Journal of Chromatography A, 979 (2002) 299–306
www.elsevier.com / locate / chroma
C
omparison of the separation of large DNA fragments in the
presence and absence of electroosmotic flow at high pH
*
Tai-Chia Chiu, Huan-Tsung Chang
Department of Chemistry, National Taiwan University, Section 4, Roosevelt Road, Taipei, Taiwan
Abstract
This paper describes the analysis of large DNA fragments at pH .10.0 by capillary electrophoresis (CE) in the presence of electroosmotic flow (EOF) using hydroxyethylcellulose (HEC) solution. HEC solution in the anodic reservoir enters the capillaries filled with high-pH buffer by EOF after sample injection. With respect to resolution, sensitivity, and speed, separation conducted under discontinuous conditions (different pH values of HEC solutions and buffer filling the capillary) is appropriate. Using HEC solution at concentrations higher than its entanglement threshold ensures a good separation of large DNA fragments in the presence of EOF at high pH. In addition to pH and HEC, the electrolyte species, dimethylamine, methylamine, and piperidine, play different roles in determining the resolution. The separation of DNA fragments ranging in size from 5 to 40 kilo base pairs was completed in 6 min using 1.5% HEC prepared in 20 mM methylamine–borate, pH 12.0, and the capillary filled with 40 mM dimethylamine–borate, pH 10.0. In comparison, this method allows faster separations of large DNA fragments compared with that conducted in the absence of EOF using dilute HEC solutions.
2002 Elsevier Science B.V. All rights reserved.
Keywords: Electroosmotic flow; Buffer composition; DNA
1
. Introduction 9], cellulose and its derivatives [10–12], polyvinyl-pyrrolidone (PVP) [13], and poly(ethylene oxide) Capillary electrophoresis (CE) using polymer so- (PEO) [14], have been tested successfully. Common-lutions has become a major technique for DNA ly, deactivated capillaries are used to minimize the sequencing and the analysis of polymerase chain variation in electroosmotic flow (EOF) and DNA reaction (PCR) products [1–3]. It has considerably adsorption on the capillary wall [15,16].
assisted the rapid progress in the Human Genome The analysis of large DNA fragments is of consi-Project, and has driven the success of completing the derable interest and importance in many respects. It draft sequence well ahead of schedule [4,5]. The helps to assess the effect of tumorigenesis on telo-success of CE in this regard is due in part to the use meres by measurement of variations in telomeric of replaceable polymer solutions with a high sieving length (7–10 kilo base tandem repeats of 59-ability. A number of hydrophilic polymers, such as TTAGGG-39 in humans) that exists between in-linear polyacrylamide (LPA) and its derivatives [6– dividuals, between chromosomes and between cells within an individual [17]. However, the analysis of small variations in large DNA fragments is not easy.
*Corresponding author. Tel.: 2362-1963; fax:
1886-2-It is known that the electrophoretic mobility of long
2362-1963.
