGGT GT82 GAAGATCCGTCAGACGATGG 20 sitA GT83 TAGTCGCGGGCCAGATAG RT03 CGTCATCCAGACCAAAGAGC 83 orf1 RT04 CCGGTTTTTCAATAAACTCGAC RT05 CGATGACCGGCTTTTTAATG 83 orf3 RT06 CTAGCGGAGATTTGGTACTGC RT07 CAGTCCACCTTTATTCCGATTG 67 orf16 RT08 AGGTACGACCCCGACTGG RT11 GGTAGGGGAGCGTTCTGTAA 67 23S rRNA RT12 TCAGCATTCGCACTTCTGAT RT17 TCAATAGCAATTAAGCACAAAAGAA 18 rmpA RT18 TTGTACCCTCCCCATTTCC RT19 AAATCATTACCCACAACTAACAAAAA 80 rmpA2 RT20 TTAGACGGCTTTTTAATTCATGG GT25 AAAACAGAATCAAATATGCTGCAA 158 rcsA GT26 CGTTGAGATTTGCGAAGTACC RT31 AAATTCACCCCGGAAAGC 120 rcsB RT32 GCAGTACTTCGCTCTCTTTCG GT27 AAACCGTCCTGGAAAACCA 84 kvgA GT28 CAACCAGCTGGATAGCATGA
study. Intriguingly, the expression of rmpA, rmpA2 and
rcsA in the Dfur strain were all reduced by iron chelation,
implying that the iron chelator Dip has a non-specific
inhibitory effect on controlling the gene transcription.
As in P
rmpA, the promoter of rmpA, putative Fur box
sequences could be found in the upstream regions of
rmpA2 and rcsA (Fig. 1b), although being less conserved in
the rmpA2 promoter. We performed an EMSA to
determine whether Fur directly affects the expression of
rmpA2 and rcsA. As shown in Fig. 1(c), the purified
recombinant His
6-Fur protein was able to bind to the
upstream regions of rmpA, rmpA2 and rcsA, but not to the
P6 DNA, which did not contain a Fur box (Cheng et al.,
2010). Addition of 200 mM EDTA to the reaction mixture
appeared to abolish the interactions (data not shown),
Table 2. cont.
Primer Sequence (5§A3§)* Enzyme/TaqMan
probeD
Target
GT13 GTATTTTTATTCGCGATGTACTGC 67 kvhR
GT14 GCCTGAACAGCGGAGAGA
*Underlining indicates the nucleotide sequence recognized by the restriction enzyme. DEnzyme refers to the restriction enzyme used in FURTA; TaqMan probe refers to qRT-PCR.
Fig. 1. Fur directly represses the expression of rmpA, rmpA2 and rcsA. (a) qRT-PCR analysis. The K. pneumoniae CG43S3 [pRK415], Dfur [pRK415] and Dfur [pfur] strains were grown overnight in LB both with and without 200 mM Dip, and the relative expression of rmpA, rmpA2, rcsA, rcsB, kvgA and kvhR in bacteria was then measured by qRT-PCR analysis. Values are mean±SDof three independent experiments. (b) DNA sequence alignment between the E. coli typical Fur box and the putative Fur boxes in the upstream regions of rmpA, rmpA2 and rcsA. The relative positions to the translational start sites are indicated. (c) EMSA of the recombinant His6-Fur and its target promoters. DNA of the upstream regions of rmpA, rmpA2 and rcsA was
incubated with an increasing amount of the His6-Fur for 30 min and then loaded onto a 5 % non-denaturing polyacrylamide gel.
indicating that formation of the Fur–Fe(II) complex was
required for the specific binding.
Fur represses CPS biosynthesis via RmpA and
RcsA
To investigate how Fur differentially regulates the
expres-sion of the three CPS regulators, double mutants with a
deletion of rmpA, rmpA2 or rcsA from the Dfur strain
background were constructed, and the effects of the
mutations on bacterial CPS biosynthesis were assessed.
