行政院國家科學委員會專題研究計畫 成果報告
(子計畫九)家禽冠狀病毒基因重組分析及其與嚴重急性呼
吸道症候群的關聯性
計畫類別: 整合型計畫 計畫編號: NSC92-3112-B-002-034- 執行期間: 92 年 05 月 01 日至 93 年 04 月 30 日 執行單位: 國立臺灣大學獸醫學系暨研究所 計畫主持人: 王金和 共同主持人: 闕玲玲 計畫參與人員: 李盺錞 黃元品 報告類型: 完整報告 處理方式: 本計畫可公開查詢中 華 民 國 93 年 5 月 14 日
前言、研究目的、文獻探討
Like most RNA viruses, coronavirus has a high frequency of mutation, which involves discontinuous transcription and polymerase jumping (10). It accumulates several point mutations during each round of RNA replication. Thus, even a plaque-purified coronavirus stock would be a population of related quasi-species rather than homogeneous virus. For example, the recombination frequency of the entire mouse hepatitis virus reaches 25% (1).
Vaccination with Massachusetts (Mass) group strains for IBV control in chickens is very common in Taiwan. These vaccine strains are suggested to be a large donor pool for IBV recombination. The important of this pool has not been determined although the recombination and mutation have been approved by several reports (7,8,9,10,18). IB has occurred frequently in Taiwan in spite of vaccination (13,14,15,16,17) since it first appeared in 1965 (13). The reason of vaccination breaks has been mostly due to the different serotypes between imported vaccine strains and Taiwan filed IBVs. However, the evidence of recombination in Taiwanese IBVs is lacking. The aims of this project are to determine the recombination frequency of the IBV in Taiwan during the recent 10 years.
研究方法 Virus strains.
One IBV strain, TP/64 isolated from a layer farm in Taipei in 1964 and 31 strains isolated from chickens with nephropathgenic IBV infection in Taiwan during 1991 to 2003 were used in this study (Fig 1). IBV 1121/91 was isolated in 1991 and the last one, IBV 3110/03 was isolated in 2003. Thirteen of them were selected for the whole S1 gene and partial N gene sequencing (Table 2). Viruses were propagated in specific-pathogen-free (SPF) chicken embryos (Animal Health Research Institute, Council of Agriculture, Tamsui). The allantoic fluid was harvested 48-72 hours after inoculation and was frozen and stored at -70 C.
Sequences of the N terminus of the S1 gene of IBVs.
For each strain from the field, the hyper variable region 1 (HVR 1) in the N terminus of the S1 gene were amplified with a primer set, rC2U/rC3L modified by Tseng (12) from C2U/C3L by Wang et al. (15,16). The sequences of these primers were rC2U (forward):
5’-TGGTTGGCA(T/C)TTACA(A/C/T)GG(A/G/T)-3’ and rC3L (reverse):
5’-(A/G)CAATGTGTAACAAA(T/C)ACT-3’. The size of the expected PCR product was 231 bp. The rC2U-rC3L fragment was from nt 114 to nt 341 in the N terminus of the S1 gene.
Complete S1 sequences of IBVs.
Thirteen strains selected from the phylogenetic tree based on the rC2U-rC3L fragment were used for sequencing the whole S1 gene of IBVs (Table 1). The primer, Oligo5’/IBVc2 primer was used for amplification the whole S1 segment, and the RT-PCR products were sequenced double from both sites (all together 4 times at least). The sequences of this primer set were
oligo5’: AAACT GAACA AAAGA CAGAC TTAG (20305-20328) IBVc2: GCCAT AACTA ACATA TGGAC AAC (22033-22011) Partial N sequences of IBVs.
The N gene of the 13 selected strains was sequenced with the following primers [13Tseng]. The sequence of this primer set for nucleocapsid (NC) gene were
NP1: GGTA G(C/T)GG(C/T) GTTCC TGATA A (nt 158-177 in N) NP2: TCATC TTGTC (A/G)TCAC CAAAA (nt 781-762 in N).
Recombination among IBVs.
The sequences of the whole S1 genes of the selected 13 IBV strains obtained by direct sequencing from PCR products except IBV 1171/92 (accession number: AF250005 in the GenBank) and IBV 1211/92 (accession number: AF250006) were used for checking
recombination between H120 and Taiwanese strains. The sequence CTT(A/T)(A/T)G found in the S1 gene of IBV, similar to the concensus leader sequence was used as the junctions of the fragments for identifying the cross over sites. Those fragments were nt. 1-30, 31-136, 137-1057, 1058-1620. The phylogenetic trees constructed from the for S and N genes were used to compare the clustering of an individual strain for inter-strain recombination.