E-mail address: changht@ccms.ntu.edu.tw(H.-T. Chang). DNA fragments becomes independent of the
molecu-0021-9673 / 02 / $ – see front matter 2002 Elsevier Science B.V. All rights reserved. P I I : S 0 0 2 1 - 9 6 7 3 ( 0 2 ) 0 1 4 3 8 - 3
lar size and DNA fails to enter the gel or is trapped 2 . Materials and methods by the sieving matrix [18]. To achieve a reasonable
resolution, a number of polymer solutions at con- 2 .1. Equipment
*
centrations below their entanglement threshold (F )
have been employed for the separation of kilo base The basic design of the separation system has been pair (kbp) DNA [19–21]. The success of these described previously [24]. Briefly, a high-voltage separations stems from the fact that DNA drags power supply (Gamma High Voltage Research, Or-along the polymer molecules it encounters during mond Beach, FL, USA) was used to drive electro-migration, with support from the dynamic formation phoresis. The entire detection system was enclosed and deformation of the U-shaped DNA conformation in a black box with a high-voltage interlock. The [18,22]. In addition, it has been suggested that the high-voltage end of the separation system was placed relative DNA / HEC size plays a significant role in in a laboratory-made Plexiglass box for safety. A determining the resolution and no a priori upper size 1.5-mW He–Ne laser with 543.6 nm output from limit to DNA by CE in a constant field is anticipated Uniphase (Mantence, CA, USA) was used for excita-[19–23]. Alternatively, pulsed-field capillary gel tion. The light was collected with a 103 objective electrophoresis (PFCGE) is employed for the analy- (numerical aperture 0.25). A RG 610 cutoff filter was sis of DNA fragments up to 1.6 Mbp, with a faster used to block scattered light before the emitted light analysis compared with pulsed-field gel electropho- reached the photomultiplier tube (Hamamatsu R928). resis [19]. However, this system is relatively compli- The amplified currents were transferred directly cated. through a 10 kV resistor to a 24-bit A / D interface at Alternatively, we have demonstrated the sepa- 10 Hz (Borwin, JMBS Developments, Le Fontanil, ration of kbp DNA at pH 8.2 in the presence of EOF France) and stored in a personal computer. Bare using PEO solution with concentrations above its fused-silica capillaries (Polymicro Technologies,
*
F [24]. However, the fact that the EOF decreases Phoenix, AZ, USA) with 75 mm I.D. and 365 mm gradually due to the dynamic coating of PEO O.D. were used.
molecules on the capillary wall needs to be carefully
considered in terms of speed and reproducibility 2 .2. Chemicals [25,26]. We have concluded that adsorption is more
profound using high concentrations of PEO solution All chemicals for preparing buffer solutions and when the capillary is filled with low concentrations polymers were from Aldrich (Milwaukee, WI, USA). of Tris–borate (TB) buffer. Although we have also EtBr was obtained from Molecular Probes (Eugene, found that PEO adsorption is less at high pH, the OR, USA). Solutions prepared from methylamine, analysis of DNA using PEO is not quite successful dimethylamine, and piperidine were adjusted with because of the hydrolysis of PEO. On the other hand, suitable amounts of boric acid to pH values ranging we also realize that the analysis of DNA at high pH from 8.0 to 12.0 and thus the prepared solutions are might be faster, due to less PEO adsorption, and called MB, DB, and PB buffers, respectively. Unless more sensitive, because of the stronger fluorescence otherwise noted, X mM MB, DB or PB buffer of DNA intercalated with ethidium bromide (EtBr). indicates an X mM amine solution adjusted with a Thus, the development of a method for the sepa- suitable amount of boric acid. These buffers were ration of large DNA fragments at high pH is worthy used to fill the capillaries when the separations were from a practical point of view. In this study, we carried out in the presence of EOF. Certain amounts tested the separation of large DNA fragments at pH of HEC (molecular mass 1 300 000 g / mol) were values ranging from 8.0 to 12.0 using HEC solution added separately to 50 mL of 20 mM MB, DB, and in the presence of EOF. We explored the effect of PB containing 5 mg / ml EtBr, and the prepared HEC buffers prepared from borate and three different solutions were used for DNA separations. For the amines on DNA separation. Unlike at pH 8.2, HEC experiments conducted in the absence of EOF, HEC
*
solutions at concentrations much greater than the F solutions were also prepared in 13 TBE, pH 8.2. had to be used for the separation of large DNA FX 174 RF DNA-HaeIII digest and KiloBase DNA
T.-C. Chiu, H.-T. Chang / J. Chromatogr. A 979 (2002) 299–306 301
(Uppsala, Sweden) and were used only when the improvements in resolution for the separation of separations were carried out in the presence of EOF. DNA fragments at neutral pH using histidine buffers
ˇ
A 5 kb DNA ladder was purchased from Gibco / BRL [29]. Bocek’s and Kuhr’s groups individually dem-(Bethesda, MD, USA). onstrated that the separation efficiency is much greater at high pH ( .10.0) [7,30,31]. Tseng and 2
.3. Separation in the presence of EOF Chang have recently developed a technique based on a pH junction for optimizing the sensitivity and Prior to analysis, capillaries were treated with 0.5 resolution of the analysis of DNA [32]. In the
M NaOH overnight [25]. After each run, capillaries present study, we further evaluated the effect of pH were washed with 0.5 M NaOH at 1 kV for 10 min in the range 10.0 to 12.0 on the separation of DNA to remove HEC solutions and to refresh the capillary using PB, DB, and MB buffers. Table 1 shows that, wall, and subsequently filled with MB, DB or PB in the presence of EOF (counter flow), the analyses buffer. This treatment was quite successful with conducted either at pH 10.0 or 11.0 were more respect to speed and reproducibility. The bulk EOF successful in terms of sensitivity, and the results are
24 2 21 21
mobilities were greater than 5.8?10 cm V s , in good agreement with the maximum quantum with relative standard deviation (RSD) values yields of intercalated DNA at about pH 11.0 [33].
,2.0%. The injection of DNA samples was carried Using these conditions, the separations were faster 24 out by electrokinetic injection at 1 kV for 10 s. ( ,13 min) due to high EOF mobilities ( .5.8?10
2 21 21
During the analysis, HEC solutions in the anodic cm V s ). At high pH, peak profiles (peak width reservoir entered the capillary by EOF and served as for the 1353-bp fragment ,0.02 min at half-height) sieving matrices for DNA separations. Owing to the for the DNA fragments were sharp, mainly due to bulk EOF mobilities being greater than the electro- weaker interactions with the capillary wall. On the phoretic mobility of DNA migrating against the other hand, the separation was not suitable at pH EOF, large DNA fragments were detected earlier 12.0, mainly because of Joule heating, weak interca-towards the cathode end. lation between EtBr and the DNA fragments, and partial denaturation of DNA [33,34]. In addition, the 24 2 21 2
.4. Separation in the absence of EOF relatively small bulk EOF ( ,3.5?10 cm V 21
s ) compared with that at pH 9.0–11.0 is proble-For the separation of the 5 kb DNA ladder in the matic for the separation of small DNA fragments. It absence of EOF, 5% PVP was used to dynamically took more than 17 min for the small DNA fragments coat the capillary overnight. Prior to separation, the ( ,72 bp) that have higher electrophoretic mobilities, capillary was washed with TB buffer and sub- and thus resolution was lost due to peak broadening sequently filled with 0.05% HEC solutions at differ- [8]. Despite the pH effect, Table 1 also shows that ent pH values. In the absence of EOF, DNA mi- the amines affected the sensitivity and resolution of grated towards the anode when a negative electric the separation of the FX 174 RF DNA-HaeIII field of 25 V/ cm was applied. After each run, the digest. As suggested by several groups [35,36], such capillary was coated with PVP to achieve optimum effects may be due to changes in DNA conformation resolution and reproducibility. (hydrogen bonding) and the interaction between
DNA and EtBr.
It is also interesting to note that the EOF
mo-3
. Results and discussion bilities are different at the same pH when using different amines to prepare the buffers. For example, 3
.1. Impact of amines and pH using 1.7% HEC prepared in 20 mM DB buffer, pH 11.0, the EOF mobilities were 7.28, 5.98, and 6.83?