Consistent with previous reports (Cheng et al., 2010; Ebel
& Trempy, 1999; Lai et al., 2003), deletion of rmpA, rmpA2
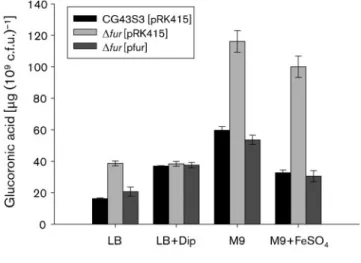
or rcsA reduced the amount of bacterial CPS (Fig. 2). By
contrast, a significant increase in the amount of CPS was
found in the Dfur strain. Interestingly, deletion of rmpA or
rcsA, but not rmpA2, suppressed the fur deletion phenotype
(Fig. 2). The results suggest that the activation of CPS
biosynthesis in the Dfur strain is mediated by RmpA or
RcsA, but not RmpA2, under the assay conditions used.
It has been reported that the K2 cps gene cluster of K.
pneumoniae Chedid contains 19 open reading frames
(ORFs) organized into three transcription units, orf1-2,
orf3-15 and orf16-17 (Arakawa et al., 1995). Analysis of the
cps promoters revealed no conserved Fur box, suggesting
that Fur exerts indirect control over the transcription of cps.
To investigate this possibility, transcripts of orf1, orf3 and
orf16 in wild-type (CG43S3), Dfur, DrmpA, DrmpA2, DrcsA,
DfurDrmpA, DfurDrmpA2, DfurDrcsA, DfurDrmpADrcsA
and DfurDrmpADrmpA2DrcsA strains were measured via
qRT-PCR. As shown in Fig. 3, all three transcripts were
differentially decreased in DrmpA, DrmpA2 and DrcsA
strains. Compared with either the rmpA or the rcsA
deletions, the deletion of rmpA2 had less effect on the
transcription of orf1, orf3 and orf16. Interestingly, deletion of
rmpA had more profound reducing effects on the
transcrip-tion of orf1 and orf16 than deletranscrip-tion of rcsA. Moreover,
expression levels of cps in DrmpA, DrmpADrcsA and
DrmpADrmpA2DrcsA were similar, suggesting a major
regulatory role of RmpA in controlling cps expression.
However, RcsA and RmpA2 may also play a major role in cps
expression under conditions that have not been identified.
Moreover, further study is needed to determine whether a
regulatory interaction exists between RmpA, RmpA2 and
RcsA.
Consistent with the results shown in Fig. 2, the deletion
effect of fur was eliminated in the DfurDrmpA or DfurDrcsA
strains when the orf1 and orf16 transcripts were expressed
(Fig. 3a, c). Deletion of rmpA from the Dfur strain
significantly decreased the level of all three cps transcripts.
The quantities of the cps transcripts in DfurDrmpADrcsA or
DfurDrmpADrmpA2DrcsA were similar to that in the
DfurDrmpA strain. These results further support the
assumption that RmpA plays a major role in the
Fur-mediated repression of cps transcription. By contrast, no
apparent difference in cps expression was observed between
Dfur and DfurDrmpA2, indicating a minor role, if any, for
RmpA2 in the Fur-mediated regulation of cps expression.
Nevertheless, the much higher expression levels of cps that
were observed in DfurDrmpADrmpA2DrcsA than in strain
DrmpADrmpA2DrcsA suggest that an unknown regulator
may be involved in the Fur-mediated control of cps
expression.
Availability of iron affects CPS biosynthesis in
K. pneumoniae
To determine whether Fur regulates gene expression in an
Fe(II)-dependent manner (Andrews et al., 2003; Escolar
et al., 1999), we analysed the effects of iron depletion and
iron repletion on CPS biosynthesis. As shown in Fig. 4, the
amount of CPS was increased in the Dfur strain when the
bacteria were grown in LB medium containing ~18 mM
iron (Abdul-Tehrani et al., 1999). The fur deletion effect
was no longer observed in the fur-complemented strain,
nor was it observed when Dip was added to the growth
medium. In addition, the addition of 60 mM FeSO
4to M9
medium caused an apparent decrease in the amount of
CPS in the type strain compared with that of
wild-type strain grown only in M9 medium. The Dfur strain
grown in M9 medium both with and without FeSO
4produced a higher amount of CPS than the wild-type
strain, indicating that an iron level of approximately 2 mM
in M9 medium (Abdul-Tehrani et al., 1999) may be
sufficient for Fur activity to repress CPS biosynthesis.
Fig. 2. Fur represses CPS biosynthesis via RmpA and RcsA.Bacterial strains were grown in LB medium at 37 6C with agitation. After 16 h growth, the bacterial glucuronic acid contents were determined. Values are mean±SDof three independent experiments.
These results suggest that iron repletion increased Fur
activity, thereby repressing the biosynthesis of CPS.
The regulatory role of Fur in iron-acquisition
systems of K. pneumoniae CG43
To assess whether Fur affects iron acquisition in K.
pneumoniae as in other bacteria, a CAS assay was
performed to analyse siderophore secretion. As shown in
Fig. 5(a), an orange halo around the colony of K.
pneumoniae Dfur strain grown on a blue CAS plate was
observed. Introduction of the complement plasmid pfur
into the Dfur strain appeared to diminish the orange halo
phenotype. A
BLASTsearch with the DNA sequences of the
iron-acquisition systems in K. pneumoniae NTUH-K2044
as templates (Hsieh et al., 2008) for the homologues in the
contig database of K. pneumoniae CG43 (unpublished
results from Dr S.-F. Tsai) was subsequently performed. As
shown in Table 3, eight putative iron-acquisition systems
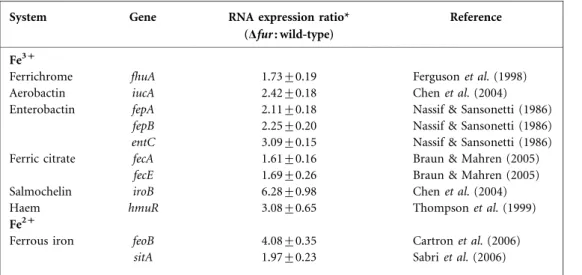
were identified. Expression of the genes (iucA, fepA, fepB,
entC, iroB, hmuR and feoB), corresponding to five
iron-acquisition systems assessed via qRT-PCR, were increased
at least twofold in the Dfur strain. Expression of the fhuA,
fecA, fecE and sitA genes was also activated in the Dfur
strain, although with a less than twofold increase (Table 3).
As shown in Fig. 5(b), sequences with similarity to the E. coli
Fur box (de Lorenzo et al., 1987) could be identified in the
putative promoters P
iroB, P
entC, P
hmuR, P
feoA, P
fecA, P
fhuAand
P
sitA. A Fur box homologue was also found in the coding
region of iucA, at position 24 to
+15 relative to the start
codon. These Fur-box-containing DNA fragments were then
cloned into pT7-7, and the resulting plasmids were
introduced individually into the E. coli indicator strain
H1717. As shown in Fig. 5(c), E. coli H1717 harbouring a
plasmid with P
iucA, P
iroB, P
entC, P
hmuR, P
feoAor P
fecAshowed
FURTA-positive phenotypes. However, the H1717 strains
harbouring pT7-7 derivatives with the upstream regions of
fhuA or sitA exhibited a FURTA-negative phenotype. The
results suggest that Fur can bind to each of the predicted Fur
Fig. 3. qRT-PCR analyses of the expression of the K2 cps genes. Bacterial strains were grown in LB medium at 37 6C with agitation and then subjected to qRT-PCR analyses for detection of orf1 (a), orf3 (b) and orf16 (c) expression. Values are mean±SDof three independent experiments.box sequences on iroB, entC, iucA, hmuR, feoB and fecA to
exert its regulatory function in vivo.