Phylogenetic analysis
DNA sequences were compiled and edited by using the Vector NTI software package. Phylogenetic trees based on the N terminus if the S gene, the whole S1 gene and partial N gene were constructed by using the neighbor Joining and bootstrap analysis (n = 100) to determine the best fitting tree for each gene. For comparison, the S1 sequences of reference strains H120 (accession number:M21970), J2/China (accession number: AF286303), and Ark99 (accession number:L10384) were obtained from the GenBank.
結果
Analysis of the N terminus of the S gene.
The phylogenetic tree based on the sequences of the N terminus of the S gene showed that Taiwanese IBVs were divided into two lineages, Taiwan Group I (TW I) and Taiwan Group II (TW II) except 1916/93, 2992/02 and 2294/02 (Fig 1). Most Taiwanese strains belonged to TW I and only a few belonged to TW II. IBV 1916/93 was considered to be a vaccine strain (15). IBV 2294/02 was a Mass group field strain. A new strain, IBV 2992/02 strain, whose S sequence was quite similar to J2/China strain was strange since the importation of animal products from China was illegal.
Comparison of the S1 Sequences of IBVs
Complete S1 sequences were determined for each of the 13 strains indicated in Table 1, including 11 strains not previous available. The 13 IBVs could be divided into two groups, TW I and TW II. All of them belonged to TW I except 2296/95, belonged to TW II. The S1 gene of those IBVs contained approximately 44.4% G+C residues. Among all strains in TW I, the S1 sequences were 1620 long and differed from one another by 5 to 15%, with IBV 2992/02 the most distant from one another.
The deduced amino acid sequences of those strain showed several substitutions (Table 2). Among TW I group, the E at the residue 55 was replaced by Q, the R at the residue 80 was replaced by T gradually, and the S at the residue 96 was replaced by L. An amino acid deletion at the position 87 was found in the strains of Mass group, and the strain 2994/02 even showed deletion of 3 amino acids at the position 60.
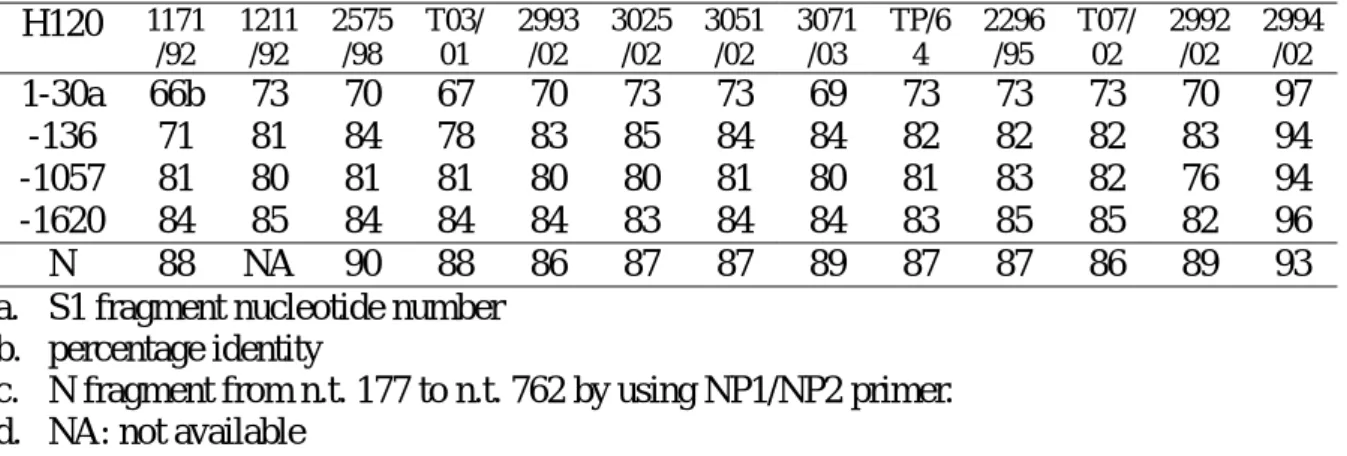
After checking the identity of the different fragments in Taiwanese IBVs and H120 strain, no recombination was found between them (Table 3) because all of them were less than 90% except those of IBV 2994/02. All the fragments of IBV 2994/02 had similar sequences with H120 (Mass group), thus, IBV 2994/02 belonged to a Mass group.
