行政院國家科學委員會專題研究計畫 成果報告
以 real time PCR 及 Western Blot 鑑定肝細胞再生時,基
因微陣列監測出免疫相關基因之變遷
計畫類別: 個別型計畫 計畫編號: NSC93-2314-B-002-263- 執行期間: 93 年 08 月 01 日至 94 年 07 月 31 日 執行單位: 國立臺灣大學醫學院外科 計畫主持人: 賴鴻緒 共同主持人: 陳炯年,李伯皇 計畫參與人員: 吳秀鵑 報告類型: 精簡報告 處理方式: 本計畫可公開查詢中 華 民 國 94 年 8 月 1 日
行政院國家科學委員會專題研究計畫成果報告
以 REAL TIME PCR 及 WESTERN BLOT 鑑定肝細胞再生時,基因微
陣列監測出免疫相關基因之變遷
VERIFICATION BY REAL TIME PCR AND WESTERN
BLOT FOR IMMUNE RELATED GENES DETECTED FROM
CDNA MICROARRAY DURING LIVER REGENERATION
計畫編號:NSC 93-2314-B-002-263
執行期限:93 年 8 月 1 日至 94 年 7 月 31 日
主持人:賴鴻緒 台大醫學院 外科
中文摘要
吾人以前研究顯示,肝臟部分切除 後,除肝細胞再生外,細胞免疫之變遷, 包括 CD4、CD8、淋巴球、自然殺手細胞 之增加,血中γ-IFN 之上升,及巨噬細胞 之增加等,均相當明顯,且發生時間在肝 細胞再生後期,是否與再生終結機轉有關 則尚不明瞭。若能研究瞭解肝細胞再生時 免疫相關基因之變遷,對將來以基因治療 控制免疫反應,來促進肝細胞再生、增加 病患存活率必有很大貢獻。 本計劃以重約200 克之 Wistar 雄性大 鼠做實驗,以基因微陣列方式監測,並以 real time polymerase chain reaction(RT-PCR) 及 Western Blot 鑑定肝細胞再生過程中, 各種免疫相關基因表現在各時段之變遷。 所有大鼠將分別接受約百分之七十及四十 之肝臟部分切除手術,各於術前及術後2、 4、6、12、24、48、72 小時及 5、7 天後犧 牲取樣,測定(1)剩餘肝臟之重量比值; (2) 剩餘肝臟之有絲分裂指標; (3)以基因微陣 列 尼 龍 膜 晶 片 、 肝 細 胞 mRNA 標號、 hybridization 及影像分析等方法,測定各種 基因表現之變遷,並詳細分析免疫相關基 因群; (4)以 RT-PCR 分析剩餘肝臟中,各種 免疫相關基因之mRNA 表現情形,並比較 與 Microarray 所監測出免疫相關基因變遷 之一致性; (5) 以 Western blot 分析剩餘肝 臟中,免疫相關基因之蛋白質產量,並比 較其與基因及 mRNA 變遷之一致性; (6) 比較 70%及 40%肝臟切除後所有免疫相關 基因、mRNA、蛋白質表現之程度、型態、 時程之差異性。 結果顯示:(1)剩餘肝臟重量比值,切 肝 40%後恢復速度較慢,但至術後 72 小 時,40%與 70%兩組幾乎到達同樣重量; (2)有絲分裂切肝 40%後 48 小時也出現, 至72 小時逐漸減少,但程度上比 70%切肝 後明顯較少;(3)共 8 種免疫相關基因在切 肝 手 術 後 , 有 三 種 型 態 明 顯 的 變 化 ; (4)histocompatibility 2, O region alpha locus gene and early B-cell factor gene 在切肝術 後早期明顯上升,表示此 2 種基因可能為 肝 細 胞 再 生 之 啟 動 機 轉 相 關 基 因 ; (5)histocompatibility 2, Q region alpha locus gene, immunoglobulin J chain precursor gene, histocompatibility 2, complement component factor gene, and histocompatibility 2, L region locus gene 在切肝術後中期,有明顯下降趨勢,至晚期又有回升現象,表示此4 種基因可能在肝細胞再生之分化機轉,扮 演了down-regulation 的角色;(6) interferon gamma receptor 2 gene and mast cell growth factor gene 在切肝術後晚期明顯上升,表示 此 2 種基因可能為肝細胞再生之終結機轉 相關基因;(7)IL-6 及 IL-10 兩種基因在 microarry 顯 示 之 基 因 變 化 極 其 相 關 m-RNA (RT-PCR 檢測)變化之量、程度及 型態均相似,表示IL-及 IL-10 兩種基因在 肝細胞再生機轉中,應扮演較重要的角 色;(8)以 Western Blotting 檢測出來之相關 蛋白質量,與基因及m-RNA 量均不符合, 可能需考慮之因素較複雜。本研究結果顯 示,免疫相關基因在肝細胞再生之啟動、 分化、終結等過程之調控機轉,均扮演了 重要之角色。 關鍵字:肝細胞再生、部分肝臟切除術、 基因微陣列、RT-PCR、Western Blot、免 疫
ABSTRACT
Cell mediated immune response, including increasing of CD4, CD8 lymphocytes, NK cells, γIFN and macrophage were noted during liver regeneration after partial hepatectomy. Whether immune response is the termination factor for regeneration is not clear. The study about the relationship between immune related gene expressions and liver regeneration could be very important for gene therapy to increase liver regeneration and survival rate.