Extracellular Fe(II) has been demonstrated to be
trans-ported into bacteria via the iron-acquisition systems
FeoABC and SitABCD (Cartron et al., 2006; Sabri et al.,
2006). As shown in Fig. 5, expression of the feo gene but
not the sit gene was affected by Fur. The feoB deletion
mutant, which was predicted to have decreased the
bacterial Fe(II) transport ability, was therefore generated
to investigate whether the Fe(II)-dependent regulation of
CPS biosynthesis is affected by the Feo system. However,
no difference in the amount of CPS between the wild-type
and DfeoB strains, grown in both LB and M9 supplemented
with various concentrations of Dip or FeSO
4, was found
(data not shown). It is possible that SitABCD or other
iron-acquisition systems are involved in the
Fur–Fe(II)-dependent regulation of CPS biosynthesis, which may then
compensate for the mutation effect of feoB.
DISCUSSION
We have demonstrated that Fur directly controls
expres-sion of the CPS regulators RmpA, RmpA2 and RcsA (Fig.
Fig. 4. Fur affects K. pneumoniae CPS biosynthesis in anFe(II)-dependent manner. Bacteria were grown in media supplemented both with and without either 200 mM Dip or 60 mM FeSO4as
indicated. After 16 h growth, the bacterial glucuronic acid contents were determined. Values are mean±SD of three independent experiments.
Fig. 5. Fur regulation of iron acquisition in K. pneumoniae CG43. (a) Deletion of fur increases K. pneumoniae siderophore produc-tion, as assessed by using a CAS assay. Each bacterial strain assayed is indicated, and the orange haloes formed around the colonies correspond to the iron-chelating activity of siderophores in the bacteria. (b) DNA sequence alignment between the E. coli typical Fur box and the putative Fur boxes in the upstream regions of the eight iron-acquisition systems. Positions identical to the consensus sequences are underlined. (c) Assessment of the binding of Fur to the DNA sequences using FURTA. E. coli H1717 strains carrying the pT7-7 derivatives are indicated. Red colonies (Lac+) are denoted FURTA-positive phenotypes; pT7-7, FURTA-negative control.
1). It has been reported previously that fur mutation does
not produce an obvious change in rmpA2 promoter activity,
as assessed by the lacZ reporter system (Cheng et al., 2010).
By contrast, qRT-PCR analysis revealed that deletion of fur
caused an approximately twofold increase in rmpA2 mRNA
(Fig. 1a). The discrepancy may be due to the dosage effect of
the plasmid-based lacZ reporter system, which is known to
overestimate b-galactosidase activity. The EMSA results
shown in Fig. 1(c) also support the direct binding of Fur to
the rmpA2 promoter. The putative Fur box in the rmpA2
promoter does not align as well as those in the rmpA and
rcsA promoters with the E. coli Fur box (Fig. 1b), and hence
it is possible that K. pneumoniae Fur exerts greater flexibility
with respect to its recognition sequences and/or differences
in its mode of action at the rmpA2 promoter.
The two homologous genes rmpA and rmpA2 are on
plasmid pLVPK, and both encode CPS regulators for the
activation of CPS biosynthesis (Chen et al., 2004; Lai et al.,
2003). Compared with RmpA, RmpA2 has an extended
N-terminal region and a different promoter sequence,
implying that the two transcriptional factors are
function-ally different. As shown in Fig. 2, the deleting effect of fur
was eliminated by the further deletion of rmpA or rcsA, but
not of rmpA2, suggesting that these genes have different
roles in the regulation of CPS biosynthesis. Further
investigation is needed to clarify the roles of the two
homologous regulators in K. pneumoniae.
Fur has been demonstrated to be a global regulator in
many bacteria (Cornelis et al., 2009; Mey et al., 2005;
Moore & Helmann, 2005). The deletion of fur in
Helicobacter pylori was shown to reduce the expression of
Lon protease (Choi et al., 2009), which can affect the
protein stability of RcsA and RmpA2 in E. coli and K.
pneumoniae (Lai et al., 2003; Trisler & Gottesman, 1984).
However, fur deletion in K. pneumoniae CG43 has no
obvious effect on the expression of lon (data not shown).
The Fur protein sequences of H. pylori and K. pneumoniae
have low identity (25.6 %), suggesting that the Fur
regulatory circuit is different in the two bacteria.
The K2 cps gene cluster is predicted to encode proteins that
are involved in the synthesis, transport, assembly and
modification of CPS (Whitfield & Roberts, 1999). As shown
in Fig. 3, the differential regulation exerted by RmpA,
RmpA2 and RcsA on cps expression affects both the amount
and the composition of CPS. Further studies are needed to
investigate whether RmpA, RmpA2 and RcsA also affect
CPS modifications, thus influencing the interactions between
bacteria and host cells. The mutant DfurDrmpADrmpA2
DrcsA had a higher level of cps expression than the mutant
DrmpADrmpA2DrcsA, indicating that one or more unknown
regulators besides RmpA, RmpA2 and RcsA may be involved
in the Fur-mediated control of cps transcription. The
com-plex regulation of cps expression in K. pneumoniae requires
further exploration.
In K. pneumoniae, Fur regulates the expression of
flavodoxin and CPS biosynthesis in addition to regulating
its
own
expression
(Achenbach
&
Genova,
1997;
Achenbach & Yang, 1997; Cheng et al., 2010). Here, we
showed that Fur serves as a repressor in the regulation of at
least eight iron-acquisition systems in K. pneumoniae
CG43, although at different levels (Table 3). Analysis of
the putative Fur boxes on iroB, entC, hmuR, iucA, feo and
fec revealed high levels of identity to the consensus
sequence (15–16 of 19 positions), whereas those of fhuA
and sitA exhibited relatively lower identities (13 of 19
positions). This suggests that a highly conserved sequence
of the 19 bp sequence is required for a positive FURTA
phenotype. During infection, differential expression of the
iron-acquisition system is anticipated to provide an
adaptive advantage because of its flexibility in responding
Table 3. qRT-PCR analyses of the expression of iron-acquisition genes in K. pneumoniae wild-type and Dfur strains
System Gene RNA expression ratio*
(Dfur : wild-type)
Reference
Fe3+
Ferrichrome fhuA 1.73±0.19 Ferguson et al. (1998)
Aerobactin iucA 2.42±0.18 Chen et al. (2004)
Enterobactin fepA 2.11±0.18 Nassif & Sansonetti (1986)
fepB 2.25±0.20 Nassif & Sansonetti (1986)
entC 3.09±0.15 Nassif & Sansonetti (1986)
Ferric citrate fecA 1.61±0.16 Braun & Mahren (2005)
fecE 1.69±0.26 Braun & Mahren (2005)
Salmochelin iroB 6.28±0.98 Chen et al. (2004)
Haem hmuR 3.08±0.65 Thompson et al. (1999)
Fe2+
Ferrous iron feoB 4.08±0.35 Cartron et al. (2006)
sitA 1.97±0.23 Sabri et al. (2006)
to various environmental stimuli (Caza et al., 2008;
Valdebenito et al., 2006). Therefore, it is suggested that
the eight iron-acquisition systems in CG43 are coordinated
differently. Whether CG43 harbours other iron-acquisition
genes remains to be investigated.
In this study, we characterized the role of Fur in the CPS
regulatory circuit of K. pneumoniae CG43, and found that
RmpA, RcsA and RmpA2 are directly regulated by Fur. We
also demonstrated that Fur regulates CPS biosynthesis via
RcsA or RmpA, but not RmpA2, in an Fe(II)-dependent
manner. Moreover, we report that a fur deletion affects the
expression of the eight iron-acquisition systems identified
in K. pneumoniae CG43.
ACKNOWLEDGEMENTS
We are grateful to Dr K. Hantke (University of Tu¨bingen, Germany) for providing the E. coli strain H1717. This work was supported by grants from the National Science Council (NSC 97-2314-B-039-042-MY2) and China Medical University CMU97-204 and CMU97-345 to C.-T. L., and NSC 97-2320-B-009-001-MY3 to H.-L. P.
REFERENCES
Abdul-Tehrani, H., Hudson, A. J., Chang, Y. S., Timms, A. R., Hawkins, C., Williams, J. M., Harrison, P. M., Guest, J. R. & Andrews, S. C. (1999).Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol 181, 1415–1428.
Achenbach, L. A. & Genova, E. G. (1997).Transcriptional regulation of a second flavodoxin gene from Klebsiella pneumoniae. Gene 194, 235–240.
Achenbach, L. A. & Yang, W. (1997).The fur gene from Klebsiella pneumoniae: characterization, genomic organization and phylogenetic analysis. Gene 185, 201–207.
Altschul, S. F., Madden, T. L., Scha¨ffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997).GappedBLASTandPSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402.
Andrews, S. C., Robinson, A. K. & Rodriguez-Quinones, F. (2003).
Bacterial iron homeostasis. FEMS Microbiol Rev 27, 215–237.
Arakawa, Y., Wacharotayankun, R., Nagatsuka, T., Ito, H., Kato, N. & Ohta, M. (1995).Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol 177, 1788–1796.
Blumenkrantz, N. & Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal Biochem 54, 484– 489.
Boddicker, J. D., Anderson, R. A., Jagnow, J. & Clegg, S. (2006).
Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect Immun 74, 4590–4597.
Braun, V. & Mahren, S. (2005). Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol Rev 29, 673–684.
Cartron, M. L., Maddocks, S., Gillingham, P., Craven, C. J. & Andrews, S. C. (2006).Feo – transport of ferrous iron into bacteria. Biometals 19, 143–157.
Caza, M., Lepine, F., Milot, S. & Dozois, C. M. (2008).Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect Immun 76, 3539–3549.
Chen, Y. T., Chang, H. Y., Lai, Y. C., Pan, C. C., Tsai, S. F. & Peng, H. L. (2004).Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198.
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C. & Peng, H. L. (2010).RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192, 3144–3158.
Choi, Y. W., Park, S. A., Lee, H. W. & Lee, N. G. (2009).Alteration of growth-phase-dependent protein regulation by a fur mutation in Helicobacter pylori. FEMS Microbiol Lett 294, 102–110.
Cornelis, P., Matthijs, S. & Van Oeffelen, L. (2009). Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22, 15–22.
de Lorenzo, V., Wee, S., Herrero, M. & Neilands, J. B. (1987).
Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169, 2624–2630.
Domenico, P., Schwartz, S. & Cunha, B. A. (1989).Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect Immun 57, 3778–3782.
Ebel, W. & Trempy, J. E. (1999).Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J Bacteriol 181, 577–584.
Escolar, L., Perez-Martin, J. & de Lorenzo, V. (1999).Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181, 6223–6229.
Favre-Bonte, S., Joly, B. & Forestier, C. (1999). Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect Immun 67, 554–561.
Ferguson, A. D., Hofmann, E., Coulton, J. W., Diederichs, K. & Welte, W. (1998). Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282, 2215–2220.
Gottesman, S. & Stout, V. (1991).Regulation of capsular polysac-charide synthesis in Escherichia coli K12. Mol Microbiol 5, 1599– 1606.
Griggs, D. W. & Konisky, J. (1989).Mechanism for iron-regulated transcription of the Escherichia coli cir gene: metal-dependent binding of Fur protein to the promoters. J Bacteriol 171, 1048–1054.
Han, S. H. (1995). Review of hepatic abscess from Klebsiella pneumoniae. An association with diabetes mellitus and septic endophthalmitis. West J Med 162, 220–224.
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166, 557–580.
Hantke, K. (1987).Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol Gen Genet 210, 135–139.
Hantke, K. (2003).Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol 11, 192–195.
Hsieh, P. F., Lin, T. L., Lee, C. Z., Tsai, S. F. & Wang, J. T. (2008).
Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 197, 1717–1727.
Keen, N. T., Tamaki, S., Kobayashi, D. & Trollinger, D. (1988).
Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70, 191–197.
Koczura, R. & Kaznowski, A. (2003).Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb Pathog 35, 197–202.
Lai, Y. C., Peng, H. L. & Chang, H. Y. (2001).Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect Immun 69, 7140–7145.
Lai, Y. C., Peng, H. L. & Chang, H. Y. (2003).RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 185, 788–800.
Lau, Y. J., Hu, B. S., Wu, W. L., Lin, Y. H., Chang, H. Y. & Shi, Z. Y. (2000). Identification of a major cluster of Klebsiella pneumoniae isolates from patients with liver abscess in Taiwan. J Clin Microbiol 38, 412–414.
Lin, J. C., Chang, F. Y., Fung, C. P., Xu, J. Z., Cheng, H. P., Wang, J. J., Huang, L. Y. & Siu, L. K. (2004). High prevalence of phagocytic-resistant capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect 6, 1191–1198.
Lin, C. T., Huang, T. Y., Liang, W. C. & Peng, H. L. (2006).Homologous response regulators KvgA, KvhA and KvhR regulate the synthesis of capsular polysaccharide in Klebsiella pneumoniae CG43 in a coordinated manner. J Biochem 140, 429–438.
Majdalani, N. & Gottesman, S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59, 379–405.
Mey, A. R., Wyckoff, E. E., Kanukurthy, V., Fisher, C. R. & Payne, S. M. (2005).Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun 73, 8167–8178.
Moore, C. M. & Helmann, J. D. (2005). Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol 8, 188–195.
Moranta, D., Regueiro, V., March, C., Llobet, E., Margareto, J., Larrate, E., Garmendia, J. & Bengoechea, J. A. (2010). Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta-defensins by airway epithelial cells. Infect Immun 78, 1135–1146.
Nassif, X. & Sansonetti, P. J. (1986).Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54, 603–608.
Podschun, R. & Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and patho-genicity factors. Clin Microbiol Rev 11, 589–603.
Sabri, M., Leveille, S. & Dozois, C. M. (2006).A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152, 745–758.
Sahly, H., Podschun, R., Oelschlaeger, T. A., Greiwe, M., Parolis, H., Hasty, D., Kekow, J., Ullmann, U., Ofek, I. & Sela, S. (2000).Capsule
impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immun 68, 6744–6749.
Schwyn, B. & Neilands, J. B. (1987).Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160, 47– 56.
Skorupski, K. & Taylor, R. K. (1996).Positive selection vectors for allelic exchange. Gene 169, 47–52.
Stojiljkovic, I., Baumler, A. J. & Hantke, K. (1994). Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol 236, 531–545.
Stout, V. (1994).Regulation of capsule synthesis includes interactions of the RcsC/RcsB regulatory pair. Res Microbiol 145, 389–392.
Tabor, S. & Richardson, C. C. (1985). A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A 82, 1074–1078.
Tang, H. L., Chiang, M. K., Liou, W. J., Chen, Y. T., Peng, H. L., Chiou, C. S., Liu, K. S., Lu, M. C., Tung, K. C. & Lai, Y. C. (2010).Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 29, 689–698.
Thompson, J. M., Jones, H. A. & Perry, R. D. (1999). Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun 67, 3879–3892.
Trisler, P. & Gottesman, S. (1984).lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J Bacteriol 160, 184–191.
Valdebenito, M., Crumbliss, A. L., Winkelmann, G. & Hantke, K. (2006). Environmental factors influence the production of enter-obactin, salmochelin, aerenter-obactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int J Med Microbiol 296, 513–520.
Wandersman, C. & Delepelaire, P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58, 611–647.
Whitfield, C. & Roberts, I. S. (1999). Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol 31, 1307–1319.
Yang, Y. S., Siu, L. K., Yeh, K. M., Fung, C. P., Huang, S. J., Hung, H. C., Lin, J. C. & Chang, F. Y. (2009).Recurrent Klebsiella pneumoniae liver abscess: clinical and microbiological characteristics. J Clin Microbiol 47, 3336–3339.