The cleavage recognition site between S1 and S2 subunits of those 13 strains was compared. The oldest IBV strain, TP/64 had RLSRR, 3 strains (IBV 2992/02, 2994/02, 3025/02) had
RRSRR, and the rest 9 strains had RRFRR sequence. The most common cleavage recognition site was neither identical to most strains reported (6), nor correlated to genotypic grouping. TW I strains contained the same cleavage site as TW II strains.
Comparison of the N Sequences of IBVs
The 177-762 fragment of the N gene had 86% to 90% nucleotide identity between H120 and Taiwanese strains (Table 3), which were much higher than those of the S1 gene (p <0.05). The
nucleotide sequences of S1 and N genes were compared for possible recombination. IBV 2992/02 was found to be a recombinant because its S1 gene was almost identical to the S1 of a Chinese strain, J2/China but its N gene belonged to Taiwan cluster. IBV 2994/02 is another recombinant because its S1 gene belonged to H120 but its N gene was similar to Ark99, a representative strain of the American group.
Comparison of the S1 Sequences of IBVs and SARS virus
Table 4 showed the percent identity between IBV and SARS virus. However, no similarity was found between these two viruses.
討論
We have demonstrated that genomic recombination events can take place in the intergenic sequences between S and N genes. Evidence of recombination was found only in the strains, in a fashion in which the 5’ segment of the recombinant sequence presented similarity with one strain and the 3’ segment had similarity with another strain. The same phenomenon was demonstrated in ovo inoculation with different strains (5). In a genome, if the strains that very resemble each other in one gene are very different in another, it suggests that inter-strain recombination may have occurred. This might be from the discontinuous transcription or replication during negative strain formation (2). The difference of the S1 gene among different IBVs varies from 1 to 40% (18) however, that of N gene was 5-15%, less than that in the S1 gene. N gene is more conserved, so homologous recombination is easy to occur.
The result in Table 3 shows that recombination has not occurred in the S1 genes of the Taiwanese IBVs during the recent 10 years in spite of the extensive use of H120 vaccine strain in the field for more than 30 years. Although recombination may be a common mechanism for genetic variation for IBV, factors which precipitate these events are not known. Recombination events could result from the polymerase jumping from one template to another in the synthesis of either the negative or the positive-strain RNA (10).
Phylogenetic analysis of the S1 gene of IBVs permits the establishment of relationships between IBVs from different locations and different times. For example, the IBVs isolated 10 years ago showed high similarity with the strains isolated recently indicates the same virus is still present after ten years (Fig. 1). The high level of sequence similarity indicates that a stable S1 gene lineage has been maintained in Taiwan, especial TW I.
An interesting finding is the intruded strain, IBV 2992/02, which is similar to J2/China strain. J2/China strain is very similar to Q1 and T3 strains despite recover from geographically different areas in China (19,20). This strain was isolated from a broiler farm build near the migratory bird wet land in Yilan. IBV vaccine strain Ma5 (Intervet, Holland) was used at one day old. Some chicks showed respiratory signs and died from the age of two weeks. Six percent of them were died due to this infection till 4 weeks old. The dead chicks showed urate deposition in both kidneys (data not shown). However, no proventricular lesion was found as in China (19). Thus, despite the similarity in the S1 gene, the pathogenesis is not the same because the N gene of 2992/02 is similar to Taiwanese strains. Since no N gene sequences of these three Chinese strains are available, the similarity between them and 2992/02 is not known. The difference in lesions might be due to the difference in genes other than the S1 gene.
The IBVs in the present study have been isolated from chickens in the infected flocks vaccinated with Mass group IBV strains. Thus, most isolated strains belong to Taiwanese clusters except one strain, 2994/02, which belongs to Mass group. This might account for the difference of IBVs isolated from Liu’s paper, in which more Mass group IBVs were isolated (11). During the recent 20 years almost all Taiwanese IBVs are nephropathogenic (21) because renal urate lesions is considered to be caused by IBV, and the chickens showed renal urate were taken to this laboratory for diagnosed. The presence of two lineages makes the control of IB in Taiwan not
very difficult. The present results indicate that most Taiwanese IBVs are still in the two
distinguish lineages, TW I and TW II as our previous reports (15,16). This means control Taiwan IB might be possible by using vaccines developed from Taiwanese strains.
REFERENCES
1. Baric, R. S., K. Fu, M. C. Schaad, and S. A. Stohlman. Establishing a genetic recombination map for murine coronavirus strains A59 complementaion groups. Virology 177:646-656. 1990. 2. Brian, D. A., and W. J. M. Spaan. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111, 1997
3. Collisson, E. W., R. L. Parr, L. Wang, and A. K. Williams. An overview of the molecular characteristics of avian infectious bronchitis virus. Poult. Sci. Rev. 4:41-55, 1992
4. Cook, J. Coronoaviridae. In: Poultry Diseases, 5th ed. F. Jordan, M. Pattison, D. Alexander, and T. Faragher, eds. W. B. Saunders, London. pp. 298-306. 2002.
5. Estervez, C., P. Villeags, and J. El-Attrache. A recombination event, induced in ovo, between a low passage infectious bronchitis virus field isolate and a highly embryo adapted vaccine strain. Avian Dis. 47:1282-1290. 2003.
6. Jackwood, M. W., D. A. Hilt, S. A. Callison, C.-W. Lee, H. Plaza, and E. Wade. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 45:366-372. 2001.
7. Jia, W, C. R., Karaca, and S. A. Naqi. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 140:257-271. 1995. 8. Kottier, S., D. Cavanagh, and P. Britton. Experimental evidence of recombination in coronavirus infection bronchitis virus. Virology 213:569-580. 1995.
9. Lai, M. M. Coronavirus: Organization, replication and expression of genome. Annu Rev Microbiol 44:303-333. 1990.
10. Lai, M. M. Genetic recombination in RNA viruses. Curr. Trop. Microbiol. Immunol. 176:21-32. 1992.
11. Liu, H. J., L. H. Lee, W. L. Shih, M. Y. Lin, and M. H. Liao. Detection of infectious bronchitis virus by multiplex polymerase chain reaction and sequence analysis. J. Virol. Methods 109:31-37. 2003.
12. Tseng, Z. M. Analysis of nucleotide and preparation of inactivated vaccine of infectious bronchitis virus isolated in Taiwan. In: Master thesis, National Chung-Hsing Univer. Taichung, Taiwan. p13. 1999.
13. Tseng, C. C., N. Z. Li, C. H. Yao, and C. H. Wang . Isolation and adaptation of infectious bronchitis virus in Taiwan from 1993 to 1995. J. Chin. Soc. Vet. Sci. 22:113-120. 1996. 14. Wang, C. H., M. C. Hsieh, and P.C. Chang. Isolation, pathogenicity and H120 protection efficacy of infecitous bronchitis viruses isolated in Taiwan. Avian Dis. 40:620-625. 1996.
15. Wang, C. H., and C. T. Tsai. Genotypic grouping for the isolates of avian infectious bronchitis virus in Taiwan. Arch. Virol. 141:1677-1688. 1996.
16. Wang, C. H., and Y. C. Huang. The relationship between serotype and genotype based on hypervariable region of S1 gene of infectious bronchitis virus. Arch. Virol. 145:291-300. 2000. 17. Wang, C. H., J. C. Hong, and J. S. Sheak. Expression of a fragment in spike gene for use as an antigen in ELISA test for the detection of antibody against infectious bronchitis virus. Vet
Microbiol 85:333-342, 2002
18. Wang, L., D. Junker, L. Hock, E. Ebiary, and E. W. Collisson. Evolutionary implications of genetic variations in the S1 gene of infectious bronchitis virus. Virus Res. 34:327-328. 1994. 19. Yu, L., Y. Jiang, S. Low, Z. Wang, S. J. Nam, W. Liu, and J. Kwang. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 45: 416-424. 2001.
20. Yu, L., Z. Wang, Y. Jiang, S. Low, and J. Kwang. Molecular epidemiology of infectious bronchitis virus isolates from China and Southeast Asia. Avian Dis. 45:201-209. 2001.
21. Ziegler, A. F., B. S. Ladman, P. A. Dunn, A. Schneider, S. Davison, P. G. Miller, H. Lu, D. Weinstock, M. Salem, R. J. Eckroade, and J. Gelb, Jr. Nephropathogenic infectious bronchitis in
Pennsylvania chickens 1997-2000. Avian Dis. 46:847-858. 2002. ACKNOWLEDGMENTS
We thank Miss M. C. Cheng of Animal Health Research Institute for providing some IBVs. The financial supports from National Science Council and Council of Agriculture, Taiwan is appreciated.
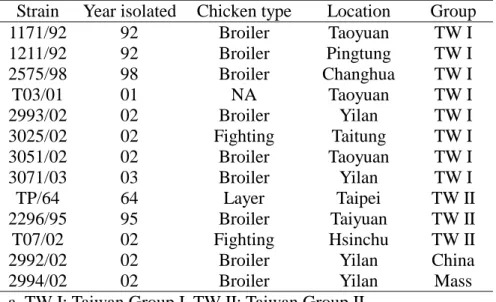
Table 1. History of the selected infectious bronchitis virus strains isolated in Taiwan in 1964 and during 1992 to 2003
Strain Year isolated Chicken type Location Group
1171/92 92 Broiler Taoyuan TW I 1211/92 92 Broiler Pingtung TW I 2575/98 98 Broiler Changhua TW I T03/01 01 NA Taoyuan TW I 2993/02 02 Broiler Yilan TW I 3025/02 02 Fighting Taitung TW I 3051/02 02 Broiler Taoyuan TW I 3071/03 03 Broiler Yilan TW I TP/64 64 Layer Taipei TW II 2296/95 95 Broiler Taiyuan TW II T07/02 02 Fighting Hsinchu TW II
2992/02 02 Broiler Yilan China
2994/02 02 Broiler Yilan Mass
a. TW I: Taiwan Group I, TW II: Taiwan Group II. b. NA: not available.
Table 2 another file
Table 3. The percent identity of the S1 gene and N gene of Taiwanese IBV strains
strain 1171 /92 1211 /92 2575 /98 T03/ 01 2993 /02 3025 /02 3051 /02 3071 /03 TP /64 2296 /95 T07/ 02 2992 /02 2994 /02 1171/92 94.4 93.6 93.9 93.5 92.9 93.1 93.4 81.0 87.1 85.7 77.3 80.0 1211/92 93.6 94.1 94.0 94.1 93.0 94.0 93.4 80.2 86.9 85.3 76.7 79.0 2575/98 92.2 91.0 94.4 98.9 97.3 98.5 98.0 80.1 86.9 86.6 77.1 80.0 T03/01 93.4 92.9 94.6 94.0 92.7 93.5 93.3 79.9 86.2 85.3 76.9 80.0 2993/02 96.2 95.7 92.4 93.8 97.3 98.7 98.1 80.5 86.5 86.4 77.0 79.5 3025/02 95.5 95.0 92.6 93.8 99.1 97.4 96.4 80.2 86.6 85.9 77.5 79.5 3051/02 95.2 92.6 94.6 94.6 95.5 95.5 97.4 80.3 86.6 86.1 77.0 80.0 3071/03 92.0 91.0 95.2 92.9 92.9 92.7 94.1 80.5 86.1 85.8 76.9 79.8 TP/64 95.2 94.6 91.2 93.3 95.8 95.0 93.8 91.3 84.6 84.1 74.1 79.6 2296/95 95.8 94.3 92.2 94.5 95.3 94.6 95.2 92.0 95.5 93.4 76.8 80.7 T07/02 95.7 95.2 92.2 93.6 99.5 99.0 95.3 92.7 95.3 94.8 76.4 80.7 2992/02 93.3 92.2 97.6 95.5 93.4 93.4 95.8 94.1 92.6 93.4 93.3 73.6 2994/02 87.9 86.7 92.7 91.5 87.5 87.7 89.8 92.2 87.7 87.5 87.4 92.0
Upper right: the whole S1 gene, lower left: the N gene.
strains H120 1171 /92 1211 /92 2575 /98 T03/ 01 2993 /02 3025 /02 3051 /02 3071 /03 TP/6 4 2296 /95 T07/ 02 2992 /02 2994 /02 1-30a 66b 73 70 67 70 73 73 69 73 73 73 70 97 -136 71 81 84 78 83 85 84 84 82 82 82 83 94 -1057 81 80 81 81 80 80 81 80 81 83 82 76 94 -1620 84 85 84 84 84 83 84 84 83 85 85 82 96 N 88 NA 90 88 86 87 87 89 87 87 86 89 93 a. S1 fragment nucleotide number
b. percentage identity
c. N fragment from n.t. 177 to n.t. 762 by using NP1/NP2 primer. d. NA: not available
Table 4. Nucleotide identity in N fragment of the S1 gene between SARS virus and Taiwanese strains
SARS 1171 1211 2296 2306 2575 2651 2992 2993 2994 3025 3051 3071 T03 T07
S1 47 46 46 47 46 46 45 46 47 45 46 46 46 46 N 50 NA 48 48 48 50 48 49 48 49 50 49 49 49 a. N fragment from n.t. 177 to n.t. 762 by using NP1/NP2 primer.
b. NA: not available
計畫成果自評部份,請就研究內容與原計畫相符程度、達成預期目標情況、研究成果之學 術或應用價值、是否適合在學術期刊發表或申請專利、主要發現或其他有關價值等,作一 綜合評估。
本計畫已被 Avian Diseases 2004 期刊接受刊登。