The C-Terminal Segment Is Essential for Maintaining
the Quaternary Structure and Enzyme Activity
of the Nitric Oxide Forming Nitrite Reductase
from Achromobacter cycloclastes
Wei-Chao Chang,* Jang-Yi Chen,* Tschining Chang,† Ming-Yih Liu,‡ William J. Payne,§ Jean LeGall,‡ and Wen-Chang Chang*,†
*Institute of Biochemical Science, National Taiwan University, Taipei, Taiwan; †Institute of Biological Chemistry,
Academia Sinica, Taipei, Taiwan; and ‡Department of Biochemistry and Molecular Biology and
§Department of Microbiology, University of Georgia, Athens, Georgia
Received July 24, 1998
We have constructed and expressed a series of mu-tated nitrite reductase (NIR) mutants based on the se-quence of NIR from Achromobacter cycloclastes. Delet-ing a pentapeptide, an undecapeptide, or a heptadeca-peptide from the C-terminus of NIR resulted in a series of C-terminal deletion mutated proteins designated as NIR-5, NIR-11, and NIR-17, respectively. A C-terminally extended mutated protein, NIR18, was also produced, which contains an extra octapeptide attached to the C-terminus of the wild-type NIR. An SDS-PAGE system using tris-tricine buffer could retain the native NIR in its trimeric form, thus offering a convenient method to check the quaternary structure of NIR analogs. By using this system it was found that NIR-5 was maintained as trimer and retained 72% of wild-type enzyme activity. However, both NIR-11 and NIR-17 behaved as monomers in the SDS-PAGE and lost all their enzyme activity. Al-though NIR18 maintained its trimeric structure it was enzymatically inactive. These results clearly indicate that the C-terminal undecapeptide is essential for main-taining the quaternary structure as well as the full en-zymatic activity, as expected from the X-ray crystallog-raphy studies. © 1998 Academic Press
There are two types of nitric oxide forming nitrite reductase: cd1 cytochrome nitrite reductase (cd1 NIR)
and copper nitrite reductase (CuNIR) (1). Both are involved in the reduction of nitrite in the denitrifying, respiratory pathway. It is known that cd1 NIR
trans-fers electrons from cytochrome c551 to nitrite ions to
perform its function, and that the active enzyme is a dimer. The CuNIR accepts electrons from azurin (Pseudomonas aureofaciens) (2), pseudoazurin
(Alcali-genes faecalis s-6) (3) or a blue copper protein
(Achro-mobacter cycloclastes) (4) and then transfers them to
the nitrite ions. A detailed account of the electron transport steps were further demonstrated by using site-directed mutagenesis of A. faecalis NIR(5). It was shown that electons transfer from reduced pseudo-azurin to the type I copper and then to the type II copper and finally to the nitrite. But the structure of active enzyme remains disputed (6,7). X-ray crystallography showed that CuNIR is organized as a trimer(8,9), and that hydrogen bonds form an extensive network both within and between subunits to maintain its quater-nary structure. Moreover, the interaction of residues 13 and 14 of one subunit with residues 336 –340 of another is one of the key packing forces maintaining the trimeric structure(9). We report herein, by using site-directed mutagenesis, that the C-terminal seg-ment of about 10 residues of a CuNIR is important for maintaining both the trimeric structure and the enzy-matic activity.
MATERIALS AND METHODS
Bacterial strains and plasmids. E.coli JM109 used for plasmid cloning was purchased from Promega Inc. E.coli, strain M15 (PREP4), derived from strain K12, was purchased from Qiagen Inc. and used for gene expression. The plasmids, pGemT, pGem3zf(1) (Promega) and pUC19, were used as cloning vector.
Media. E.coli, strains were cultured in L-broth aerobically at 37°C. The concentration of ampicillin was 50mg/ml for blue/white colony selection. IPTG, 0.5 nM, and X-Gal, 40mg/ml (final concentra-tion), were added to the medium. For the expression of NIR analog proteins under control of E.coli T5 promotor, two lac operator se-quences and 2 mM IPTG were added.
Chemicals and enzymes. All restriction endonucleases and other DNA modification enzymes were purchased from Boehringer Mann-heim or Promega Inc. [a-32P]dATP and [a-35S]dATP were purchased
from Amersham Inc. and the recombinant gene expression and the protein purification kits from Qiagen Inc. The gradient gel, tris-BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS250, 782–785 (1998)
ARTICLE NO. RC989316
782 0006-291X/98 $25.00
Copyright © 1998 by Academic Press All rights of reproduction in any form reserved.
glycine, tris-tricine buffer and sample buffer for SDS-PAGE were purchased from Novel Experimental Technology.