The main purpose of this project was to find out the variation of immune related genes during liver regeneration after partial hepatectomy. Male Wistar rats around 200g were used as subject. Partial hepatectomy around 70% and 40% were be performed and they will be sacrificed at 2, 4, 6, 12, 24, 48, 72 hours and 5, 7days after hepatectomy. We measured: (1) weight of remnant liver; (2) mitotic index; and (3) genomic survey of the expression for many proto-oncogenes by
cDNA microarray on nylon membrane, labeling of liver mRNA hybridization and image analysis of the cluster of immune related genes; (4) the mRNA expression of immune related genes by real time polymerase chain reaction (RT-PCR), and compare the compatibility between the genes and the mRNA expression ; (5) the protein product of immune related genes by Western Blot analysis, and compare the compatibility between the genes expressions and the protein products; (6) Compared the difference of all positive immune related gene expression in the variation degree, changing pattern, specific timing and genes subgrouping cluster between 70% and 40% partial hepatectomy.
The results were: (1) the remnant liver weight recovered slower, but can reach 90% in 72h after partial hepatectomy; (2) the mitosis of hepatocytes also increased markedly at 48h although not so high as 70% group rats, and also decreased at 72h after partial hepatectomy; (3) there were 8 immune related genes identified with marked changes in 3 different patterns after partial hepatectomy; (4) histocompatibility 2, O region alpha locus gene and early B-cell factor gene showed a marked elevated peak at early PH period, suggesting as initiative genes for regeneration; (5) histocompatibility 2, Q region alpha locus gene, immunoglobulin J chain precursor gene, histocompatibility 2, complement component factor gene, and histocompatibility 2, L region locus gene showed a down-regulated pattern at mid-term PH period, suggest as differentiative genes for regeneration; (6) interferon gamma receptor 2 gene and mast cell growth factor gene showed a late increasing peak suggesting as terminated genes for regeneration; (7) the changing curves of IL-6 and IL-10 genes cloning by microarray and their related m-RNA detected by RT-PCR were similar after 70% vs 40% partial hepatectomy, suggesting IL-6 and IL-10 genes play more important roles in regeneration mechanism; (8) protein products detected by Western Blotting showed no
similar changes when compared with microarray and RT-PCR, suggesting protein products were affected by more factors. It is suggested that some immune related genes did play important roles in the mechanism of initiation, differentiation and termination during liver regeneration after partial hepatectomy.
Key words: liver regeneration, partial
hepatectomy, microarray, RT-PCR, Western Blot, immune
INTRODUCTION
Although the liver contains good regeneration ability, hepatic failure can still occur after massive hepatic injury or hepatectomy. Because of the high incidence of hepatitis B carrier and so as to many hepatoma patients, the mortality rate of hepatic disease (including hepatoma cases) is one of the tope ten causes of death in Taiwan. It indeed thretoned the people’s health of our country. The capacity of the mammalian liver to regenerate in response to numerous stimuli has been well documented[1,2]. Although there are much controversy continues on the initiation, differentiation metabolic changes, and termination of liver regeneration after partial hepatectomy that will initiate proliferation of the remaining hepatocytes, several factors, such as hormones, growth factors, nutritional components, and pharmacological agents, have been demonstrated to directly or indirectly affect liver regeneration[3-5]. Partial hepatectomy may raise the possibility of changes in local or systemic immunological response. During liver regeneration after partial hepatectomy, cell-mediated immunity is activated, and as a result, autoreactive suppressor T cells[6,7], and natural killer (NK) cells[8,9] are activated against the allogeneic response. In our previous study, we have focused on immunological response during liver regeneration, and found that CD4, CD8 cells, γ-IFN and NK cells were activated markedly and might play important roles in the regulation of liver regeneration after partial
hepatectomy.[10] It is believed that genetic regulation for immune reaction should play an important role in hepatic cell regeneration.
In recent study, the critical roles of some proto-oncogenes were noted in the control of cell proliferation, differentiation, and apoptosis by the new technique of complementary DNA microarray. Arora et al reported that c-Myc antisense limits rat liver regeneration by regulating cytochrome p-450 3A activity[11]. Ozeki and Tsukamoto found that retinoic acid can repress c-fos and c-jun expression and induce apoptosis in regenerating liver[12]. Some proto-oncogenes about immunologic reaction such as IL-6R/IL-6 complex[13], CD4 and CD8 cells[14] were also detected to be involved in the regulation during the process of liver regeneration. Mass survey for gene expression variations by the method of cDNA microarray was reported to be a useful instrument.[15,16] However, the detailed analysis of the immune related proto-oncogene expression about immune related genes detected by cDNA microarry method, and comparison on the difference of degree, pattern and timing are still not reported. Furthermore, verifying the mRNA of these immune related genes by real time polymerase chain reaction (RT-PCR) and the protein product by Western Blot is also necessary for identify the true roles of these immune related genes in the mechanism of liver regeneration.
We have demonstrated that most T-lymphocyte subpopulations (including NK cells) in peripheral blood increased significantly to reach their peaks on the fifth and seventh post-hepatectomized day[10]. Serum r-IFN and the liver-resident NK cells also had the same elevated changing curve. We also found that neopterin concentrations, both in serum and urine, increased significantly and reached their peaks between the fifth and seventh days after hepatectomy, and that the increase in the neopterin concentration in urine was more pronounced than that in serum[17,18]. It indicated that
either these immunologic responses were induced by liver regeneration or the immunologic responses just acted as a regulating factor for termination of the liver regeneration from the time sequence.
MATERIALS AND METHODS
Experimental Protocol
Sixty male Wistar rats (purchased from Charles River, Osaka, Japan) weighing approximately 200g were used as subjects. All of them received 40% partial hepatectomy and they were sacrificed before and 2, 4, 6, 8, 12, 24, 48, 72 hours and 5, 7 days after hepatectomy. Six were sacrificed each time and the remnant livers were removed immediately for further tests.
Surgical Procedures
All rats are anesthetized by intraperitoneal pentobarbital (10mg/kg) injection. A midline laparotomy was performed. Partial hepatectomy was then carried out by means of aseptic extirpation of the left lateral lobes (around 40%) according to the procedure of Higgins and Anderson. [19] The removed liver sample was immediately weighed. Laparotomies with manipulation of liver were done in the sham operated rats. All of the surgeries were performed between 8 am and 11 am to reduce the influence of diurnal variation.
Measurements
(1)Evaluation of the remnant liver
Observation of the liver surface and color. Then weighing the liver immediately after sacrifice, and the ratio of remnant liver weight/body weight will be calculated.
(2)Mitotic index of remnant liver
The small pieces of liver tissue for hisopathological examination at certain postoperative period will be fixed in 10%
neutral formalin, embedded in paraffin, sectioned and stained with hematoxylin-eosin for microscpic observation. The mitotic index will be determined by counting the number of parenchymal cells undergoing mitosis in 50 randomly-selected fields under magnification ×400. The results will be expressed as the mitotic index (the total number of mitoses per 50 different fields examined).
(3) Genomic survey of remnant liver by cDNA microarray
A. Non-isotopic labeling of liver mRNA Total RNA will be extracted from remnant livers of sacrificed rats in each postoperative time sequence. The tissue will be homogenized in 3 ml of solution A containing 4M guanidine thiocyanate[20], 0.5% sarcosyl, 25 mM sodium citrate, and 0.1M betamercaptoethanol at pH 7.0, followed by phenol extraction, isopropanol precipitation, and ethanol precipitation. Quality of RNA will be examined by agarose gel electrophoresis. Messenger RNA was purified from total RNA by using mRNA Isolation Kit(Qiagen Oligotex extraction Kit, Qiagen, Germany) according to the manufacturers’ protocol.
B. Microarray system
The whole system included tool pin, adapter, washing slots, sampling slots, sample distribution region, program and automatically changing plate system. Those treated pins will be fitted into multiple-pore adapter, which is designed specially to make the pins elastic while spotting samples on membrane. The adapter could be changed for single or multiple pins system according to experimental need. The PCR-amplified cDNA in V-bottomed 96-well microtiter plates were distributed onto a positively charged nylon membrane with spots spaced 150 ~ 500 µm apart by an arraying machine. The arraying machine is a computer controlled XYZ translation system outfitted
with Teflon-coated tool pins for sample delivery. It was designed and constructed by Wittech company (Taipei, Taiwan) with position resolution and repeatability better than ± 5 µm. Samples will be held at the tip of the pins for delivery by the action of surface tension, and the delivery volume was governed by the diameter of the tips. Every time after DNA was spotted on membranes, by means of movement of translation stage, the solid pin will be immersed into washing slots, and washed by 95% ethanol, and deionized water respectively with sonicating. The microarray chip carrying 20,544 PCR-amplified cDNA fragments were prepared by arraying machine built in house. The cDNA microarray membrane contains 6,144 non-redundant expressed sequence tag (EST) clones with putative gene names selected from the IMAGE consortium mouse cDNA libraries based on the insert length and sequence of the EST clones in the Unigene database
(4) Real time PCR for verifying the immune-related gene detected by cDNA microarray
A. Quantitative PCR kit (DA0005) with positive control primer set
5.0 µg total RNA of each sample was extracted and the cDNA was obtained by RNA Isolation and Reverse Transcription Kit (Cat. No. DA003). The PCR reactions were performed by PCR Kit (Cat. No. DA004). Amplification cycles were carried out at 94°C for 3 min, followed by 35 cycles of 94°C for 40 sec, 58°C for 40 sec, 72°C for 40 sec, concluded by 7 min elongation at 72°C. 10µL amplicons were separated by 3% agarose electrophoresis. Amplification with GAPDH primers was performed using the above procedure to confirm the efficacy of the reverse transcription process for each RNA-extracted cell.
B. LightCycler real-time PCR
For LightCycler reaction a mastermix of
the following reaction components was prepared to the indicated end-concentration: 13 µl water, 2.4 µl MgCl2 (4 mM), 0.8 µl
forward primer (0.4 µM), 0.8 µl reverse primer (0.4 µM) and 2.0 µl LightCycler (Fast Start DNA Master SYBR Green I; Roche Diagnostics). LightCycler mastermix (19 µl) was filled in the LightCycler glass capillaries and 1 µl cDNA was added as PCR template. Capillaries were closed, centrifuged and placed into the LightCycler rotor. The following LightCycler experimental run protocol was used: denaturation program (95°C for 10 min), amplification and quantification program repeated 30~40 times (95°C 15 s, 60°C for 10 s, 72°C for 60 s with a single fluorescence measurement), melting curve program (60-98°C with a heating rate of 0.1°C per second and a continue fluorescence measurement) and finally a cooling step to 40°C. For the mathematical model it is necessary to determine the crossing points (CP) for each transcript. CP is defined as the background fluorescence. ‘Fit Point Method’ must be performed in the LightCycler software 3.3 (Roche Diagnostics), at which CP will be measured at constant fluorescence level.
(5) Western Blot Analysis A. Protein Extraction
1. Cut about 0.5cm3 tissue and cut to pieces. 2. Add 0.5ml lysis buffer and mix.
3. Homogenize the tissue pieces to be liquid form by high-speed homogenizer, then keep on ice 10 min.
4. Transfer the tissue liquid to micro centrifuge tube, 12000rpm 10min. 5. Collect the supernatant, pipette a few
protein solution for determination of protein concentration, the other keep in -80 .℃
B. Protein Quantitation
1. Protein Quantitation is by Bradford Method.
with 4 parts distilled, deionized water with a standard or sample. Incubate at room temperature for at least 5 minutes. 3. Absorbance will increase over time;
sample should not incubate at room temperature for more than 1 hour. 4. Measure absorbance at 595 nm and make
a standard curve.
5. Dilute your samples and measure in the same way. Calculate the concentration through the standard curve by
absorbance.
C. Electrophosis and Transfer
1. Prepare Electrophoresis Buffer and Transfer Buffer
2. Dilute 6X Sample Buffer to 1X with protein sample solution. Heat at 95℃ for 10 min.
3. After gel packing, set up the gel running tank.
4. Load more than 30µg total protein to each well. Run gel with rainbow molecular weight markers.
5. Run gel in Electrophoresis Buffer at 80mV for 40min and 115mV for 2-3 hr. 6. Fix gel in the Transfer Buffer for 15min. 7. Fix the PVDF membrane in methanol for
15min then transfer to Transfer Buffer for 15 min.
8. Set up the transfer tank: add buffer, insert sandwiches, and run 100 mA 30 V for 1.5~3hr.
9. Mark the molecular weight markers locations on the PVDF membrane. D. Western Blotting
1. hybridization with blocking buffer at 37 for 1 hr℃
2. incubation at 37 with primary antibody ℃ (INFgR2 IgG, I7662-17N, United States Biological) diluent for 1 hr
3. wash off excess primary antibody with TTBS buffer for 10 min, and repeat 3 time
4. incubation at 37 with secondary ℃ antibody (Goat gati-rabbit IgG-HRP, sc-2004, Santa Cruz) diluent for 1 hr
5. wash off excess secondary antibody with TTBS buffer for 10 min, and repeat 3 time
6. add 1 ml ECL mixture (A:B=1:1), and expose at X-ray film
RESULTS
The rats were living well during experimental stage. After sacrifice, the remnant liver weight increased to 90% in 72h after partial hepatectomy. However, the growing speed is not so fast as 70% group rats when 40% partial hepatectomy was done (Fig 1.). The mitosis index showed that the mitosis of hepatocytes in the remnant liver increased to 64±8.8 at 48h, then decreased to 20±4.6 at 72h after 40% partial hepatectomy. The elevation was also not as high as that in 70% group rats (Table 1).
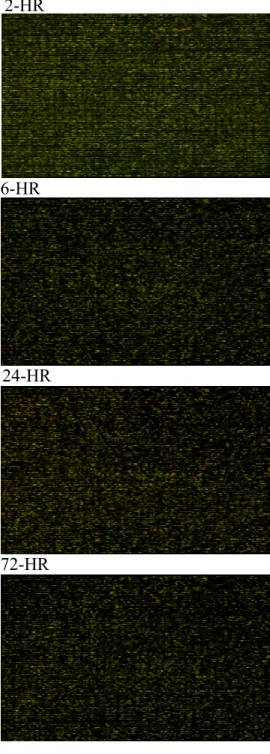
When the microarray chip was analyzed by a flatted scanner and the GenePix in each time sequence, the variations of all the 20,500 proto-oncogenes expression could be classified into 72 different patterns as previous report (Fig 2.). The various patterns of the genes expressions include the pattern containing a single peak which occurred at 2h, 4h, 6h, 12h, 48h, 72h, 5d, and 7d. In addition, typical double-peaks patterns occurred at different time sequence such as 12h, and 5d after partial hepatectomy. Moreover, patterns showing increasing trend since 4h or patterns illustrating a decreasing trend since 4h were also noticed in some proto-oncogenes expressions. Beyond that, the protruding types of patterns from 12h to 24h and from 6h to 72h, or the excavated types of patterns from 4h to 5d and 6h to 5d were found. Mixed types of time-dependent curves were detected as other unclassified variation patterns of genes expressions. The changing patterns are about the same when compared 40% vs 70% groups rats, however, the changing degrees was not so high in 40% partial hepatectomized rats.
Among the immune related genes, the changing degrees in 40% group rats were not so marked but more stable when compared
with 70% group rats. Histocompatibility locus class II region was highly elevated at 3d after 40% partial hepatectomy that was high at 6h and 7d, but not 3d after 70% partial hepatectomy. That means this gene might be not directed related to the control mechanism of liver regeneration.
There were 8 immune related genes identified with marked changes in 3 different patterns after partial hepatectomy (Fig 3.). Histocompatibility 2, O region alpha locus gene and early B-cell factor gene showed a marked elevated peak at early PH period, suggesting as initiative genes for regeneration. Histocompatibility 2, Q region alpha locus gene, immunoglobulin J chain precursor gene, histocompatibility 2, complement component factor gene, and histocompatibility 2, L region locus gene showed a down-regulated pattern at mid-term PH period, suggest as differentiative genes for regeneration. Interferon gamma receptor 2 gene and mast cell growth factor gene showed a late increasing peak suggesting as terminated genes for regeneration.
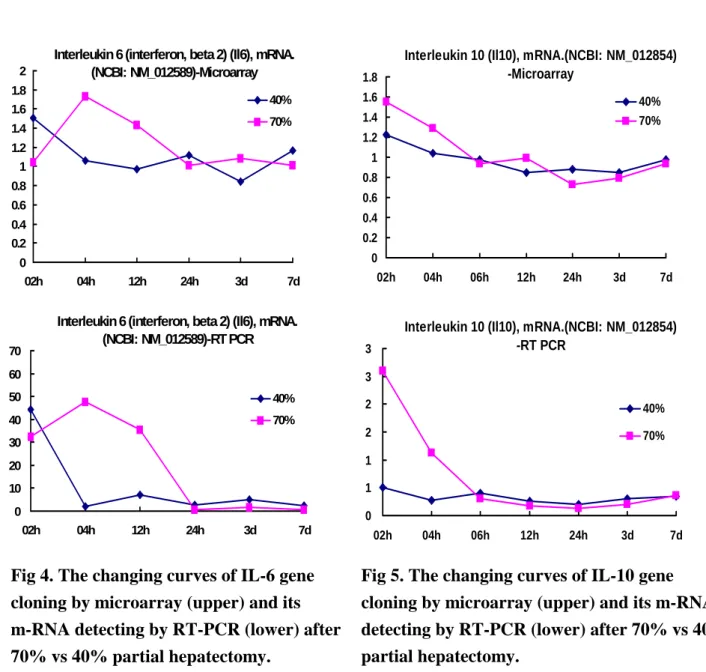
As for IL-6 and IL-10 genes, the time sequence changes and changing patters are similar in 40% vs 70% groups rats, with a milder degree in 40% group rats (Fig 4. and Fig 5.). The changing curves of IL-6 and IL-10 genes cloning by microarray and their related m-RNA detected by RT-PCR were similar after 70% vs 40% partial hepatectomy, suggesting IL-6 and IL-10 genes play more important roles in regeneration mechanism. Protein products detected by Western Blotting showed no similar changes when compared with microarray and RT-PCR, suggesting protein products were affected by more factors (Fig 6.). It is suggested that some immune related genes did play important roles in the mechanism of initiation, differentiation and termination during liver regeneration after partial hepatectomy.
DISCUSSION
The phenomenon of liver regeneration is
one of the most rapid forms of tissue growth occurring in mammals. It happened in the very early phase after partial hepatectomy, as was proven by previous studies
here[10,17,18,21-25] and in many other centers[1-5].
Our previous study demonstrated that the DNA synthetic rate increase abruptly, peaking at 24h, the mitotic index reach a maximum at 48h, the remaining lobes had doubled in size by 48h and have attained around 90%of the normal liver weight at 72h after partial hepatectomy in rats[21]. In recent studies, most of them put their focus on the factors that can promote and increase liver regeneration. Several factors, such as hormones, growth factors, nutritional components, and pharmacological agents, have been demonstrated to directly or indirectly affect liver regeneration. We have proved that high concentrated glucose or branched chain amino acid can promote liver regeneration[21]. Combination of Insulin, glucagon, and epidermal growth factors can also promote liver regeneration[22]. We also found that energy substrate insulin, glucagon, epidermal growth factor, and mitochondria carnitine palmitoyl-transferase play important roles during liver regeneration[23-25].
The regenerating liver is a system in which the relationships between proto-oncogene expression and cell replication should be examined during a
physiologic growth response. Proto-oncogene expression after partial hepatectomy is specific, sequential, and highly regulated. As measured by levels of mRNAs, the changes have been detected in the expression of c-fos, c-myc, p53, and the ras gene family (c-Ha-ras, c-Ki-ras, and N-ras)[26-28]. In contrast, expression of c-src and c-abl does not change after partial hepatectomy while c-mos transcripts cannot be detected in normal or regenerating liver[29].
From English medical journal, several proto-oncogenes with some chemical agents involved in mechanism during liver
regeneration were reported[24,26-29]. Arora et al reported that c-Myc antisense limits rat liver regeneration by regulating cytochrome p-450 3A activity[11]. Ozeki and Tsukamoto found that retinoic acid can repress c-fos and c-jun expression and induce apoptosis in regenerating liver[12]. Some proto-oncogenes about immunologic reaction such as IL-6R/IL-6 complex1[13] and CD4, CD8 cells1[14] were also detected to be involved in the regulation during the process of liver regeneration. Liver regeneration might be regulate by expression of immune related proto-oncogenes concerned about T cell, lymphokine, macrophage changes[9].
Mass survey for immune related genes by microarray method is very important for detecting the liver regeneration related genes. Many immune related genes were found to be changed markedly after partial hepatectomy. However some changes are not regular and not according time sequence may mean nothing. Even the eight genes we have detected in this study, the genes may be involved in the initiation, differentiation and termination control mechanism, it is important to be verified by further examination of their related m-RNA by RT-PCR and their protein products by Western Blot. In this study, we can’t prove all of these relationships among the genes, m-RNAs, and protein products. Speaking of protein products, we can’t prove their relationships even the gene and mRNA were matched in IL-6 and IL-10 genes. The true reason must be detected in the future study, however, many complicated factors including technique, liver cell characteristics and liver regeneration itself may all involved. Further study for more detailed time sequence, different method for protein quantification, and antiagent administration for increasing liver regeneration proofs may be needed.
REFERENCES
1. Chamuleau RAFM, Bosman DK: Liver
regeneration. Hepatogastroenterol 35:309-12,1988
2. Michalopoulos GK, DeFrances MC:
Liver regeneration. Science 276:60-6,1997.
3. Rigotti P, Peter JC, Tranberg KG, et al: Effects of amino acid infusion on liver regeneration after partial hepatectomy in the rat. JPEN 10:17020,1986.
4. Ferrero M, Desiderio MA, Martinotti A, et al: Expression of a growth arrest specific gene (gas-6) during liver regeneration: molecular mechanisms and signaling pathways. J Cell Physiol 158:263-9,1994.
5. Wang X, Quail E, Hung NJ, et al: Increased level of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regeneration liver. Proceed Nation Acad Sci USA 98:11468-73,2001.
6. Flye MW, Yu S: Augmentation of cell-mediated cytotoxicity following 50% partial hepatectomy. Transplantation 49:581-7,1990.
7. Okovityi SV, Gaivoronskaia VV: Immune mechanisms of hepatoprotector effects of etomersol and thymogen. Eksperimentalnaia i Klinicheskaia Farmakologiia 65:44-6,2002.
8. Olthoff KM: Molecular pathways of regeneration and repair after liver transplantation. World J Surg 26:831-7 2002.
9. Geissler M, Mohr L, Weth R, et al: Immunotherapy directed against alpha-fetoprotein results in autoimmune liver disease during liver regeneration in mice. Gastroenterology 121:931-9,2001. 10. Lai HS, Chen WJ, Chen KM: Changes in
T-lymphocyte subpopulations and serum lymphokine concentrations after partial hepatectomy in rats. Nutrition 12:700-5,1996.
11. Arora V, Knapp DC, Smith BL, et al: c-Myc antisense limits rat liver regeneration and indicates role for c-Myc in regulating cytochrome P-450 3A activity. J Pharmacol Exp Therap 292:921-8,2000.
12. Ozeki A, Tsukamoto I: Retinoic acid repressed the expression of c-fos and c-jun and induced apoptosis in regenerating rat liver after partial hepatectomy. Biochem Biophy Acta 1450:308-19,1999.
13. Jones SA, Rose-John S: The role of soluble receptors in cytokine biology: the agonistic properties of the sIL-6R/IL-6 complex. Biochim Biophy Acta. 1592:251-63, 2002.
14. Geissler M, Mohr L, Weth R, et al: Immunotherapy directed against alpha-fetoprotein results in autoimmune liver disease during liver regeneration in mice. Gastroenterology 121:931-9, 2001. 15. Makoto Arai, Osamu Yokosuka,
Tetsuhiro Chiba, et al: Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. J Biol. Chem 278: 29813-8,2003.
16. Fukuhara, Yasuyuki; Hirasawa, , et al: Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol 38:784-92,2003. 17. Lai HS, Chen Y, Chen WJ: Carnitine
contents in remnant liver, kidney and skeletal muscle after partial hepatectomy in rats: randomized trial. World J Surg 22:42-7, 1998.
18. Lai HS, Chen WJ: Changes of serum and urinary neopterin concentrations in rats after partial hepatectomy. Eur J Surg 165:604-8,1999.
19. Higgins GM, Andersan RM: Experimental pathology of the liver. Arch Pathol 12:186-92,1931.
20. Chomezynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156-9,1987.
21. Lai HS, Chen WJ, Chen KM: Liver regeneration after partial hepatectomy. J
Formosan Med Assoc 89:1045-51,1990. 22. Lai HS, Chen WJ, Chen KM: Energy
substrate for liver regeneration after partial hepatectomy in rats: effects of glucose vs fat. JPEN 16:152-6,1992. 23. Lai HS, Chen WJ: Alteration of remnant
liver carnitine palmitoyl-transferase I activity and serum carnitine concentration after partial hepatectomy in rats. J Surg Res 59:754-8,1995.
24. Lai HS, Chen WJ, Chen KM. Alteration of high-energy phosphate, serum energy substrate and their metabolites after partial hepatectomy in rats. J Formosan Med Assoc 90:621-5,1991.
25. Lai HS, Chung YC, Chen WJ: Liver regeneration after partial hepatectomy in rats: effects of insulin, glucagon, and epidermal growth factors. J Formosan Med Assoc 91:685-90,1992.
26. Arora V, Knapp DC, Smith BL, et al: c-Myc antisense limits rat liver regeneration and indicates role for c-Myc in regulating cytochrome P-450 3A activity. J Pharmacol Exp Therap 292:921-8,2000.
27. Iwao K, Tsukamoto I: Quercetin inhibited DNA synthesis and induced apoptosis associated with increase in c-fos mRNA level and the upregulation of p21WAF1CIP1 mRNA and protein expression during liver regeneration after partial hepatectomy. Biochem Biophy Acta 1427:112-20,1999.
28. Lee SM, Li ML, Tse YC, et al: Paeoniae Radix, a Chinese herbal extrac, inhibit hepatoma cells growth by inducing apoptosis in a p53 independent pathway. Life Science 71:2267-77,2002.
29. Yaswen P, Goyette M, Shank PR, et al: Expression of c-ki-ras, c-Ha-ras and c-myc in specific cell types during hepatocarcinogenesis. Mol Cell Biol 5:780,1985.
Time
Group
6H 24H
48H
72H
40% PH
0
Rare
64±8.8
20.6±4.6
70% PH
0
Rare
104.1±12.2*
24.2±6.2
C 0
Rare 0
0
Mean±SD; Sum of 50 randomized field under 400x; C: control; H: hour;
PH: Partial hepatectomy; *: vs 40%PH, p<0.05
Time
Group
0H 4H 12H 3d 7d
70%
IL-6 37.23
30.62
36.29
36.81
43.07
GAPDH 62.77
69.38
63.71
63.19
56.93
IL-6/GAPDH(%) 59.31 44.13
56.96
58.25
75.66
40%
IL-6 37.23
33.75
31.17
36.04
35.51
GAPDH 62.77
66.26
68.83
63.97
64.49
IL-6/GAPDH(%) 59.31 50.93
45.29
56.34
55.07
70%/40% 1
0.87
1.26
1.03
1.37
Time
Group
0H 2H 4H 6H 12H 24H 3d 7d
70%
IL-10 23.92 23.66 23.90 23.83 23.67 26.87 27.93 27.75
GAPDH 76.09 76.34 76.10 76.17 76.34 73.13 72.07 72.25
IL-10/GAPDH(%) 31.43 31.00 31.40 31.28 31.00 36.74 38.75 38.40
40%
IL-10 23.92 25.46 34.14 32.88 25.69 23.69 23.45 25.88
GAPDH 76.09 74.54 55.03 67.12 74.31 76.31 76.55 74.12
IL-10/GAPDH(%) 31.43 34.15 62.04 48.98 34.58 31.05 30.63 34.92
70%/40%
1 0.91 0.51 0.64 0.90 1.18 1.27 1.10
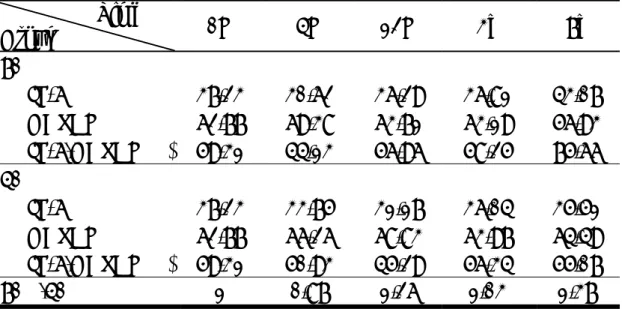
Table 1. Mitotic Index after 40% and 70% PH
Table 2. Western Blotting Results for IL-6 (interferon beta 2) Gene Related Protein Analysis
Fig 3. Eight immune related genes detected by cDNA microarray
demonstrated marked changes in the changing curve according to the time sequence after 70% partial hepatectomy. Fig 1. Remnant Liver Weight/Body Weight Ratio after 40% and 70% partial
Hepatectomy.
2-HR
6-HR
Fig 2. The colorimetric image of cDNA microarray hybridization chips with 20,500 genes showed uneven changed patterns before 2, 6, 24, and 72 hours after 70% partial hepatectomy.
(microarray image of sample with Cy5 to a reference RNA with Cy3)
B
1 2 3 4 5 6 7 8 9
histocompatibility 2, Q region locus immunoglobulin j chain precurso histocompatibility 2, complement component factor histocompatibility 2, L regio
C
1 2 3 4 5 6 7 8 9
Interferon gamma receptor 2
D
1 2 3 4 5 6 7 8 9
mast cell growth facto
A
1 2 3 4 5 6 7 8 9 histocompatibility 2, O region alpha locus early B-cell facto
Normal Rat 4.31% 0 1 2 3 4 5 0 6 24 48 72 Hours % 70% 40% 72-HR 24-HR
Fig 4. The changing curves of IL-6 gene cloning by microarray (upper) and its m-RNA detecting by RT-PCR (lower) after 70% vs 40% partial hepatectomy.
Interleukin 10 (Il10), mRNA.(NCBI: NM_012854) -RT PCR 0 1 1 2 2 3 3 02h 04h 06h 12h 24h 3d 7d 40% 70%
Interleukin 10 (Il10), mRNA.(NCBI: NM_012854) -Microarray 0 0.2 0.4 0.6 0.8 1 1.2 1.4 1.6 1.8 02h 04h 06h 12h 24h 3d 7d 40% 70%
Fig 5. The changing curves of IL-10 gene cloning by microarray (upper) and its m-RNA detecting by RT-PCR (lower) after 70% vs 40% partial hepatectomy.
Fig 6. Western Blot electrophoresis of IL-6 (Abcam ab6672 1:400 dilute 26kd) gene related protein product. *GAPDH 1:1000 dilute as control agent.
Interleukin 6 (interferon, beta 2) (Il6), mRNA. (NCBI: NM_012589)-RT PCR 0 10 20 30 40 50 60 70 02h 04h 12h 24h 3d 7d 40% 70%
Interleukin 6 (interferon, beta 2) (Il6), mRNA. (NCBI: NM_012589)-Microarray 0 0.2 0.4 0.6 0.8 1 1.2 1.4 1.6 1.8 2 02h 04h 12h 24h 3d 7d 40% 70%