24 2 21 21
Although pH affects the electrophoretic mobility 10 cm V s when the capillary was filled with of DNA, intercalation, the quantum yield of interca- 40 mM PB, MB, and DB buffers, pH 11.0, respec-lated DNA, and the stability of the capillary coating, tively. Owing to slight differences in the conduc-only a few reports deal with its impact on DNA tivity ( ,20 mA), the viscosity change due to Joule separation in CE [26–28]. Magnusdottir et al. found heating should be neglected. In the presence of HEC,
T .-C . Chiu , H .-T . Chang / J. Chromatogr . A 979 (2002) 299–306 Table 1 a Effect of pH and background electrolyte on the separation of FX 174 RF DNA-HaeIII digest in the presence of EOF
b b
Buffer Peak height (V) Bandwidth (w1 / 2, min) Resolution (1353 / 1078)
(40 mM) filling the pH 10.0 pH 11.0 pH 12.0 pH 10.0 pH 11.0 pH 12.0 pH 10.0 pH 11.0 pH 12.0 capillary PB MB DB PB MB DB PB MB PB MB DB PB MB DB PB MB PB MB DB PB MB DB PB MB 10.0 PB 0.33 0.82 0.05 0.38 N.D. 0.18 N.D. N.D. 0.02 0.01 0.01 0.02 N.D. 0.03 N.D. N.D. 2.48 2.72 2.02 3.24 N.D. 1.13 N.D. N.D. 11.0 0.62 0.42 0.11 0.58 0.11 0.37 N.D. N.D. 0.03 0.01 0.02 0.03 0.02 0.02 N.D. N.D. 2.31 2.69 0.62 2.90 1.38 1.40 N.D. N.D. c 12.0 N.D. 0.67 0.05 0.17 0.03 0.87 0.45 0.42 N.D. 0.02 0.02 0.03 0.03 0.02 0.03 0.02 N.D. 2.36 0.23 1.02 1.49 1.38 0.62 1.92 10.0 MB 0.26 1.75 0.09 0.17 N.D. 0.06 N.D. N.D. 0.03 0.01 0.02 0.05 N.D. 0.03 N.D. N.D. 2.10 2.93 1.66 2.16 N.D. 0.59 N.D. N.D. 11.0 0.34 1.59 0.24 0.38 N.D. 1.89 1.00 N.D. 0.02 0.01 0.02 0.05 N.D. 0.02 0.02 N.D. 2.26 2.73 1.94 1.12 N.D. 2.06 1.52 N.D. 12.0 0.72 0.84 0.10 0.42 0.41 0.70 1.18 N.D. 0.04 0.03 0.02 0.03 0.02 0.03 0.03 N.D. 0.78 1.45 1.77 2.18 1.61 1.77 0.59 N.D. 10.0 DB 1.74 1.23 0.26 0.29 N.D. 1.02 N.D. N.D. 0.01 0.01 0.02 0.02 N.D. 0.02 N.D. N.D. 1.35 2.80 2.66 2.66 N.D. 1.54 N.D. N.D. 11.0 1.61 1.78 1.04 0.43 N.D. 1.42 N.D. N.D. 0.01 0.01 0.01 0.02 N.D. 0.02 N.D. N.D. 2.38 2.90 2.16 2.18 N.D. 2.24 N.D. N.D. 12.0 1.41 0.76 0.48 0.12 1.35 1.65 1.40 N.D. 0.03 0.02 0.02 0.04 0.02 0.02 0.02 N.D. 1.13 2.36 2.06 0.80 1.89 2.07 1.97 N.D.
Buffer (20 mM ) used to prepare 1.7% HEC solutions. a
Other conditions as in Fig. 1. b
For the 1353-bp fragment. c
T.-C. Chiu, H.-T. Chang / J. Chromatogr. A 979 (2002) 299–306 303
the magnitude of the EOF does not simply relate to 3 .2. Effect of HEC ionic strength and the extent of dissociation of SiOH
on the capillary wall, because the species adsorbed Unlike for the separation of small DNA fragments, on the capillary wall not only directly affect the 40 mM DB buffer, pH 12.0, and 20 mM MB buffer, EOF, but also affect the EOF by changing the extent pH 10.0, were used to fill the capillary and prepare of HEC adsorption [25,26]. In these studies, we also the HEC solution, respectively, for the separation of showed that HEC adsorption decreases with increas- the kbp DNA marker. Using these conditions, a ing ionic strength and pH, and is less on a hydro- greater resolving power was achieved, mainly be-philic wall. It is known that DNA interactions with cause DNA migrated against a smaller EOF mobility
24 2 21 21
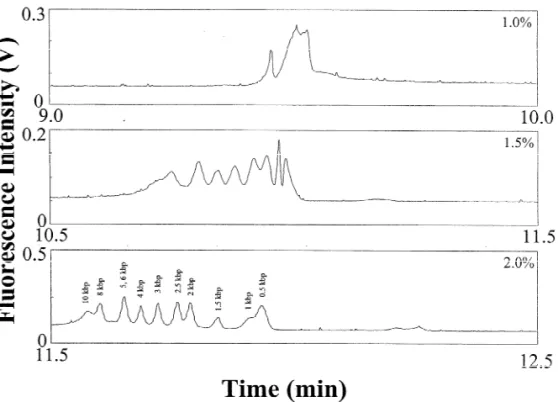
the capillary wall decrease, and thus reproducibility (3.5?10 cm V s ), leading to a longer sepa-and resolution improve, when using a capillary ration time. Fig. 1 shows the striking result that the coated with HEC [21]. Thus the buffer species also large DNA fragments were only separated at higher affects the peak profiles and resolution, as shown in HEC concentrations at high pH, which is in contrast Table 1. Overall, we suggest that, on the basis of to the results obtained by others [20] and to our sensitivity, two conditions are more appropriate: (1) previous result using PEO at pH 8.0 [24]. With the capillary is filled with 40 mM DB, pH 11.0, increasing HEC concentration, both the EOF and the when using 1.7% HEC prepared in 20 mM MB, pH diffusion coefficient of DNA decreased. As a result, 10.0; and (2) the capillary is filled with 40 mM MB, the selectivity (longer residence times) and efficiency pH 11.0, when using 1.7% HEC prepared in 20 mM improved, leading to better resolution. Our reasoning DB, pH 11.0. Regarding the resolution, the former is is supported by the fact that an electropherogram superior for the separation of large DNA fragments. showing 10 peaks was only achieved using 2.0%
Fig. 1. Separation of a 25 mg / mL kbp DNA marker in the presence of EOF at 250 V/ cm using different HEC solutions containing 5 mg / mL EtBr prepared in 20 mM MB, pH 10.0. Capillary: 60 cm total length and 50 cm effective length, filled with 40 mM DB buffer, pH 12.0. Electrokinetic injection was conducted at 16.7 V/ cm for 10 s.
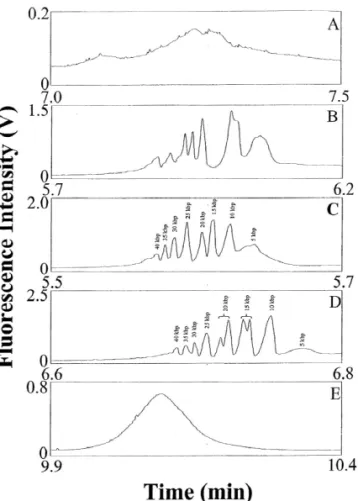
HEC, but not 1.7% HEC. Because the concentration pH 12.0 and the capillary filled with 40 mM DB of HEC used was much higher than its entanglement buffer, pH 8.0 or 9.0, the separations also failed, as threshold (0.37%) [20], we suggest that sieving of shown in Fig. 2A and B. This is probably due to DNA by HEC solutions takes place. Joule heating because of partial protonation of the amine and the existence of relatively large amounts 3
.3. Separation of the 5 kb DNA ladder of boric acid in the system. For example, 0.78, 0.44, 0.20, 0.10 and 0.01 g boric acid were added to Next we tested the separation of the 5 kb DNA prepare 40 mM DB buffers, pH 8.0, 9.0, 10.0, 11.0 ladder (the longest DNA fragment is 40 kbp) in the and 12.0, respectively. Fig. 2C shows that eight presence of EOF using 1.5% HEC prepared in peaks were resolved in 6 min using a capillary filled 20 mM MB buffer at different pH values and the with 40 mM DB, pH 10.0. The electropherogram in capillary filled with 40 mM DB buffer, pH 8.0–12.0. Fig. 2D shows that 10 peaks were resolved when the The separations were unsuccessful when using 1.5% capillary was filled with 40 mM DB, pH 11.0, HEC at pH ,11.0 (not shown), mainly because of indicating that the sample contained more than the the high EOF (shorter separation time), thereby eight DNA fragments suggested by the manufacture. causing poor resolution. When using 1.5% HEC at This indicates that our proposed method provides either high resolving power or greater sensitivity at high pH. The success of the separation might also be due to changes in DNA conformation (e.g. from J to I shape), presumably because of the existence of the EOF and the high viscosity of HEC matrices with small pore sizes [37]. Changes in DNA conformation due to a fast flow-rate and viscosity have also been investigated and tested for the separation of large DNA fragments ( .5 kbp) in slalom chromatography [38,39]. We point out that the separations were unsuccessful under isocratic conditions using the same buffers to prepare HEC solutions and to fill the capillary at pH 8.2–12.0, indicating that pH changes play an important role in improving the resolution. This might be due to conformation changes when DNA migrates in environments of different pH and background electrolytes.
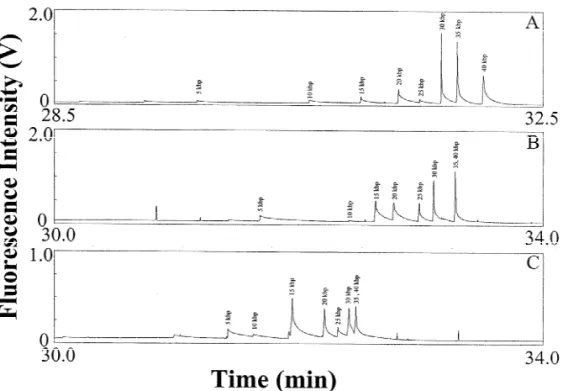
For comparison, we performed separations of the same DNA sample in the absence of EOF using a PVP-coated capillary and 0.05% HEC at pH 10.0, 11.0, and 12.0, separately. Fig. 3A shows that the separation was only successful at pH 10.0, which is similar to that at pH 8.2 and 9.0 (not shown). Unlike separation in the presence of an EOF, DNA was only separated at extremely low concentrations (e.g.
*
0.05%) of HEC, far below its F [20]. Note that the migration order is reversed compared with that in
Fig. 2. Separation of the 5 mg / mL 5 kb DNA ladder in the Fig. 2. It should be pointed out that the loss of presence of EOF at 125 V/ cm using 1.5% HEC solutions resolution between the 35 / 40 kbp DNA pairs containing 5 mg / mL EtBr prepared in 20 mM MB, pH 12.0.
occurred at pH 11.0 and 12.0, as shown in Fig. 3B
Capillary: 40 cm total length and 30 cm effective length, filled
and C. These results suggest that the separation of
with 40 mM DB buffer at (A) pH 8.0, (B) 9.0, (C) 10.0, (D) 11.0,
T.-C. Chiu, H.-T. Chang / J. Chromatogr. A 979 (2002) 299–306 305
Fig. 3. Separation of a 25 mg / mL 5 kb DNA ladder in the absence of EOF at 225 V/ cm using 0.05% HEC solutions prepared in 20 mM MB at (A) pH 10.0, (B) 11.0 and (C) 12.0, containing 5 mg / mL EtBr. Capillary: 40 cm total length and 30 cm effective length, filled with 40 mM DB buffers, pH 10.0, 11.0, and 12.0, respectively. Other conditions as in Fig. 1.
which DNA drags along the polymer molecules is and 0 at 375 V/ cm, respectively. As the electric field problematic at high pH. The loss of resolution at pH increases, dispersion proportional to the DNA size is 12.0 was also partially due to denaturation of DNA. predicted and thermal effects may dominate over In comparison, we conclude that separations in the diffusion [41]. In addition, shorter differential migra-presence of an EOF using higher concentrations of tion times contribute to the loss of resolution at high HEC solutions are faster. To this end, we suggest electric field strengths. Overall, the optimum electric that low concentrations of HEC are appropriate in field strength is 125 V/ cm, as shown in Fig. 2D. The the absence of an EOF, while higher concentrations results also suggest that voltage gradient techniques of HEC are superior for the separation of large DNA are promising for optimum resolution when separat-fragments at high pH. ing DNA fragments with a wide range of sizes.
To minimize the electric dispersion of DNA, which is proportional to the contour length of DNA
[40], the separation of large DNA fragments has 4 . Conclusions commonly been conducted at low electric field
strengths (several tens V/ cm). To further investigate The effects of pH and background electrolyte the impact of the electric field strength, we separ- prepared from three different amines on the sepa-ately conducted separations at 75, 125, 250, and 375 ration of DNA in the presence of an EOF have been V/ cm. At 75 V/ cm, all peaks co-migrated at a time investigated. Separations of large DNA fragments at of 10.9 min. DNA fragments less than 25 kbp were pH .10.0 have been demonstrated, with improve-resolved in a shorter period of time, while DNA ments in sensitivity (about five- to eight-fold). Un-fragments greater than 30 kbp co-migrated at 250 or like for separation using 13 TBE buffer, the use of 375 V/ cm. For example, the resolution values for HEC solution with concentrations much greater than 5 / 10, 20 / 25, and 35 / 40 kbp were 1.5, 1.7, and 0.4 at its entanglement threshold is essential for optimum 125 V/ cm, 4.9, 1.8, and 0 at 250 V/ cm, and 7.7, 1.9, resolution in the separation of large DNA fragments
[13] Q. Gao, E.S. Yeung, Anal. Chem. 20 (1998) 1382.
in the presence of an EOF at high pH. The resolution
[14] H.-T. Chang, E.S. Yeung, J. Chromatogr. B 669 (1995) 113.
increases with increasing HEC concentration at high
´
[15] J. Horvath, V. Dolnık, Electrophoresis 22 (2001) 644.
pH, mainly because of the decrease in diffusion and
[16] P.G. Righetti, C. Gelfi, B. Verzola, L. Castelletti,
Electro-the change in DNA conformation. In addition, DNA phoresis 22 (2001) 603.
migrating against the EOF should be one of the most [17] R.K. Moyzis, J.M. Buckingham, L.S. Cram, M. Dani, L.L. important contributors for better resolution. Com- Deaven, M.D. Jones, J. Meyne, R.L. Ratliff, J.-R. Wu, Proc.
Natl. Acad. Sci. USA 85 (1988) 6622.
pared with separation in the absence of an EOF, this
´
[18] L. Mitnik, L. Salome, J.L. Viovy, C. Heller, J. Chromatogr.
method allows faster separation. Unlike when using
A 710 (1995) 309.
pressure, the difficulty of filling the capillary with
[19] Y. Kim, M.D. Morris, Anal. Chem. 67 (1995) 784.
viscous polymer solutions no longer exists. One
[20] A.E. Barron, W.M. Sunada, H.W. Blanch, Electrophoresis 17
other advantage of using this approach for the (1996) 744.
analysis of large DNA fragments is the possibility of [21] A.E. Barron, H.W. Blanch, D.S. Soane, Electrophoresis 15
separating large volumes of DNA samples using a (1994) 597.
[22] X. Shi, R.W. Hammond, M.D. Morris, Anal. Chem. 67
viscous polymer solution [42–44]. Thus our future
(1995) 1132.
experimental aims will focus on applying this
meth-[23] A.E. Barron, W.M. Sunada, H.W. Blanch, Electrophoresis 16
od to the recovery of large-sized DNA such as
(1995) 64.
important genes from biological samples. [24] H.-S. Chen, H.-T. Chang, Electrophoresis 19 (1998) 3149.
[25] H.-S. Chen, H.-T. Chang, Anal. Chem. 71 (1999) 2033. [26] W.-L. Tseng, M.-M. Hsieh, S.-J. Wang, H.-T. Chang, J.
Chromatogr. A 894 (2000) 219. A
cknowledgements
[27] J. Ren, P.M. Ueland, Hum. Mutat. 13 (1999) 458. [28] T. Nock, J. Dove, B. McCord, D. Mao, Electrophoresis 22
This work was supported by the National Science
(2001) 755.
Council of Taiwan under contract number NSC 90- [29] S. Magnusdottir, C. Gelfi, M. Hamdan, P.G. Righetti, J. 2113-M002-058. Chromatogr. A 859 (1999) 87.
´ ´ ´ ˇ
[30] Z. Mala, K. Kleparnık, P. Bocek, J. Chromatogr. A 853 (1999) 371.
[31] Y. Liu, W.G. Kuhr, Anal. Chem. 71 (1999) 1668. R
eferences
[32] W.-L. Tseng, H.-T. Chang, Electrophoresis 22 (2001) 763. [33] J.-B. LePecq, C. Paoletti, J. Mol. Biol. 27 (1967) 87. [1] H. Zhou, A.W. Miller, Z. Sosic, B. Buchholz, A.E. Barron, L.
[34] P.H. Johnson, L.I. Grossman, Biochemistry 16 (1977) 4217. Kotler, B.L. Karger, Anal. Chem. 72 (2000) 1045.
[35] N.C. Stellwagen, A. Bossi, C. Gelfi, P.G. Righetti, Anal. [2] J. Zhang, M. Yang, X. Puyang, Y. Fang, L.M. Cook, N.J.
Biochem. 287 (2000) 167. Dovichi, Anal. Chem. 73 (2001) 1234.
[36] S.M. Clark, R.A. Mathies, Anal. Chem. 69 (1997) 1355. [3] H. Tan, E.S. Yeung, Anal. Chem. 70 (1998) 4044.
[37] M. Ueda, H. Oana, Y. Baba, M. Doi, K. Yoshikawa, Biophys. [4] T.K. Christopoulos, Anal. Chem. 71 (1999) 425R.
Chem. 71 (1998) 113. [5] Y. Baba, J. Chromatogr. B 687 (1996) 271.
[38] E. Peyrin, Y.C. Guilaume, A. Villet, A. Favier, Anal. Chem. [6] O. Salas-Solano, E. Carrilho, L. Kotler, A.W. Miller, W.
72 (2000) 853. Goetzinger, Z. Sosic, B.L. Karger, Anal. Chem. 70 (1998)
[39] J. Hirabayashi, K. Kasai, J. Chromatogr. A 893 (2000) 115. 3996.
ˇ ´ ˇ
[40] S. Popelka, Z. Kabatek, J.-L. Viovy, B. Gas, J. Chromatogr.
´ ´ ´ ˇ
[7] K. Kleparnık, Z. Mala, P. Bocek, Electrophoresis 22 (2001)
A 838 (1999) 45. 783.
[41] C. Heller, Electrophoresis 22 (2001) 629. [8] M. Chiari, S. Riva, A. Gelain, A. Vitale, E. Turati, J.
[42] M.-M. Hsieh, W.-L. Tseng, H.-T. Chang, Electrophoresis 21 Chromatogr. A 781 (1997) 347.
(2000) 2904. [9] C. Heller, Electrophoresis 20 (1999) 1962.
[43] W.-L. Tseng, M.-M. Hsieh, S.-J. Wang, C.-C. Huang, Y.-C. [10] R.W. Hammond, H. Oana, J.J. Schwinefus, J. Bonadio, R.J.
Lin, P.-L. Chang, H.-T. Chang, J. Chromatogr. A 927 (2001) Levy, M.D. Morris, Anal. Chem. 69 (1997) 1192.
179. [11] D.H. Atha, Electrophoresis 19 (1998) 1428.
[12] G. Raucci, C.A. Maggi, D. Parente, Anal. Chem. 72 (2000) [44] C.-C. Huang, M.-M. Hsieh, T.-C. Chiu, H.-T. Chang,