Cloning of NIR and its mutated genes. The cloning of the NIR gene (pQNIR) was described previously (10). For cloning of the C-terminally extended or deleted NIRs, the following primers were used: 5 9GATGAC-GATGACCAAAGCAGCCGGTGC39 (N-terminal primer, for every mu-tated genes) 59CATCGACGCCGGCTTGACGAC39 (C-terminal extended primer). 59CTAGGTCACCTTGAAGTGGCCGGC39 (for NIR-5), 59CTA-ATCATCGTTCCATTCGCCGGT39 (for NIR-11) and 59CTAGACGAC-CGATGTCATCAGATC39 (for NIR-17). Polymerase chain reaction (PCR) was performed using the above primers and pQNIR recombinant plasmid as template to produce those modified coding sequences.
RESULTS AND DISCUSSION
The recombinant NIRs expressed in E.coli were eas-ily purified and refolded to the native states as judged by absorption spectrum, SDS PAGE and activity assay as described earlier(10). NIR-11 and NIR-17 were ex-pressed at low levels for unknown reasons (data not shown). These two proteins, although purified by affin-ity column and further purified with HPLC gel filtra-tion column, still contained some minor impurities (Fig.1, lanes 5, 6, 11 and 12).
The NIRs behaved quite differently in the SDS-PAGE systems using tris-glycine as opposed to those employing tricine as running buffer. In the tris-glycine buffer system, NIR appears as monomeric and dimeric forms under non-denaturing condition and without heating before loading on the gel. But the enzyme migrated as the trimeric molecule in the tris-tricine buffer system under the same conditions (Fig.2). In fact, this trimer is rather stable in the tricine buffer system. After heating at 80°C in the
tricine system more than 5% of the NIR remained in the trimeric form (data not shown). It is clear that the tricine buffer, but not the glycine, could preserve the form of NIR observed in crystals. Apparently the non-covalent bonds holding the subunits together in the native states were not broken by SDS in this tricine buffer system at room temperature. Tris-tricine is thus a useful and convenient tool for checking the quater-nary structures of NIR and its analogs.
By using this tricine SDS-PAGE without heating of samples before loading, we found that all of NIR analogs with an N-terminal extension-peptide, MRGSHHHHH-HGSSRDDDDK, which was derived from the oligonucle-otide spanning the ribosome-binding sequence and the codon for the N-terminus of mature NIR, migrated as the dimer and the monomer and could not assemble into active trimers (Fig.3 lanes 6,12,18,24,and 30). After di-gestion with enterokinase to remove that N-terminal
ex-TABLE 1
The Subunit Molecular Weight and Enzyme Activity of NIR Analogs
Analogs Mol. wt. (kDa) Rel. activity (%)
NIR 37.1 100 NIR18 38.0 0 RNIR* 37.1 108 NIR-5 36.6 72 NIR-11 36.0 0 NIR-17 35.2 0
* Recombinant wild-type NIR.
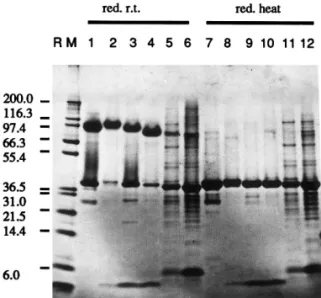
FIG. 1. The effect of heating on the protein profile in the tris-tricine SDS-PAGE system. Lanes 1– 6: samples treated with reduc-ing agent together with SDS and kept at room temperature for 5 min migrated as trimer form. Lanes 7–12: samples reduced and heated at 95°C for 5 min migrated as monomer. Lanes 1 and 7: NIR purified from A. c. Lanes 2 and 8: NIR18. Lanes 3 and 9: E. coli expressed NIR. Lanes 4 and 10: NIR-5. Lanes 5 and 11: NIR-11. Lanes 6 and 12: NIR-17. RM: protein molecular weight maker.
FIG. 2. The behavior of NIR in the tris-glycine and tris-tricine SDS-PAGE (left panel), and its calibrated molecular weight (right panel) in the tris-tricine system. R/h: samples treated with reducing agent and heated for 5 min at 95°C; nR/rt: non-reduced samples kept at room temprature for 5 min. M: the protein molecular weight makers (kDa): aprotinin: 6; lysozyme: 14.4; trypsin inhibitor: 21.5; carbonic anhydrase: 31; lactate dehydrogenase: 36.5; glutamate de-hydrogenase: 55.4; bovine serum albumin: 66.3; phosphorylase b: 97.4;b-galactosidase: 116.3; myosin: 200.
