A Polymerase Chain Reaction for Detecting Herpesvirus anguillae
in Asymptomatic Eels
Hsiu-Hui Shih(1)
(Manuscript received 29 September, 2003; accepted 28 November, 2003)
ABSTRACT: The present study describes a polymerase chain reaction (PCR) assay for detection of
Herpesvirus anguillae (HVA) DNA. PCR primers (622F and 622R) were developed based on a HindIII-1550 bp DNA fragment from HVA genomic library. Templates of HVA DNA yielded an evident amplification product showing the expected mobility of a 622-bp DNA fragment. The assay is rapid, sensitive and specifically detects HVA DNA including both the Taiwanese and the Japanese isolates. HVA persistent infections in asymptomatic farmed eels at Taiwan were revealed successfully in the present study.
KEY WORDS: Herpesvirus anguillae, HVA, PCR, Anguilla eel.
INTRODUCTION
A disease outbreak manifested by dermal haemorrhagic lesions was reported from farmed eels in Taiwan, where a herpesvirus was isolated (Ueno et al., 1992; Shih et al., 1993). Before this, a herpesvirus isolated from diseased eel was characterized and designated as Herpesvirus
anguillae (HVA) from Japan (Sano et al., 1990). The results of biological and serological
comparisons with the Japanese isolate showed that the Taiwanese isolate possesses similar virological characteristics and antigenic relations to the HVA (Ueno et al., 1996).
According to a recent observation, a HVA latent infection was established by a Dutch isolate that was isolated repeatedly from farmed European eel of an outwardly healthy stock and much greater virus release was provoked by dexamethasone treated fish (Van Nieuwstadt
et al., 2001). The latent HVA was activated by exogeneous or endogeneous stimuli, and could
subsequently be shed without clinical signs. An indirect ELISA was successfully developed to detect HVA-specific antibodies in eel sera and applied for tracing HVA carriers (Van Nieuwstadt et al., 2001).
To permit the detection of lower titers of virus that may be encountered in asymptomatic carriers or latent viruses, a more sensitive and rapid PCR technique was developed in this study. Primers (HVA622F/R) were designed on a basis of the DNA sequence of a cloned Taiwanese HVA 1550-bp HindIII DNA fragment in a recombinant plasmid, pH1-1 (Shih et
al., 2003). The PCR was then used to investigate the HVA carriers in two cultivated eel
populations of Anguilla japonica and A. rostrata in Taiwan.
MATERIALS AND METHODS
Viruses and cells
Tilapia ovary (TO-2) cells (Chen et al., 1983) were grown in L-15 medium (GIBCO/BRL, New York, USA) supplemented with 10% fetal bovine serum (FBS), penicillin (100 IU/mL),
__________________________________________________________________________________________ 1. Department of Life Science and Institute of Zoology, National Taiwan University, Taipei 106, Taiwan. E-mail:
and the Japanese isolate were cultured in TO-2 cells at 20℃ supplemented with 2% FBS. Channel catfish virus (CCV) was cultured in brown bullhead (BB) cells, which were grown in Eagle's MEM medium (GIBCO/BRL), supplemented with 2% FBS. The Taiwanese HVA isolate were purified as described previously (Shih et al., 2003) and stored at -20℃ until use. Eels
Two independent populations of eels were used in this study. Juvenile Japanese eel (average weight 2.6 g), A. japonica, and American eel (average weight 5.5 g), A. rostrata, were collected from two culture ponds located at central or southwestern Taiwan (Table 1) from March to May, 1999. Samples were all healthy by clinical examination. Eel cell DNA was extracted from pieces of the liver, spleen and kidney collected and applied as templates for PCR screening.
Table 1. A survey of Herpesvirus anguillae (HVA) carriers from farmed eels using polymerase chain reaction.
Eel Source No. of eels examined No. of HVA- positive
eels Percentage of carrier Anguilla japonica A. rostrata Lu-Kang Yung-Lin 409 27 69 4 16.9 14.8 DNA extraction
Total DNA was extracted from purified Taiwanese HVA, Taiwanese HVA infected TO-2 cells, Japanese HVA infected TO-2 cells, uninfected TO-2 cells, and CCV infected BB cells, using a commercial genomic DNA isolation kit (Geno Tech, USA). The phenol-chloroform extraction and ethanol precipitation protocols described previously (Maniatis et al., 1982) were routinely carried out for the eel tissues processing. DNA concentrations were measured with a GeneQuant II RNA/DNA calculator (Pharmacia Biotech, Uppsala, Sweden).
PCR conditions
Oligonucleotide primes (HVA622F and HVA622R) were used for the amplification of HVA DNA fragments. The following are the sequences of the primers: HVA622F, 5'-TTG AGG TTG TTG TCG TGC C-3'; HVA622R, 5'-CTC TCA TGT CAT CCA GAC GG-3'. With this primer set, a 622-bp fragment is expected to be amplified from HVA genomic DNA.
The deproteinized DNA samples were used for amplification in a 10 µL reaction mixture containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2 mM MgCl2, 200 µM each of dNTP,
1 µmol each of primers, 4 units of Taq DNA polymerase (Biotools, USA). The amplification was performed in a RapidCycler Thermal programmer (Idaho Technology, USA) using 10-µL glass capillaries. Twenty five amplification cycles were run, each cycle consisting of a denaturation step at 94℃ for 0 s, annealing at 55℃ for 0 s, extension at 72℃ for 25 s. Since the temperature of a totally 10 µL reaction mixture was easily equilibrium inside a glass capillary as it reached the setting point during PCR, the period of both denaturation and annealing steps were not necessarily prolonged. Thus a very rapid reaction processing for only half hour was achieved.
Amplification products were analysed by electrophoresis through 1% agarose gels and ethidium bromide staining. A 100-bp DNA ladder standard (Pharmacia Biotech) was used as markers.
RESULTS AND DISCUSSION
Sensitivity and specificity of HVA PCR assays
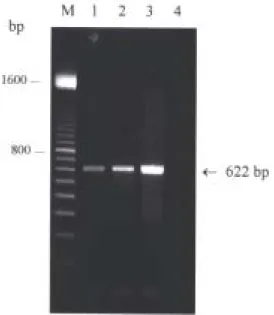
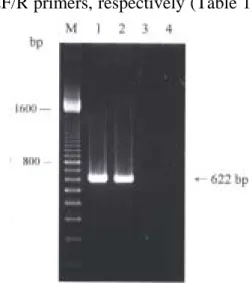
To assess the sensitivity of the assay, the HVA PCR was conducted using 10-fold dilutions of purified HVA genomic DNA mixed with 1.0 µg of total cell DNA derived from a juvenile eel as the DNA template. The expected 622-bp product was easily detected in samples with concentrations as low as 1 picograms (pg) of HVA DNA by agarose gel electrophoresis and ethidium bromide staining (Fig. 1). The assay is sensitive, being capable of detecting pg concentrations of HVA DNA in the presence of 106 times as much eel DNA. To confirm the specificity of this assay, the PCR protocols were run with DNA templates including infected cell DNA from the Taiwanese HVA infected TO-2 cells and DNA from the Japanese HVA infected TO-2 cells. The amplification results showed that the size of PCR products from these 2 isolates was identical and the expected size of 622 bp was shown (Fig. 2). While DNA template from BB cells infected by another fish herpesvirus, CCV, was used in parallel as control, there was not any PCR product amplified (Fig. 2, lane 3). The results suggest that the PCR developed here is specific for detecting HVA including both the Taiwanese and the Japanese isolates.
The PCR assay is rapid, since a totally 10 µL for each reaction mixture was applied with a fast thermo-conducting glass capillary in this protocol. The results are available 5-6 h after the samples were collected while a commercial genomic DNA isolation kit is used along.
PCR technology offers an advance over traditional methods of diagnosis of HVA infections, which were based on clinical signs and histopathology, and identifying HVA isolates. The isolation of infectious HVA in cell culture is an important diagnostic approach (Ueno et al., 1992; Lee et al., 1999), but requires a few days to propagate the virus, and other diagnostic assays to identify HVA as the isolated agent will be still needed. A serological test to detect HVA-specific antibodies in eel was developed and applied for tracing virus carriers (Van Nieuwstadt et al., 2001). However, its value as a diagnostic assay for HVA is limited since a mouse-derived monoclonal antibody specific for the heavy chain of eel immunoglobulin used is not commercially available. In addition to DNA hybridization, which was successful for detecting HVA DNA in infected eels (Shih et al., 2003), a rapid, sensitive and specific PCR protocol is developed to detect HVA DNA in the present
Fig. 1. Sensitivity of HVA DNA detection using the HVA622F/R primer set. Agarose gel electrophoresis was used to analyse PCR products from reactions including the HVA 622F/R primer set and 10-fold dilutions of purified genomic HVA DNA mixed with 1.0 µg of total eel cell DNA. The amount of HVA DNA in each reaction was 1.0 picograms (pg, lane 1), 0.01 ng (lane 2), 0.1 ng (lane 3), or no HVA DNA (lane 4). M: 100-bp markers. Size of DNA markers given in base pairs (bp).
study. The large-scale investigation of HVA infections in field as presented below could be easily processed than previous assays.
As screening the presence of HVA DNA within asymptomatic eels from 2 cultivated eel populations by PCR, it could be shown that HVA DNA was present in 16.9% of Japanese eels and 14.8% of American eels examined with the HVA622F/R primers, respectively (Table 1). Since outbreak of the herpesviral disease did not
occur and HVA carriers were detected using PCR, a persistent infection is suggested to be present in these two stocks.
The latency of HVA can be activated by exogeneous or endogeneous stimuli, and the virus can subsequently be shed without clinical signs (Van Nieuwstadt et al., 2001). Treatment with dexamethasone was used successfully to prove that European eels from an outwardly healthy stock were infected with HVA. When stressed, carrier eels can release HVA (Van Nieuwstadt et al., 2001). Although the mechanisms or stresses that might induce the eel herpesviral disease outbreak are not probed in this study, the PCR assay developed is proven to be useful to diagnose the disease. Since the high stocking density in recirculation systems with biofilters is common practice in eel farming recently, the spreading of HVA infections and the outbreaks of disease will be frequently encountered once the virus has been introduced into an eel farm. This assay may be useful for the monitoring of the HVA carriers and control of the eel herpesviral disease epizootics.
Fig. 2. Specificity of HVA622F/R primer set. DNA products from PCR amplification using primer set HVA622F/R were revealed by electrophoresis through a 1% agarose gel and staining with ethidium bromide. DNA templates used in this assay including infected cell DNA from the Taiwanese HVA infected TO-2 cells (lane 1), from the Japanese HVA infected TO-2 cells (lane 2), from the CCV infected BB cells (lane 3), and cell DNA from uninfected TO-2 cells (lane 4). M: 100-bp markers.
ACKNOWLEDGEMENTS
This work was supported by Council of Agriculture under Grant No. 87-AST-1.4-FID-06 (10). I would like to thank Dr H. Fukuda (Tokyo University of Fisheries) for his kindly providing HVA Japanese isolate, CCV and BB cells; Ms L. Yu and I. W. Liu for assistance in PCR.
LITERATURE CITED
Chen, S.-N., S.-C. Chi, Y. Ueno and G.-H. Kou. 1983. A cell line derived from tilapia ovary. Fish Pathology 18: 13-18.
Lee, N.-S., J. Kobayashi and T. Miyazaki. 1999. Gill filament necrosis in farmed Japanese eels, Anguilla japonica (Temminck & Schlegel), infected with Herpesvirus anguillae. Journal of Fish Diseases 22: 457-463.
Maniatis, T., E. F. Fritsch and J. Sambrook. 1982. Molecular Cloning. Cold Spring Harbor Laboratory, USA. pp. 458-459.
from eel. Pathology of Marine Science 1: 15-31.
Shih, H.-H., C.-W. Hu and C.-S. Wang. 2003. Detection of Herpesvirus anguillae infection in eel using in situ hybridization. Journal of Applied Ichthyology 19: 99-103.
Shih, H.-H., C.-C. Lu and S.-N. Chen. 1993. Eel herpesvirus in Formosa: a herpesvirus from cultured Japanese eel. Reports on Fish Disease Research 13: 86-96 (in Chinese, with English abstract).
Ueno, Y., T. Kitao, S.-N. Chen, T. Aoki and G.-H. Kou. 1992. Characterization of a herpes-like virus isolated from cultured Japanese eels in Taiwan. Fish Pathology 27: 7-17.
Ueno, Y., J.-W. Shi, T. Yoshida, T. Kitao, M. Sakai, S.-N. Chen and G.-H. Kou. 1996. Biological and serological comparisons of eel herpesvirus in Formosa (EHVF) and
Herpesvirus anguillae (HVA). Journal of Applied Ichthyology 12: 49-51.
Van Nieuwstadt, A. P., S. G. Dijkstra and O. L. M. Haenen. 2001. Persistence of herpesvirus of eel Herpesvirus anguillae in farmed European eel Anguilla anguilla. Diseases of Aquatic Organisms 45: 103-107.