Plant Pathology (2007) 56, 183 –189 Doi: 10.1111/j.1365-3059.2006.01485.x
Blackwell Publishing Ltd
Zantedeschia mild mosaic virus, a new widespread virus
in calla lily, detected by ELISA, dot-blot hybridization and
IC-RT-PCR
C.-H. Huang
a, W.-C. Hu
a, T.-C. Yang
band Y.-C. Chang
a*
aDepartment of Plant Pathology and Microbiology, National Taiwan University, Taipei; and bPropagation Technology Section, Taiwan Seed
Improvement and Propagation Station, the Council of Agriculture, Hsinshe, Taichung County, Taiwan
A new potyvirus, Zantedeschia mild mosaic virus (ZaMMV), was recently identified in calla lily (Zantedeschia spp.) in Taiwan. The sequenced 3′-terminal region indicated a unique amino acid sequence of ZaMMV, the N terminus of the capsid protein (CP), containing 39 glutamine residues before the DAG motif. In order to obtain antiserum for subsequent studies, a recombinant ZaMMV CP without polyglutamine was successfully expressed in Escherichia coli and used as the antigen. The specificity of ZaMMV antiserum was confirmed by immunoblot and ELISA analyses. Three detection methods, ELISA, dot-blot hybridization and immunocapture reverse transcription PCR (IC-RT-PCR), were developed and were able to detect ZaMMV successfully. During 2003–2004 field surveys, the results demonstrated that ZaMMV, as well as Zantedeschia mosaic virus (ZaMV), are prevalent in calla-growing areas. In contrast, infection by Dasheen mosaic virus (DsMV), the most important virus of aroid plants, decreased dramatically because DsMV-free calla lily seed-lings were grown in the field. Accordingly, the detection methods developed in this study will be useful in studying ZaMMV and also in producing ZaMMV-free calla lilies.
Keywords: Araceae, Zantedeschia mosaic virus, Dasheen mosaic virus, potyvirus, virus detection, Zantedeschia spp.
Introduction
Calla lily (Zantedeschia spp.), originating from Africa, has been imported into Taiwan from New Zealand, the Netherlands and the USA for breeding and production of cut flowers. Bacterial soft rot and viral diseases are the limiting factors for calla lily production in Taiwan, as well as in other countries. Many viruses have been reported to infect calla lily including Dasheen mosaic virus (DsMV), the most important virus of aroid plants (Rana et al., 1983; Chen et al., 2001), Cucumber mosaic virus (CMV) (Mokra & Gotzova, 1994), Konjak mosaic virus (KoMV),
Bean yellow mosaic virus (BYMV), Tomato spotted wilt virus (TSWV), Impatiens necrotic spot virus (INSV),
Alfalfa mosaic virus (AMV), Arabis mosaic virus (ArMV),
Potato virus X (PVX), Tobacco rattle virus (TRV) (Pham
et al., 2002), Turnip mosaic virus (TuMV) (Chen et al., 2003) and Zantedeschia mosaic virus (ZaMV) (Chang
et al., 2001; Kwon et al., 2002). A new calla lily-infecting potyvirus, Zantedeschia mild mosaic virus (ZaMMV) was recently identified in Taiwan (Huang & Chang, 2005). After cloning and sequencing the 3′-terminal
region of the virus, a unique polyglutamine sequence was discovered spanning the N-terminal region of the capsid protein (CP) (Huang & Chang, 2005).
In order to obtain antiserum for further studies, bacteria-expressed CP was used to produce ZaMMV antiserum, due to the problem of purification of virus particles from aroid plants, described by Li et al. (1998). The CP gene of ZaMMV without the polyglutamine sequence was cloned into an expression vector, successfully expressed in
Escherichia coli and used as an antigen to immunize rabbits. The specificity of the antiserum was tested by means of immunoblot and indirect ELISA (I-ELISA) assays. Moreover, three detection methods, I-ELISA, dot-blot hybridization and immunocapture reverse transcription PCR (IC-RT-PCR), were developed and used to detect ZaMMV in calla lilies from the market and field. Based on field surveys during 2003–2004, ZaMMV and ZaMV were widespread in calla lilies but DsMV became less important due to planting DsMV-free seedlings.
Materials and methods
Virus source and sample collection
The ZUN isolate of ZaMMV, originally from calla lily cv. Black Magic (Huang & Chang, 2005), was successfully *E-mail: ycchang@ntu.edu.tw
184 C.-H. Huang et al.
transferred to Philodendron selloum by mechanical inoc-ulation and then maintained on P. selloum and calla lily. Similarly, the ZAN isolate of DsMV (Chen et al., 1998)
and the ZAN isolate of ZaMV (Chang et al., 2001)
derived from calla lilies were also maintained on these two aroid plants. Virus-infected leaf tissues of P. selloum and calla lily were used as controls in immunoblot and detec-tion experiments. Seedlings of calla lily purchased from the Taiwan Seed Improvement and Propagation Station (TSIPS) and a local flower market were used to compare the detection methods. For the disease survey, calla lily plants were collected from three different fields in central Taiwan during 2003–2004.
Construction and expression of the recombinant ZaMMV CP gene
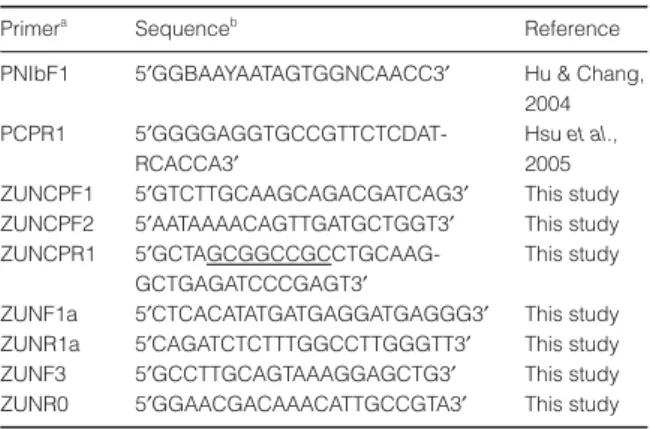
Two pairs of primers, ZUNCPF1/ZUNCPR1 and ZUNCPF2/ZUNCPR1 (Table 1), were used to amplify the CP gene of ZaMMV (GenBank accession number AY626825) from a cDNA clone, pZUN7 (Huang & Chang, 2005). The primers are located at nt −9 to +12 of
the CP gene for ZUNCPF1, nt +139 to +159 for
ZUNCPF2 and nt +948 to +969 for ZUNCPR1. PCR reactions contained 5 µL of plasmid DNA, 12·5 pmol for-ward primer, 12·5 pmol reverse primer, 10 nmol dNTPs, 0·5 U VentR® DNA polymerase (NEB Inc.) and 1x
Ther-moPol reaction buffer (NEB Inc.) in a total volume of
50 µL. The PCR reaction was performed using
Gene-Amp® PCR system 2400 (Applied Biosystems) with an initial denaturation at 94°C for 2 min, 35 cycles of 94°C/ 30 s, 55°C/30 s, 72°C/1 min, and a final extension at
72°C for 5 min. The PCR products were purified by
GFX™ PCR DNA and Gel Band Purification Kit (Amer-sham Pharmacia Biotech). After digestion with NotI, the fragments were further purified for ligation with the pET-29a(+) expression vector (Novagen) pretreated with NotI and EcoRV. The clones were first screened with enzyme digestion and then confirmed by sequencing. Selected clones were used to transform into E. coli BL21 (DE3) and express the CP of ZaMMV with a His tag fused to the C terminus and 30 amino acids from the vector fused to the N terminus. Five hundred microlitres of bacterial sus-pension was collected hourly, after 1 mm isopropyl-β -D-thiogalactopyranoside (IPTG) induction, and centrifuged at 1764 g for 10 min. The supernatant was discarded and the pellets were resuspended in 50 µL of SDS sampling
buffer and 15 µL was analyzed by 12% SDS-PAGE
(Sambrook & Russell, 2001). The recombinant CPs were purified using Ni-NTA agarose following the protocol of the QIAexpressionist™ (QIAGEN).
Immunoblot analysis
Crude plant extracts were prepared by grinding in 10 volumes of 0·01 M sodium phosphate buffer (pH 7·2). The supernatants were collected after centrifugation at 10 160 g for 5 min and used for immunoblot assays. The bacterial lysates and total proteins of plant tissue were
separated by 12% SDS-PAGE, in duplicate. One gel was stained with Coomassie brilliant blue and the other was transferred to PVDF membrane (Osmonics). After block-ing with 0·3% non-fat milk solution, the membrane was incubated in 1x gelatin-NET (0·25% gelatin, 0·15 M
NaCl, 5 mm EDTA, 50 mm Tris-HCl, 0·05% Tween-20,
pH 8·0) containing anti-ZaMMV antiserum diluted 1 : 8000 at room temperature for one hour. Finally, the membrane was probed with a goat anti-rabbit secondary antibody conjugated with alkaline phosphatase and then reacted with the colorimetric substrate, NBT/BCIP (Roche Applied Science).
Indirect ELISA
Antisera of DsMV and ZaMV were previously prepared in the laboratory using recombinant CPs as the antigens (Huang & Chang, 2000). Indirect ELISA (I-ELISA) was performed according to the protocol of Agdia Inc. Each sample had duplicate wells. The values of OD405 of each
sample were measured by the Spectra MAX 340 (Molec-ular Devices Co.) after 30 min incubation. A sample was considered positive if the absorbance value was greater than twice the mean of the healthy controls.
Dot-blot hybridization
Specific DNA probes of ZaMMV were labeled with Digoxigenin (DIG) by PCR reaction. A plasmid clone, pZUN7 was used as template, and 1175-, 311- and 251-bp cDNA fragments were separately amplified by PNIbF1/PCPR1, ZUNF1a/ZUNR1a and ZUNF3/ ZUNR0 primer pairs (Table 1). The PCR reaction solu-tion was set up as before except dNTPs were replaced by a 1x PCR DIG labeling mix (Roche Applied Science). The
PCR program included a denaturing step at 94°C for
2 min, 35 cycles of 94°C/30 s, 53°C/30 s, 72°C/45 s, and a final extension at 72°C for 5 min. The PCR products were analyzed by electrophoresis in a 1·5% agarose gel Table 1 Oligonucleotide primer sequences used in this study Primera
Sequenceb
Reference PNIbF1 5′GGBAAYAATAGTGGNCAACC3′ Hu & Chang,
2004 PCPR1 5
′GGGGAGGTGCCGTTCTCDAT-RCACCA3′
Hsu et al., 2005 ZUNCPF1 5′GTCTTGCAAGCAGACGATCAG3′ This study ZUNCPF2 5′AATAAAACAGTTGATGCTGGT3′ This study ZUNCPR1 5
′GCTAGCGGCCGCCTGCAAG-GCTGAGATCCCGAGT3′
This study ZUNF1a 5′CTCACATATGATGAGGATGAGGG3′ This study ZUNR1a 5′CAGATCTCTTTGGCCTTGGGTT3′ This study ZUNF3 5′GCCTTGCAGTAAAGGAGCTG3′ This study ZUNR0 5′GGAACGACAAACATTGCCGTA3′ This study
a
F and R indicate forward and reverse primers, respectively.
b
Nucleotide at degenerate positions are represented by a single letter code; R = A and G; Y = C and T; B = C, G and T; D = A, G and T; N = A, C, G and T. The created NotI site is underlined.
ZaMMV detected by ELISA, dot hybridization and IC-RT-PCR 185
and quantified by Kodak Digital Science™ 1D image ana-lysis software (Eastman Kodak Company). To compare the specificity and sensitivity of three ZaMMV probes, ZaMMV, DsMV and ZaMV plasmid clones previously obtained in the laboratory were used to prepare 10-fold serially diluted samples. These samples were immobilized onto nylon membrane and then separately hybridized with the same quantity of each probe. For dot-blot hybridiza-tion analysis, 5 µL of crude plant sap prepared with indi-rect sample extraction buffer (Agdia Inc.) was transferred onto the nylon membrane (Millipore) presoaked in 20x SSC. After UV cross-linking, the membranes were prehy-bridized, hyprehy-bridized, washed and detected for hybridiza-tion signals as previously described (Hsu et al., 2005). Immunocapture-RT-PCR (IC-RT-PCR) with specific primers
Plant extracts were prepared by grinding fresh tissue in 10 volumes of general extract buffer (Agdia Inc.). After centrifugation at 11 925 g for 10 min, the supernatant was kept on ice until use. Reaction tubes were coated with
anti-ZaMMV antiserum, incubated at 37°C for 1 hour, and then washed three times with PBST buffer. The coated
reaction tubes were incubated with 100 µL of plant
extracts at 37°C for 1 hour, and then washed three times with PBST buffer. After drying, the coated reaction tube was vortexed with 5 µL of 0·5% Triton X-100 that was preheated to 65°C in order to release the viral RNA from the trapped virus particles (Wetzel et al., 1992). The RT reaction was carried out with the ZUNR1a primer at 42°C for 40 min in a total volume of 12·5 µL. The RT product was used as a template in the subsequent PCR reaction with ZUNF1a/ZUNR1a primers and the PCR condition was the same as before. The PCR products were analyzed by electrophoresis in a 1·5% agarose gel.
Results
Construction and expression of the recombinant ZaMMV CP gene
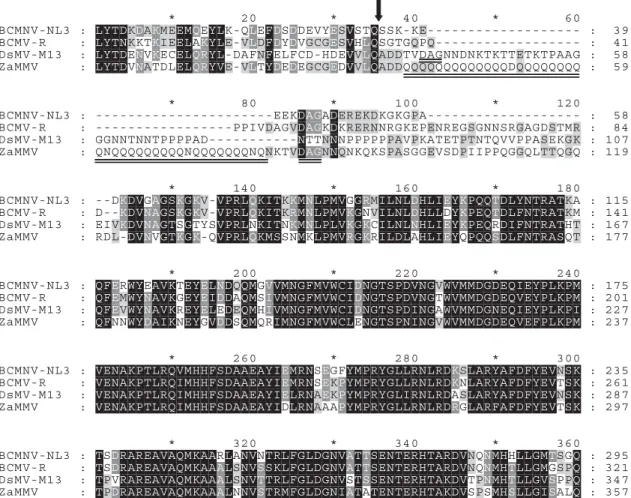
Initially, a 980-bp fragment encompassing the intact CP gene with polyglutamine sequence (Fig. 1) was amplified Figure 1 Alignment of the partial amino acid sequences of Zantedeschia mild mosaic virus (ZaMMV) and three closely related potyviruses as previously described (Huang & Chang, 2005). BCMNV-NL3: NL3 strain of Bean common mosaic necrosis virus (U19287); BCMV-R: R isolate of Bean common mosaic virus (AJ312437); DsMV-M13: M13 isolate of Dasheen mosaic virus (AJ298033) and ZaMMV (AY626825). The predicted cleavage site between NIb and CP is indicated by the arrow. The polyglutamine sequence and the DAG motif in the CP of ZaMMV are underlined.
186 C.-H. Huang et al.
by primers ZUNCPF1/ZUNCPR1 and then cloned into a pET-29a(+) expression vector. Six independent clones were identified by enzyme digestion, and one clone desig-nated pZUNCP7–4 was sequenced from both directions. After transformation and IPTG induction, the predicted 41·2-kDa recombinant protein was not expressed in E.
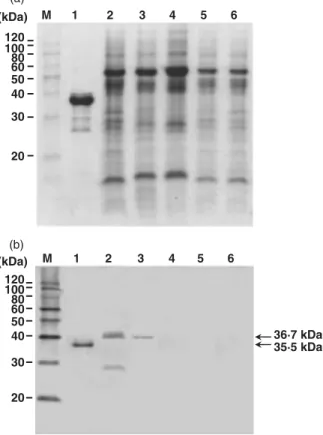
coli BL21 (DE3) by pZUNCP7–4 nor in the remaining
five clones (data not shown). Another forward primer, ZUNCPF2, was therefore used together with ZUNCPR1 to produce a PCR fragment of 830 bp that excluded the polyglutamine sequence from the N terminus of the CP gene of ZaMMV. Two correct clones were obtained and confirmed by enzyme digestion. One sequenced clone, pZUNCPF2-7-1, was used to transform E. coli BL21 (DE3). The predicted 35·5-kDa recombinant protein was successfully expressed in E. coli after induction for 1 h by IPTG and gradually increased for another 3 h (Fig. 2). The large quantities of expressed protein were purified as antigens and the antiserum of ZaMMV was prepared by a local private company.
Specificity of ZaMMV antiserum analyzed by immunoblot and ELISA assays
To test the specificity of the ZaMMV antiserum, the expressed recombinant CP of ZaMMV and total proteins of healthy and diseased plants were assayed by immuno-blot analysis. The DsMV- and ZaMV-infected plants were chosen for analysis because these potyviruses are frequently found to infect calla lilies in Taiwan (Chen
et al., 1998; Chang et al., 2001). The results showed that
ZaMMV antiserum only reacted with the CP of ZaMMV in the crude extracts from ZaMMV-infected calla lily and
P. selloum plants and did not react with any proteins from
other diseased or healthy plants (Fig. 3). The predicted molecular weight of recombinant ZaMMV CP (35·5 kDa), although containing 30 amino acids of expression
vector and a His tag, was still smaller than that of native CP (36·7 kDa) due to its lack of the polyglutamine region (equivalent to 46 amino acids) (Fig. 3). A smaller protein ∼27 kDa in size, possibly a degraded product of the CP, also reacted to ZaMMV antiserum. A recent immunoblot assay indicated that CPs of TuMV and BYMV were not recognized by ZaMMV antiserum (data not shown). According to the results of I-ELISA, ZaMMV antiserum did not react to DsMV, ZaMV, TuMV or BYMV but did react to ZaMMV-infected samples (data not shown). The ZaMMV antiserum is apparently highly specific and can be used for detection and diagnosis.
Comparison of three specific DNA probes of ZaMMV The probe 1175 was prepared by PCR with PNIbF1/ PCPR1 primers, and spans the entire 3′ region of the NIb gene and the 5′ region of the CP gene. The probe 311 Figure 2 Bacterial lysates of Escherichia coli BL21 (DE3) transformed
with pZUNCPF2-7-1 were analyzed by electrophoresis in a 12% SDS-polyacrylamide gel. Lane M, Mark12™ MW Standard (Invitrogen); lane 1, no IPTG induction; lane 2, one hour after IPTG induction; lane 3, three hours after IPTG induction; lane 4, four hours after IPTG induction. Molecular weights of the markers are indicated on the left. The position of the expressed protein is indicated by an arrow and the number represents the deduced molecular weight.
Figure 3 Specificity of Zantedeschia mild mosaic virus (ZaMMV) antiserum by immunoblot analysis. Expressed protein and crude extracts of diseased and healthy plants were separated by 12% SDS-PAGE and either stained with Coomassie brilliant blue (a) or transferred to a PVDF membrane and reacted with anti-ZaMMV antiserum (b). Lane M, MagicMark™ Western Standard (Invitrogen); lane 1, purified recombinant protein of ZaMMV CP; lanes 2 – 3, crude extracts of ZaMMV-infected calla lily and Philodendron selloum, respectively; lane 4, crude extract of DsMV-infected calla lily; lane 5, crude extract of ZaMV-infected calla lily; lane 6, healthy calla lily. Molecular weights of the markers are indicated on the left. The positions of native and recombinant proteins of ZaMMV CP are indicated by arrows, and the numbers represent the deduced molecular weights.
ZaMMV detected by ELISA, dot hybridization and IC-RT-PCR 187
covering the junction region of the NIb and CP genes, the sequence of which is variable among potyviruses, was amplified with ZUNF1a/ZUNR1a primers. The probe 251 corresponding to the 3′UTR of ZaMMV was PCR-amplified with ZUNF3/ZUNR0 primers. The cDNA clones of ZaMMV, DsMV and ZaMV were used to compare the specificity and sensitivity of the probes. The results indicated that all probes reacted with the ZaMMV cDNA clone only (Fig. 4). Probe 311 and probe 251 could detect at least 0·45 ng of ZaMMV plasmid DNA, whereas probe 1175 had a detection limit ten times lower (Fig. 4). Because probe 311 showed better sensitivity than probe 251 due to its stronger signals (Fig. 4), it was used to per-form dot-blot hybridization to detect ZaMMV in field samples. A recent result confirmed the high specificity of probe 311, which did not hybridize with TuMV or BYMV cDNA clones (data not shown).
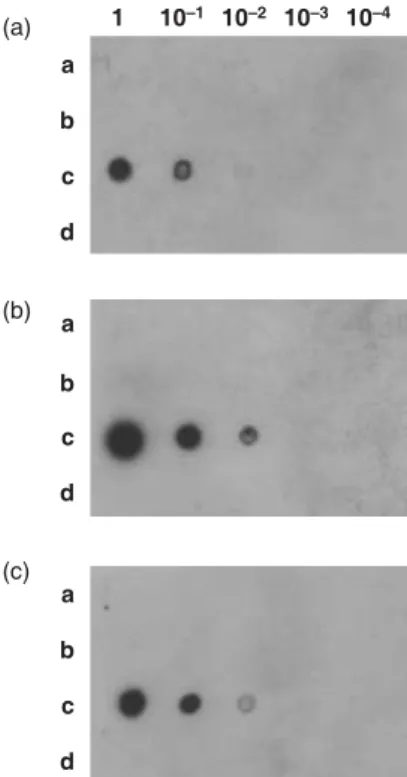
Detection of ZaMMV in calla lilies by I-ELISA, dot-blot hybridization and IC-RT-PCR
To see whether ZaMMV already exists in some areas of Taiwan, calla lilies were collected from the field in central Taiwan, purchased from the flower market and the TSIPS, and were tested with ZaMMV antiserum by I-ELISA. The numbers of infected calla lilies were 10/13 from the field, 4/24
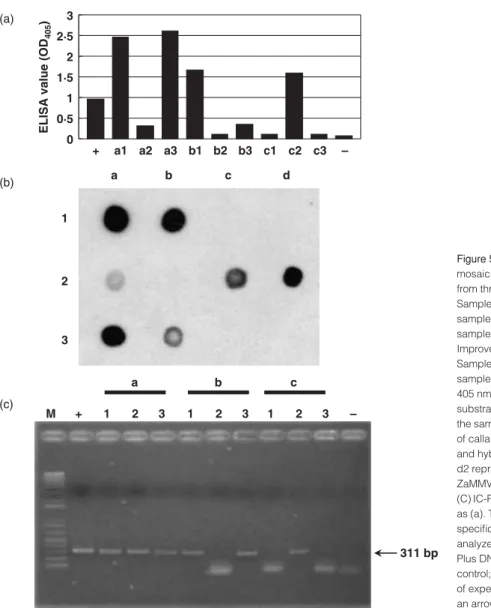
from the market, and 2/19 from TSIPS. This result indicates that 10·5–77% of samples tested were infected by ZaMMV. Calla lilies from the same three sources were randomly selected to test ZaMMV by I-ELISA, dot-blot hybridiza-tion and IC-RT-PCR at the same time. All three samples from the field (Fig. 5a, a1-a3), two samples from the market (Fig. 5a, b1 and b3) and one sample from TSIPS (Fig. 5a, c2) reacted with antiserum to ZaMMV. Crude saps of the same samples were checked by dot-blot hybridization with probe 311 and also by IC-RT-PCR with ZaMMV-specific primers, ZUNF1a and ZUNR1a. The result of IC-RT-PCR was similar to that of dot-blot hybridization (Fig. 5b and c), and also agreed with the result of I-ELISA. Some samples (Fig. 5, a2 and b3) having weak signals in dot-blot hybridization and low ELISA values produced the same amount of ZaMMV cDNA fragment in the IC-RT-PCR assay.
Occurrence of ZaMMV, DsMV and ZaMV in calla lilies To evaluate the disease incidence caused by ZaMMV, DsMV and ZaMV in calla lilies in Taiwan, two field sur-veys were done during 2003 and one during 2004. A total of 101 samples collected from three fields on different dates were tested by I-ELISA using ZaMMV, DsMV and ZaMV antisera simultaneously. The results indicated that 55% and 38·5% of calla lilies in 2003 and 42·9% in 2004 detected positive for ZaMMV (Table 2). Among samples tested, 72·5% and 65·4% in 2003 and 60% in 2004 were infected by ZaMV (Table 2). Although 27·5% and 30·8% of calla lilies were infected by DsMV in the 2003 survey, none of the samples tested positive in 2004 (Table 2). According to the ELISA data, only 20–34·6% of sampled calla lily plants did not react to any of the antisera. In addition, 38–63% of infected plants contained two or even three kinds of viruses (Table 2).
Discussion
The purification of virus particles from aroid plants is dif-ficult and resulted in the failure to produce good quality antiserum (Li et al., 1998). A similar situation arose when purifying virus particles from calla lilies and P. selloum, probably due to the mucilage in the plant tissues. Expres-sion of the recombinant CP in E. coli solved this problem and the purified protein was a good antigen to produce antiserum (Li et al., 1998). A similar approach was there-fore employed to produce ZaMMV antiserum, which had very good specificity. Bacteria-expressed viral CP can therefore be used as an alternative method to produce antiserum for viruses that are difficult to purify.
The dominant epitopes of potyvirus virions are the var-iable terminal parts of the CP, especially the N-terminal part. It has been reported that about 30 amino acids of the N terminus of the CP were exposed on the surface of the virus particle (Shukla & Ward, 1989). Therefore, the unique repetitive sequence of ZaMMV CP is composed of 39 glutamine residues which are likely to be exposed on the surface of the virion, if not degraded or processed Figure 4 Dot-blot hybridization assay for comparison of the specificity
and sensitivity of three Zantedeschia mild mosaic virus (ZaMMV) probes. The membranes were hybridized with three ZaMMV DNA probes, probe 1175 (a), probe 311 (b), and probe 251 (c), respectively. Plasmid DNAs of DsMV (a), ZaMV (b) and ZaMMV (c) clones were 10-fold serially diluted and dotted on the membranes together with H2O (d) as
188 C.-H. Huang et al.
(Huang & Chang, 2005). It has been shown that incorpo-ration of polyglutamine results in protein oligomeriza-tion, probably because the glutamine repeats form polar zippers for protein-protein interaction (Stott et al., 1995). In the current study, pZUNCP7–4 and five similar recom-binant clones that failed to express recomrecom-binant protein in
E. coli have polyglutamine at the N-terminus of ZaMMV
CP. If the recombinant protein expressed from them aggregated and then interfered with the transcription of
E. coli, it may explain why this recombinant protein
cannot be overexpressed in E. coli.
In addition to DsMV (Chen et al., 1998), ZaMV (Chang
et al., 2001) and TuMV (Chen et al., 2003), ZaMMV is
another potyvirus found to infect calla lily in Taiwan (Huang & Chang, 2005). The original source of ZaMMV might be the imported tubers of calla lily from other coun-tries. Some imported tubers collected directly from the international airport by HsinChu Branch Office of The Bureau of Animal and Plant Health Inspection and Quar-antine (BAPHIQ) were shown to be infected with ZaMMV by this laboratory (data not shown). According to TSIPS,
Figure 5 Detection of Zantedeschia mild mosaic virus (ZaMMV) in calla lilies collected from three sources. (a) Indirect-ELISA assay. Samples a1-a3, calla lilies from the field; samples b1-b3 calla lilies from the market; samples c1-c3, calla lilies from the Taiwan Seed Improvement and Propagation Station (TSIPS). Sample +, ZaMMV-infected calla lily; sample –, healthy calla lily. The absorbance at 405 nm was measured 30 min after addition of substrate. (b) Dot-blot hybridization assay of the same set of samples as (a). Crude extracts of calla lilies were dotted onto the membrane and hybridized with probe 311. Dots d1 and d2 represent total RNA of healthy and ZaMMV-infected calla lilies, respectively. (C) IC-RT-PCR assay of the same set of samples as (a). The DNA products amplified by ZaMMV specific primers, ZUNF1a/ZUNR1a, were analyzed in a 1·5% agarose gel. Lane M, 1 kb Plus DNA Ladder (Invitrogen); lane +, positive control; lane −, negative control. The position of expected amplified product is indicated by an arrow.
Table 2 Number of samples (% in parenthesis) of calla lilies collected from different fields testing positive by I-ELISA using DsMV, ZaMV and ZaMMV antisera simultaneously
ELISA resulta Field Ab 18 April 2003 Field Bb 30 May 2003 Field Cb 26 December 2004 ZaMMV 3 (7·5) 0 5 (14·3) DsMV 0 0 0 ZaMV 9 (22·5) 7 (26·9) 11 (31·4) ZaMMV + DsMV 0 0 0 ZaMMV + ZaMV 9 (22·5) 2 (7·7) 10 (28·6) DsMV + ZaMV 1 (2·5) 0 0 ZaMMV + DsMV + ZaMV 10 (25) 8 (30·8) 0 ND 8 (20) 9 (34·6) 9 (25·7) Total 40 (100) 26 (100) 35 (100) a
OD405 value greater than twice the mean of the healthy controls was
considered a positive sample. ND: sample did not react to three kinds of antisera.
b
Field A located in Hsinshe, Taichung County; field B located in Peitou, Changhua County; field C located in Tanzih, Taichung County.
ZaMMV detected by ELISA, dot hybridization and IC-RT-PCR 189
the imported calla lilies were only screened with anti-DsMV and CMV antibodies, and then used for micropropaga-tion. This may explain why the tissue-cultured seedlings purchased from TSIPS were still infected by ZaMMV.
In order to survey the prevalence of ZaMMV in Tai-wan, three detection methods were developed and were able to detect the virus successfully. I-ELISA is the most convenient method. Dot-blot hybridization is also suitable for handling large amounts of samples. Although IC-RT-PCR is not appropriate for large scale screening, it showed better sensitivity than the other two methods as previously described (Wetzel et al., 1992), and may be valuable for mother stock indexing of calla lily. So far, ZaMMV seems to infect only members of the Araceae family (Huang & Chang, 2005) but other non-aroid hosts cannot be excluded. Although ZaMMV alone caused mild mosaic on calla lilies, it was frequently found together with ZaMV and/or DsMV in the field based on this study (Table 2). It was not determined whether the infection of these three viruses was synergistic.
Although importation of ZaMMV into Taiwan may have been accidental, ZaMMV appears to be prevalent in the field. Zettler & Hartman (1986; 1987) have success-fully removed DsMV by tissue culture, but tissue culture alone cannot guarantee the seedlings will be free of virus unless sensitive detection methods are available. This explains why the tissue-cultured calla lilies from TSIPS grown in the field had a lower percentage (27·5% and 30·8%) of DsMV infection in 2003 and no infection in 2004, since the DsMV antibody has been routinely used in micropropagation of calla lily by TSIPS. Because ZaMV and ZaMMV are two new viruses found in calla lily, the detection methods for these two viruses are not yet included in the TSIPS system. Therefore, the frequencies of ZaMV and ZaMMV detected by ELISA were higher than DsMV (Table 2). DsMV and ZaMV were shown to have a significant influence on the production of calla lil-ies. Although the significance of the newly found ZaMMV has not been determined, it was commonly observed to be the causal agent of complex infection in calla lilies. The importance of ZaMMV in the calla lily industry should not be underestimated and because calla lily has become a popular cut flower in Taiwan, a virus-free certification scheme, using the described detection methods, needs to be established.
Acknowledgements
We thank Dr Ching-Chung Chen for helping to collect calla lily samples from Peitou, Changhua County and Dr Yung-An Lee for critically reviewing the manuscript. This study was partially supported by a grant from the Council of Agriculture (92AS-1·1·5-SS-X3(2-1), Executive Yuan, Taiwan, R. O. C.
References
Chang YC, Chen YL, Chung FC, 2001. Mosaic disease of calla lily caused by a new potyvirus in Taiwan. Plant Disease 85, 1289.
Chen CC, Chao CH, Chen CC, Yeh SD, Tsai HT, Chang CA, 2003. Identification of Turnip mosaic virus isolates causing yellow stripe and spot on calla lily. Plant Disease 87, 901–5. Chen J, Chen J, Chen J, Adams MJ, 2001. Molecular
characterization of an isolate of Dasheen mosaic virus from
Zantedeschia aethiopica in China and comparisons in the
genus Potyvirus. Archives of Virology 146, 1821–9. Chen YL, Chung FC, Chang YC, 1998. Molecular cloning and
characterization of dasheen mosaic potyvirus from aroid plants in Taiwan. Plant Pathology Bulletin 7, 226–7. Hsu YC, Yeh TJ, Chang YC, 2005. A new combination of RT-PCR
and reverse dot blot hybridization for rapid detection and identification of potyviruses. Journal of Virological Methods 128, 54–60.
Hu WC, Chang YC, 2004. A new mosaic disease of Amazon lily in Taiwan. Plant Pathology 53, 240.
Huang CH, Chang YC, 2005. Identification and molecular characterization of Zantedeschia mild mosaic virus, a new calla lily-infecting potyvirus. Archives of Virology 150, 1221–30.
Huang WZ, Chang YC, 2000. Detection of Dasheen mosaic
potyvirus and Zantedeschia mosaic potyvirus with different
kinds of methods. Plant Pathology Bulletin 9, 199. Kwon SB, Ha JH, Yoon JY, Ryu KH, 2002. Zantedeschia
mosaic virus causing leaf mosaic symptom in calla lily is a new potyvirus. Archives of Virology 147, 2281–9. Li RH, Zettler FW, Purcifull DE, Hiebert E, 1998. The
nucleotide sequence of the 3′-terminal region of dasheen mosaic virus (Caladium isolate) and expression of its coat protein in Escherichia coli for antiserum production. Archives
of Virology 143, 2461–9.
Mokra V, Gotzova B, 1994. Identification of virus infections in
Dieffenbachia and Zantedeschia in Czechoslovakia. Acta Horticulturae 377, 361–2.
Pham K, Langeveld SA, Lemmers MEC, Derks AFLM, 2002. Detection and identification of potyviruses in Zantedeschia.
Acta Horticulturae 568, 143–8.
Rana GL, Vovlas C, Zettler FW, 1983. Manual transmission of dasheen mosaic virus from Richardia to nonaraceous hosts.
Plant Disease 67, 1121–2.
Sambrook J, Russell DW, 2001. Molecular Cloning, A
Laboratory Manual, 3rd edn. Cold Spring Harbor, NY,
USA: Cold Spring Harbor Laboratory Press.
Shukla DD, Ward CW, 1989. Structure of potyvirus coat proteins and its application in the taxonomy of the potyvirus group. Advances in Virus Research 36, 273–314.
Stott K, Blackburn JM, Butler PJG, Perutz M, 1995. Incorporation of glutamine repeats makes protein oligomerize: implications for neurodegenerative disease.
Proceedings of the National Academy of Sciences, USA
92, 6509–13.
Wetzel T, Candresse T, Macquaire G, Ravelonandro M, Dunez J, 1992. A highly sensitive immunocapture polymerase chain reaction method for plum pox potyvirus detection. Journal of
Virological Methods 39, 27–37.
Zettler FW, Hartman RD, 1986. Dasheen mosaic virus and its
control in cultivated aroids. Taipei, Taiwan, Republic of
China: Food and fertilizer technology center for Asia and Pacific region: FFTC book no. 33.
Zettler FW, Hartman RD, 1987. Dasheen mosaic virus as a pathogen of cultivated aroids and control of the virus by tissue culture. Plant Disease 71, 958–63.