Vol. 250, No. 3, 1998 BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS
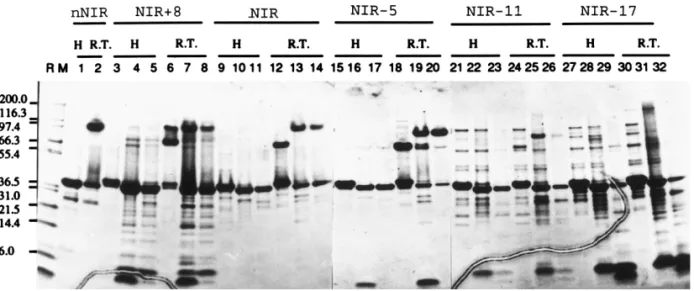
tra peptide, NIR18 (Fig. 3, lanes 7 and 8), wild-type NIR (Fig.3, lanes 13,14) and NIR-5 (Fig.3, lanes 19 and 20) could assemble into trimer form. However, those NIR
with deletion of longer peptides at C-terminus, i.e., NIR-11 and NIR-17 (Fig.3, lanes 25,26,31 and 32) could not form trimers. Thid experiments demonstrated the
FIG. 4. The optical spectra of native, recombinant, and mutated NIRs.
FIG. 3. The protein profile of the NIR analogs in the tris-tricine SDS-PAGE. RM: protein molecular weight maker; H: samples treated with reducing agent and heating; R.T.: samples treated with reducing agent and kept at room temperature for 5 min. Lanes 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30: samples before enterokinase digestion. Lanes 4, 7, 10, 13, 16, 19, 22, 25, 28, and 31: samples treated with enterokinase. Lanes 5,8,11,14,17,20,23,26,29, and 32: samples treated with enterokinase and dialyzed against Cu12-containing buffer.
Vol. 250, No. 3, 1998 BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS
importance of the C-terminal segment for maintaining the trimeric structure. As expected, the enzyme assay showed that NIR-5 retained 72% of the native enzyme activity. Both NIR-11 and NIR-17 mutants were not en-zymatically active (Table 1). Fig. 4 shows the optical spectra of native, recombinant and mututed NIR. Al-though the ratio A280/A458is not very much affected (not
shown), it is clear that the overall shape shows much distortion in the mutated proteins with no detectable NIR activity (NIR-11 and NIR-17). However, although the NIR18 mutated protein shows what appears a typi-cal spectrum this protein exhibits no enzymatic activity either. The capacity to form a trimer is apparently not sufficient for maintaining an active enzyme.
The X-ray crystallographic study by Godden et al. (8) shows that three factors determine maintenance the NIR trimer structure seen in the cryatal: 1. the inter-action between His306 and the type II copper; 2. the inter-face between two nearby subunits; and 3. the C-terminal polypeptide from position 324 to 340 extend from one subunit to another. They proposed that the third is the major factor. In this report, we show that not only the extension of the C-terminus will decrease the enzyme activity, but also the deletion will have the same effect. Deletion of more than ten residues in the C-terminus abolishes NIR activity completely, whereas deletion of five permits retention of two thirds of the activity. This result is consistent with the conclusion that the C-terminal segment interacts with another
subunit to maintain both the quaternary structure and the activity of the enzyme.
ACKNOWLEDGMENT
This study was supported by a grant from National Science Coun-cil of the Republic of China (NSC-85-2311-B-001-007-B15).
REFERENCES
1. Payne, W. J. (1985) Denitrification in the Nitrogen Cycle (Golte-man, H. Ly, Ed.), Plenum, New York.
2. Zumft, W. G., Gotzmann, D. J., and Kroneck, P. M. H. (1987) Eur. J. Biochem. 168, 301.
3. Kakutami, T., Watanabe, H., Arima, K., and Beppu, T. (1981) J. Biochem. 89, 463– 472.
4. Liu, M. Y., Liu, M. C., Payne, W. J., and Le Gall, J. (1986) J. Bacteriol. 166, 604 – 608.
5. Kukimoto, M., Nishiyama, M., Murphy, M. E. P., Turley, S., Adman, E. T., Horinouchi, S., and Beppu, T. (1994) Biochemistry
33, 5246 –5252.
6. Masako, M., Iwasaki, H., Saturai, T., Suzuki, S., and Nakahara, A. (1984) J. Biochem. 69, 859 – 868.
7. Nishiyama, M., Suzuki, J., Kukimoto, M., Ohouki, T., Horinouch, S., and Beppu, T. (1993) J. Gen. Microbiol. 139, 725–733. 8. Godden, J. W., Turky, T., Teller, D. C., Adman, E. T., Liu, M. Y.,
Payne, W. J., and Le Gall, J. (1991) Science 253, 438 – 442. 9. Adman, E. T., Godden, J. W., and Turley, S. (1995) J. Biol. Chem.
270, 27458 –27474.
10. Chen, J.-Y., Chang, W.-C., Chang, T., Chang, W.-C., Liu, M.-Y., Payne, W. J., and Le Gall, J. (1996) Biochem. Biophy. Res. Comm. 219, 423– 428.
Vol. 250, No. 3, 1998 BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS