doi: 10.4014/jmb.0811.624 First published online 15 April 2009
Analysis of the Involvement of Chitin-Binding Domain of ChiCW in
Antifungal Activity, and Engineering a Novel Chimeric Chitinase with High
Enzyme and Antifungal Activities
Huang, Chien-Jui, Shu-Huei Guo, Shu-Chun Chung, Yu-Ju Lin, and Chao-Ying Chen
*
Department of Plant Pathology and Microbiology, National Taiwan University, Taipei 10617, Taiwan, Republic of China Received: November 17, 2008 / Revised: February 26, 2009 / Accepted: March 7, 2009
An antifungal chitinase, ChiCW, produced by Bacillus cereus 28-9 is effective against conidial germination of Botrytis elliptica, the causal agent of lily leaf blight. ChiCW as a modular enzyme consists of a signal peptide, a catalytic domain, a fibronectin type-III-like domain, and a chitin-binding domain. When two C-terminal domains of ChiCW were truncated, ChiCW∆FC (lacking the chitin-binding domain and fibronectin type III-like domain) lost its antifungal activity. Since ChiCW∆C (lacking the chitin-binding domain) could not be expressed in Escherichia coli as ChiCW∆FC did, a different strategy based on protein engineering technology was designed to investigate the involvement of the chitin-binding domain of ChiCW (ChBDChiCW) in antifungal activity in this study. Because ChiA1 of Bacillus circulans WL-12 is a modular enzyme with a higher hydrolytic activity than ChiCW but not inhibitory to conidial germination of Bo. elliptica and the similar domain composition of ChiA1 and ChiCW, the C-terminal truncated derivatives of ChiA1 were generated and used to construct chimeric chitinases with ChBDChiCW. When the chitin-binding domain of ChiA1 was replaced with ChBDChiCW, the chimeric chitinase named ChiAAAW exhibited both high enzyme activity and antifungal activity. The results indicate that ChBDChiCW may play an important role in the antifungal activity of ChiCW.
Keywords: Chitinase, chitin-binding domain, antifungal activity, protein engineering, hybrid enzyme
An antifungal chitinase, ChiCW, is produced by Bacillus cereus 28-9 isolated from the lily rhizosphere and is effective
against conidial germination of Botrytis elliptica, the causal agent of lily leaf blight [9]. ChiCW, as a modular endochitinase, consists of a signal peptide, a catalytic domain (CatD), a fibronectin type-III-like domain (Fn3D), and a chitin-binding domain (ChBD). Based on peptide sequence similarity of the catalytic domain, ChiCW is categorized as a member of the family 18 chitinases [6, 7, 9]. In previous studies, the ChBD of a family 19 chitinase, ChiC, from Streptomyces griseus has been demonstrated to be involved in chitinase and antifungal activities [12, 13]. Hence, we tried to investigate the function of the C-terminal region of ChiCW on enzyme and antifungal activities. However, only ChiCW∆FC (lacking ChBD and Fn3D) was successfully expressed and purified for biochemical characterization, but ChiCW∆C (lacking ChBD) was not [10]. Therefore, an alternative approach was designed to investigate the involvement of ChBDChiCW in the antifungal activity of ChiCW.
Protein engineering technology has been developed to create a novel protein that possesses an improved or novel property by changing the amino acid sequence of an existing protein and has become an essential tool of basic research for protein biochemistry [1, 19, 21]. One obvious application of this technology is to engineer enzymes with improved properties, which can be achieved by constructing chimeric or hybrid enzymes from pre-existing elements [1, 19, 21]. In this study, an approach based on protein engineering technology was designed to investigate the involvement of ChBDChiCW in the antifungal activity of ChiCW. As is well known, ChiA1 of Bacillus circulans WL-12, as a modular chitinase, consists of a signal peptide, a CatD, two Fn3D, and a ChBD [23]. ChiA1 exhibits higher catalytic activity than ChiCW [8, 24], but ChiA1 was not able to inhibit conidial germination of Bo. elliptica in our preliminary study. Thus, the C-terminal truncated derivatives of ChiA1 were used to construct chimeric chitinases with ChBDChiCW. Antifungal activities of constructed
*Corresponding author
Phone: +886-2-33665207; Fax: +886-2-23657735; E-mail: cychen@ntu.edu.tw
Present address: Department of Plant Pathology and Microbiology, National Taiwan University, No. 1, Sec. 4, Roosevelt Rd., Taipei 10617, Taiwan, Republic of China.
chimeric chitinases were analyzed to investigate the involvement of ChBDChiCW in the antifungal activity of ChiCW.
MATERIALSAND METHODS Strains, Plasmids, and Media
All strains and plasmids used in this study are listed in Table 1. Escherichia coli strains XL1-Blue (Stratagene) and DH5α [5] were used as hosts for gene cloning and expression of recombinant protein, respectively. All bacterial strains were maintained on Luria-Bertani (LB) agar plate (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar) supplemented with appropriate antibiotics. Bo. elliptica B061, the causal agent of lily leaf blight, was cultured on V-8 juice agar [20% V-8 juice (Campbell Soup Co.), 0.3% CaCO3, 1.8% agar] at 25oC for sporulation [9].
Construction of Recombinant Plasmids
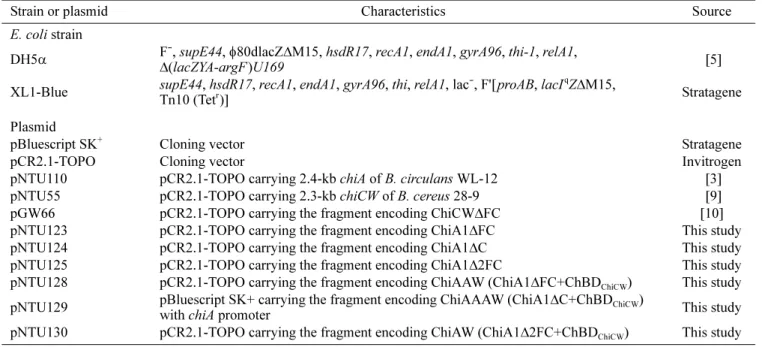
Six plasmids were constructed for expression of recombinant chitinases. Primers used in this study are listed in Table 2 and a schematic
diagram of constructed fragments encoding recombinant chitinases is shown in Fig. 1. DNA fragments were amplified by polymerase chain reaction (PCR) with the primer pairs 215/307, 215/306, and 215/305 from pNTU110. The amplified fragments were ligated into pCR2.1-TOPO (Invitrogen) to construct plasmids pNTU124, pNTU123, and pNTU125 for expression of ChiA1∆C, ChiA1∆FC, and ChiA1∆2FC, respectively.
For expression of chimeric chitinase ChiAAAW, plasmid pNTU129 was constructed. The fragment amplified by PCR with primer pair 44/219 was ligated into pCR2.1-TOPO. This recombinant plasmid was digested with XhoI and KpnI for subcloning the XhoI- and KpnI-digested fragment encoding ChBDChiCW amplified by PCR with primer pair 220/221.
For expression of chimeric chitinase ChiAAW, plasmid pNTU128 was constructed. The fragment amplified by PCR with primer pair 215/218 was digested with XbaI and XhoI and subcloned into the XbaI and XhoI sites of pBluescript SK+(Stratagene). This recombinant plasmid was digested with XhoI and KpnI for subcloning the XhoI-and KpnI-digested fragment encoding ChBDChiCW amplified by PCR
with primer pair 220/221.
pNTU125 pCR2.1-TOPO carrying the fragment encoding ChiA1∆2FC This study
pNTU128 pCR2.1-TOPO carrying the fragment encoding ChiAAW (ChiA1∆FC+ChBDChiCW) This study pNTU129 pBluescript SK+ carrying the fragment encoding ChiAAAW (ChiA1∆C+ChBDChiCW)
with chiA promoter This study
pNTU130 pCR2.1-TOPO carrying the fragment encoding ChiAW (ChiA1∆2FC+ChBDChiCW) This study
Table 2. Primers used in this study.
Primer Sequencea Restriction site
44 AGCGGCTGGAGGCGGCTATACGGC 215 CCGGTCTAGACAGAGGAGGTTGTGTATTAAATG XbaI 218 CCGGCTCGAGCAGATTGCCTGCGGCATC XhoI 219 CCGGCTCGAGCACGTTGCCTGCCGCATC XhoI 220 CCGGCTCGAGAAATCACAACCTACCGCTC XhoI 221 CCGGGGTACCCTAGTTTTCGCTAATGACGGT KpnI 222 CCGGGAGCTCAAATCACAACCTACCGCTC SacI 305 TTAGAGCTCCCAGAACATCGC SacI 306 TTACAGATTGCCTGCGGCATC 307 TTACACGTTGCCTGCCGCATC
For expression of chimeric chitinase ChiAW, plasmid pNTU130 was constructed. The DNA fragment encoding ChBDChiCW was amplified by PCR with primer pair 222/221. The amplified fragment was digested with SacI and KpnI and subcloned into the same sites of pNTU110 [3].
Bacterial Expression and Crude Extraction of Recombinant Chitinases
The recombinant plasmids were introduced into E. coli DH5α for expression of recombinant chitinases. E. coli strains were cultured in 50 ml of LB broth supplemented with ampicillin (50 ppm) on a rotary shaker at 37oC for 14 h to express recombinant chitinases. E. coli cells were harvested by centrifugation (8000 ×g, 10 min at 4o
C) and periplasmic proteins of E. coli cells were prepared according to the method of Manoil and Beckwith [17]. Periplasmic proteins were used to assay chitinase and antifungal activities.
Chitinase Activity Measurements and Protein Concentration Determination
A fluorometric assay was used to determine chitinase activity using 4-methylumbelliferyl-N,N',N''-chitotriose (4-MU-(GlcNAc)3; Sigma)
as a substrate [10]. When colloidal chitin was used as a substrate, chitinase activity was measured by using the method of Imoto and Yogishita [11]. Protein concentration was determined by the Bradford method [2] using bovine serum albumin as a standard.
Bioassay of Antifungal Activity
Antifungal activity was assayed by the method of Huang et al. [9] with a slight modification. Assay mixtures contained 10µl of a conidial suspension of Bo. elliptica B061 (1×105conidia/ml) and an
equal volume of an enzyme solution. In the control, sterile distilled water was used instead of a test solution. Conidial germination was examined under a light microscope and the percentage of inhibition was calculated after incubation of the prepared assay mixtures at 25oC for 12 h. Each assay was triplicated.
Purification of ChiAAAW and Partial Purification of ChiA1∆C All purification steps were performed at 4oC. To purify ChiAAAW, the periplasmic proteins of E. coli DH5α(pNTU129) were precipitated by ammonium sulfate at 70% saturation. The precipitate was dissolved in 25 mM potassium phosphate buffer (KPB, pH 6.0) and dialyzed against the same buffer. The dialysate was applied onto a Q-ceramic Hyper-D column (Sigma) equilibrated with 25 mM KPB (pH 6.0) for anion-exchange chromatography and eluted stepwise by 0.1, 0.3, and 0.5 M NaCl in 25 mM KPB (pH 6.0). The fractions with chitinase activities were pooled and dialyzed against 25 mM sodium citrate buffer (SCB, pH 4.0). The dialysate was applied onto a S-ceramic Hyper-D column (Sigma) equilibrated with 25 mM SCB (pH 4.0) for cation-exchange chromatography and eluted stepwise by 0.1, 0.3, and 0.5 M NaCl in 25 mM SCB (pH 4.0). ChiAAAW was eluted by 0.3 M NaCl in 25 mM SCB (pH 4.0) and the fractions with chitinase activities were pooled.
On the other hand, ChiA1∆C was partially purified from the periplasmic proteins of E. coli DH5α(pNTU124). The periplasmic proteins of E. coli DH5α(pNTU124) were precipitated by ammonium sulfate at 70% saturation. The precipitate was dissolved in 25 mM KPB (pH 6.0) and dialyzed against the same buffer. The dialysate was applied onto a Q-ceramic Hyper-D column (Sigma) equilibrated with 25 mM KPB (pH 6.0) for anion-exchange chromatography and eluted stepwise by 0.1, 0.3, and 0.5 M NaCl in 25 mM KPB (pH 6.0).
Fig. 1. Schematic diagram of the constructed chimeric chitinases and truncated derivatives used in this study.
Primers used for construction are indicated by arrows and recombinant plasmids used for expression of recombinant chitinases are presented. The designated names and molecular weights of recombinant chitinases are given.
(SDS-PAGE) and Chitinolytic Zymography Assay
SDS-PAGE and chitinolytic zymography assay were performed using Laemmli’s method with modification as previously described [9].
RESULTS
Chitinase and Antifungal Activities of ChiCW,
ChiCW∆FC, and ChiA1
As shown in Table 3, the chitinase and antifungal activities
of crude extracts of ChiCW, ChiCW∆FC, and ChiA1
against Bo. elliptica were analyzed. Compared with ChiCW, ChiCW∆FC did not inhibit conidial germination of Bo. elliptica, although the crude extract of ChiCW∆FC had a higher specific activity than that of ChiCW. Thus, ChiCW lost its antifungal activity when its C-terminal region was removed. This result indicates that the C-terminal region of ChiCW may play an important role in antifungal activity.
The effect of crude extract of ChiA1 against conidial germination of Bo. elliptica was also investigated (Table 3). The crude extract of ChiA1 had a higher specific activity
involved in the antifungal activity of ChiCW.
Vector Construction and Expression of Recombinant Chitinases in E. coli
Fig. 1 shows schematic diagrams, designated names, and calculated molecular weights of recombinant chitinases. In this study, six recombinant plasmids were constructed to express three C-terminal domain-truncated derivatives of
ChiA1 (ChiA1∆C, ChiA1∆FC, ChiA1∆2FC) and three
chimeric chitinases (ChiAAAW, ChiAAW, ChiAW). ChiAAAW was engineered by replacing ChBD of ChiA1
with ChBDChiCW. ChiAAW and ChiAW were engineered
by grafting ChBDChiCW to ChiA1∆FC and ChiA1∆2FC. The expressions of the three C-terminal domain-truncated derivatives of ChiA1 and three chimeric chitinases in E. coli DH5α cells were investigated (Table 4). Comparing the specific activities of the crude extracts, ChiA1∆C was moderately expressed in E. coli DH5α cells but ChiA1∆FC and ChiA1∆2FC were lowly expressed. When ChBDChiCW was fused with the three derivatives of ChiA1, chimeric chitinases ChiAAAW and ChiAAW were highly expressed Table 4. Chitinase and antifungal activities of crude extracts of parental, mutant, and chimeric chitinases.
Extract of E. coli straina Expressed chitinase Specific activity (U/mg)c Inhibition of conidial germination (%)d
DH5α(pNTU110) ChiA1 7.6×10-1 16±4 DH5α(pNTU124) ChiA1∆C 1.3×10-1 16±4 DH5α(pNTU123) ChiA1∆FC 5.4×10-3 4±3 DH5α(pNTU125) ChiA1∆2FC 3.8×10-4 16±6 DH5α(pNTU129) ChiAAAW 1.2 40±1 DH5α(pNTU128) ChiAAW 1.9×10-1 39±3 DH5α(pNTU130) ChiAW 1.0×10-3 61±10 DH5α(pNTU55) ChiCW 2.2×10-1 58±1 DH5α(pCR2.1-TOPO)b - 1.6×10-3 0±0
aCrude periplasmic extracts of E. coli strains expressing parental and chimeric chitinases were prepared to analyze chitinase and antifungal activities. bE. coli DH5α(pCR2.1-TOPO) as the control without expression of recombinant chitinase.
cOne unit=1µmole of 4-methylumbelliferone released per minute at 37oC. dEach treatment included 10µl of a crude extract and 10 µl of a conidial suspension.
in E. coli DH5α cells, especially ChiAAAW, but ChiAW was lowly expressed. Similar results were observed by SDS-PAGE and chitinolytic zymography (data not shown). Antifungal Activities of C-Terminal Domain-Truncated Derivatives of ChiA1 and Three Chimeric Chitinases As shown in Table 4, the antifungal activities of crude extracts of the three C-terminal domain-truncated derivatives of ChiA1 and the three chimeric chitinases were investigated. The C-terminal domain-truncated derivatives of ChiA1 did not exhibit antifungal activities to inhibit conidial germination of Bo. elliptica. Surprisingly, when ChBDChiCW was used to
replace ChBD of ChiA1 or to fuse to ChiA1∆FC and
ChiA1∆2FC, the three chimeric chitinases exhibited higher antifungal activities than that of ChiA1, ChiA1∆FC, and ChiA1∆2FC. The results implicate that ChBDChiCW is involved in the antifungal activity of chitinase and strongly suggest that ChBDChiCW contributes to the antifungal activity of ChiCW.
Purification of ChiAAAW and Partial Purification of ChiA1DC
According to the results of expression and antifungal effects of chimeric chitinases (Table 4), the expression of
ChiAAAW in E. coli DH5α cells was highest among
the three chimeric enzymes, and ChiAAAW effectively inhibited conidial germination of Bo. elliptica. Therefore, we focused on further study of ChiAAAW.
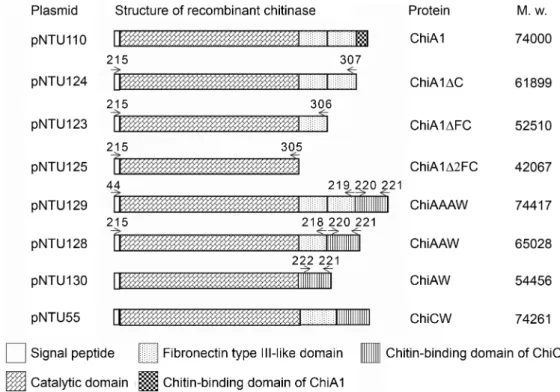
ChiAAAW was purified by ammonium sulfate precipitation, anion-exchange chromatography, and cation-exchange chromatography from the periplasmic fraction of E. coli DH5α(pNTU129). By SDS-PAGE analysis and chitinolytic zymography assay, ChiAAAW was purified to apparent homogeneity (Fig. 2). The molecular size of ChiAAAW was estimated to be 74 kDa, which closely corresponded to the value calculated from the peptide sequence of mature ChiAAAW (74.416 kDa). From a 1,200 ml culture, 0.06 mg of ChiAAAW was purified. The result of purification of ChiAAAW is summarized in Table 5.
In addition, ChiA1∆C was partially purified for further assays. Although the specific activity of crude ChiA1∆C extract was relatively high (Table 4), the amount of ChiA1∆C expressed in E. coli cells was not as high as
expected (data not shown). Therefore, ChiA1∆C was
Fig. 2. Purification of ChiAAAW. Sample from each step was analyzed by SDS-PAGE and Coomassie blue G-250 staining (A) and chitinolyic zymography (B).
Lane 1, crude periplasmic extract of E. coli DH5α(pNTU129); lane 2, ammonium sulfate precipitated proteins; lane 3, fraction of anion-exchange chromatography; lane 4, fraction of cation-exchange chromatography; lane M, low molecular weight protein marker (GE Healthcare).
Table 5. Purification of ChiAAAW.
Purification step Total protein (mg) Total activity (U)a Specific activity (U/mg) Recovery rate (%) Purification fold
Crude extract 1.44 3.15 2.19 100 1
Ammonium sulfate precipitation 0.36 2.20 6.11 70 3
Anion-exchange chromatography 0.18 1.64 9.11 52 4
Cation-exchange chromatography 0.06 2.31 38.50 73 18
aOne unit=1µmole of 4-methylumbelliferone released per minute at 37oC.
Fig. 3. SDS-PAGE and zymography assay of partially purified ChiA1∆C.
Partially purified ChiA1∆C was analyzed by SDS-PAGE and Coomassie blue G-250 staining (lane 1) and chitinolytic zymography (lane 2). Lane M, low molecular weight protein marker (GE Healthcare).
hardly purified and 0.02 mg of partially purified ChiA1∆C
was obtained from a 1,200 ml culture. By SDS-PAGE
analysis and chitinolytic zymography assay (Fig. 3), the molecular size of ChiA1∆C was estimated to be 62 kDa, which closely corresponded to the value calculated from the peptide sequence of mature ChiA1∆C (61.899 kDa). Hydrolysis of Colloidal Chitin and Antifungal Activity of Purified Chitinase
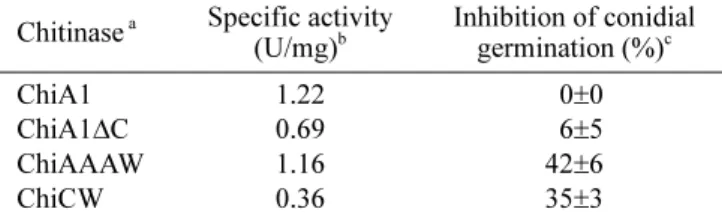
Table 6 shows the hydrolytic activities towards colloidal chitin and the antifungal activities of purified ChiA1,
ChiAAAW, ChiCW, and partially purified ChiA1∆C. In
accordance with the previous study [24], ChiA1 exhibited excellent hydrolytic activity towards colloidal chitin, and ChiA1∆C exhibited less hydrolytic activity than ChiA1. In addition, ChiCW had lower hydrolytic activity than ChiA1 but the chimeric chitinase ChiAAAW exhibited similar hydrolytic activity to ChiA1.
The antifungal activities of purified ChiA1, ChiAAAW, ChiCW, and partially purified ChiA1∆C were examined (Table 6). Although ChiA1 exhibited excellent hydrolytic activity towards colloidal chitin, ChiA1 did not inhibit conidial germination of Bo. elliptica. When ChBD of ChiA1 was deleted, ChiA1∆C as well as ChiA1 did not show inhibitory effect to Bo. elliptica. On the other hand, ChiCW exhibited antifungal activity against Bo. elliptica.
When ChBDChiCW replaced ChBD of ChiA1, ChiAAAW
exhibited antifungal activity against Bo. elliptica and the antifungal activity of ChiAAAW was slightly higher than that of ChiCW.
DISCUSSION
ChBDs involved in enzyme activity and substrate binding have been demonstrated in bacterial modular chitinases of family 18 and 19 [12, 13, 18, 22, 24]. However, correlation between ChBD and antifungal activity of bacterial chitinase has only been investigated in ChiC of S. griseus HUT6037,
encoding ChBDChiCW was subcloned into the pQE30 vector for expression and the purified recombinant ChBDChiCW did not inhibit conidial germination of Bo. elliptica (unpublished data). When ChBD of ChiA1 was replaced with ChBDChiCW to generate ChiAAAW, this chimeric chitinase showed antifungal activity (Table 6). Compared with ChiA1 and ChiCW, ChiAAAW exhibited high hydrolytic activity towards colloidal chitin and high antifungal activity (Table 6). This result indicates that using ChBDChiCW to replace ChBD of ChiA1 could engineer a new chitinase with high enzyme and antifungal activities. Thus, we suggest that ChBDChiCW plays an important role in the antifungal activity of ChiCW, and using ChBDChiCW to engineer chitinases probably can obtain new enzymes with enhanced antifungal activity.
Engineering enzymes to improve their properties is one of the obvious goals of biotechnology [1, 19, 21]. Two chitinases, Chit33 and Chit42, from Trichoderma harzianum CECT 2413 are considered to play an important role in the biocontrol activity of this strain against plant pathogens, and both Chit33 and Chit42 lack a chitin-binding domain [4, 14]. These two enzymes were engineered by addition of binding domains to increase their substrate-binding capacity and specific activity [15, 16]. According to these studies, the substrate-binding and chitinase specific activities of these chimeric chitinases were increased, but the antifungal activities of these chimeric chitinases were not reported [15, 16]. In this study, a new chimeric chitinase, ChiAAAW, was engineered by replacing ChBD of ChiA1 with ChBDChiCW and the purified ChiAAAW was demonstrated to exhibit not only high enzyme activity but also high antifungal activity. In our previous study [9], peptide sequences of ChBDs of ChiCW and ChiA1 were compared and very low similarity between the two ChBDs was observed. The result of this study shows that chitinases with improved properties can be engineered by manipulating those substrate-binding domains.
In this study, we designed an approach based on protein engineering technology to demonstrate that ChBDChiCW plays an important role in the antifungal activity of ChiCW,
and a new chimeric chitinase, ChiAAAW, with high enzyme and antifungal activities was created. This study may be helpful to understand the relationship between ChBD and the antifungal activity of the family 18 chitinase.
Acknowledgment
This research was supported by the National Science Council of the Republic of China.
REFERENCES
1. Beguin, P. 1999. Hybrid enzymes. Curr. Opin. Biotechnol. 10: 336-340.
2. Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248-254. 3. Chen, C. T., C. J. Huang, Y. H. Wang, and C. Y. Chen. 2004.
Two-step purification of Bacillus circulans chitinase A1 expressed in Escherichia coli periplasm. Protein Expr. Purif. 37: 27-31.
4. Garcia, I., J. M. Lora, J. De la Cruz, T. Benitez, A. Llobell, and J. A. Pintor-Toro. 1994. Cloning and characterization of a chitinase (CHIT42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr. Genet. 27: 83-89.
5. Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557-580.
6. Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280: 309-316.
7. Henrissat, B. and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293: 781-788.
8. Huang, C. J. and C. Y. Chen. 2005. High-level expression and characterization of two chitinases, ChiCH and ChiCW, of Bacillus cereus 28-9 in Escherichia coli. Biochem. Biophys. Res. Commun. 327: 8-17.
9. Huang, C. J., T. K. Wang, S. C. Chung, and C. Y. Chen. 2005. Identification of an antifungal chitinase from a potential biocontrol agent, Bacillus cereus 28-9. J. Biochem. Mol. Biol. 38: 82-88.
10. Huang, C. J. and C. Y. Chen. 2006 Functions of the C-terminal region of chitinase ChiCW from Bacillus cereus 28-9 in substrate-binding and hydrolysis of chitin. J. Microbiol. Biotechnol. 16: 1897-1903.
11. Imoto, T. and K. Yogishita. 1971. A simple activity measurement of lysozyme. Agric. Biol. Chem. 35: 1154-1156.
12. Itoh, Y., T. Kawase, N. Nikaidou, H. Fukada, M. Mitsutomi, T. Watanabe, and Y. Itoh. 2002. Functional analysis of the
chitin-binding domain of a family 19 chitinase from Streptomyces griseus HUT6037: Substrate-binding affinity and cis-dominant increase of antifungal function. Biosci. Biotechnol. Biochem. 66: 1084-1092.
13. Itoh, Y., J. Watanabe, H. Fukada, R. Mizuno, Y. Kezuka, T. Nonaka, and T. Watanabe. 2006. Importance of Trp59 and Trp60 in chitin-binding, hydrolytic, and antifungal activities of Streptomyces griseus chitinase C. Appl. Microbiol. Biotechnol. 72: 1176-1184.
14. Limon, M. C., J. A. Pintor-Toro, and T. Benitez. 1999. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology 89: 254-261. 15. Limon, M. C., E. Margolles-Clark, T. Benitez, and M. Penttila.
2001. Addition of binding domains increases substrate-binding capacity and specific activity of a chitinase from Trichoderma harzianum. FEMS Microbiol. Lett. 198: 57-63. 16. Limon, M. C., M. R. Chacon, R. Mejias, J. Delgado-Jarana, A.
M. Rincon, A. C. Codon, and T. Benitez. 2004. Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl. Microbiol. Biotechnol. 64: 675-685.
17. Manoil, C. and J. Beckwith. 1986. A genetic approach to analyzing membrane protein topology. Science 233: 1403-1408. 18. Morimoto, K., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J. Bacteriol. 179: 7306-7314.
19. Nixon, A. E., M. Ostermeier, and S. Benkovic. 1998. Hybrid enzymes: Manipulating enzyme design. Trends Biotechnol. 16: 258-264.
20. Ohno, T., S. Armand, T. Hata, N. Nikaidou, B. Henrissat, M. Mitsutomi, and T. Watanabe. 1996. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT6037. J. Bacteriol. 178: 5065-5070.
21. Ulmer, K. M. 1983. Protein engineering. Science 219: 666-671. 22. Wang, F. P., Q. Li, Y. Zhou, M. G. Li, and X. Xiao. 2003. The C-terminal module of Chi1 from Aeromonas caviae CB101 has a function in substrate binding and hydrolysis. Proteins 53: 908-916.
23. Watanabe, T., K. Suzuki, W. Oyanagi, K. Ohnishi, and H. Tanaka. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J. Biol. Chem. 265: 15659-15665.
24. Watanabe, T., Y. Ito, T. Yamada, M. Hashimoto, S. Sekine, and H. Tanaka. 1994. The roles of the C-terminal domain and type III domains of chitinase Al from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 176: 4465-4472.