行政院國家科學委員會補助專題研究計畫成果報告
非小細胞肺癌組織中經由差異表現法確認之
Transmembrane GTPase 之研究
計畫類別:個別型計畫 計畫編號:NSC90-2320-B-039-023 執行期間:90 年 8 月 1 日至 91 年 7 月 31 日 計畫主持人:何恆堅 中國醫藥學院醫學系生化學科 共同主持人:鍾景光 中國醫藥學院醫學系微生物學科 葉坤土 彰化基督教醫院病理科 計畫參與人員:葉雅雯 中國醫藥學院醫學系生化學科 王毓芬 彰化基督教醫院病理科 執行單位:中國醫藥學院醫學系生化學科 中 華 民 國 九 十 年 十 二 月 十 二 日Abstr act
The technique of differential display was previously used to profile the gene
expression patterns of NSCLC and several genes differentially expressed were thus
identified. In this report, we demonstrate that a DNA fragment of 347-bp length,
up-regulated in tumor tissues, showed 100% sequence similarity to human cDNA
FLJ20693 for a 370-residue protein. The gene product of cDNA FLJ20693 was
postulated to be a shorter isoform of transmembrane GTPase, termed TG370, based
upon the results of searching for sequence homology. The nucleotide sequence
alignment also indicated that the cDNA FLJ20693 and the cDNA for 741-residue
human mitofusin 1 (TG741) possibly resulted from the event of alternative splicing
from which a 127-bp region was retained in the latter. Analysis of the genome
sequence confirmed the speculation that both cDNAs were mapped to the same
chromosomal position composing of 18 exons, of which the 127-bp region of TG741
constituted exon 11. The alternative splicing in all lung cancer cell lines was also
observed to occur nearly in all tissue specimens examined. The up-regulated
expression of transmembrane GTPase was subsequently found in tumor tissues from
at least 5 out of 7 NSCLC patients. Also, a distinct PCR product was initially
detected in cell line H520 and further sequence analysis identified the presence of the
evaluate the retention of 86-bp region, it was found that, besides the predicted 486-bp
product, an unexpected 332-bp product was concomitantly observed and identified as
the result of exon 8 deletion. The expression and subcellular localization of the
full-length TG741 and other shorter isoforms were detected by flow cytometry using
three polyclonal antibodies. It was concluded that the full-length TG741 located at
plasma membrane with its N-terminal domain exposed extracellularly and the shorter
isoforms retained at cytosol. Finally, the up-regulation of transmembrane GTPase in
Introduction
Lung cancer has become the major cause of mortality, more than 5000 cases
annually, in Taiwan (1). The five-year survival rate, 65-75%, for patients going
through stage I NSCLC (non-small cell lung cancer), exhibits no significant change
during the past twenty years (2). Actually, there have been very few
well-documented prognostic factors to evaluate survival and supplement the stage
designation. Therefore, it is extremely important to identify novel markers for
detecting high-risk and early-stage patients, who could potentially benefit from more
aggressive treatment approaches.
Advances in molecular biology have accelerated the illustrations that several
differentially expressed genes were implicated in lung cancer tissues or cell lines
using differential display technique developed by Liang et al. (3). They included a
member of the NF2/ERM/4.1 superfamily (4), laminin beta3 and gamma2 chains (5),
semaphorin E (6) and RAB5A (7). We also employed this approach to profile the
gene expression patterns of NSCLC patients and thus identified several differentially
expressed genes. Among these, the overexpression of dihydrodiol dehydrogenase 1
(DD1) in tumor tissues as a prognostic factor had been reported as the first illustration
in lung cancer (8).
lung cancer tissues, as transmembrane GTPase. Based upon the cDNA sequences
reported by different groups and analysis of the genome sequence, two isoforms of
transmembrane GTPase were speculated to result from the mRNA alternative splicing.
The detail of this ubiquitous alternative splicing occurring in lung cancer tissues and
different cell lines will be thoroughly described in the text. The study also indicates
that the events of the alternative splicing will further generate several distinct cDNA
isoforms. Moreover, detection and localization of the gene products is carried out by
Mater ials and Methods
Tissue Specimens and Cell Lines. The primary cancer tissues and the
pair-wise normal tissues were surgically resected and used for research purpose with
the permission from all NSCLC patients. Nine cases of squamous cell carcinoma
(SCC), used for mRNA differential display, were obtained from China Medical
College Hospital within the time period of experimental design. Thirty-six cases of
SCC and adenocarcinoma used for immunohistochemistry were collected from
Changhua Christian Hospital, Changhua. Tissue specimens were immediately
snap-frozen and stored in liquid nitrogen until use. All patients were also subjected
to radical N2 lymph nodes dissection. Tumor size, lymph node number,
differentiation, vascular invasion and mitotic number were also evaluated.
Several lung cancer cell lines were used in the study, including H23, H125, H226,
H838, A549, H661 and H520, to illustrate the alternative splicing of cDNA. All cell
lines examined were separately cultured in RPMI 1640 (Life Technologies)
containing 10% FBS (fetal bovine serum) (Life Technologies) and 2% penicillin
(10000 U/ml)-streptomycin (10 mg/ml). These cells were then placed into 75 cm3
tissue culture flasks and grown at 37℃ under a humidified 5% CO2 until they
reached the amounts of 5x106 cells for further use.
and gene expression, were all listed in Table 1.
RNA Isolation, mRNA Differential Display and Gene Identification. Total
RNA of tumor and the pair-wise normal tissue of NSCLC patient was isolated using
TRIzol reagent (Life Technologies, Grand Island, NY, USA) according to the
instruction manual. For mRNA differential display (RNAimage kit, GenHunter
Corporation, Nashville, TN, USA), the conducting procedures were exactly the same
as those suggested by the manufacturer and described previously (8). Arbitrary
primer H-AP76 and reverse primer H-T11A were used in the study. After being
precipitated and filled in at both ends with T4 DNA polymerase, the re-amplified
cDNA fragments were subcloned into the EcoRV site of the vector pZErO-2.1
(Invitrogen Corporation, San Diego, CA, USA). The cDNA insert was then
sequenced using primers (5’-GTAAAACGACGGCCAG-3’ and
5’-CAGGAAACAGCTATGAC-3’). [α-35S]-dATP (10 mCi/ml, specific
activity>1000 Ci/mmol) was from Amersham Pharmacia Biotech.
3´ RACE. Based upon the sequence of differentially expressed cDNA, two
specific primers were designed to proceed 3´ RACE (rapid amplification of cDNA
ends). Briefly, about 3.5 µg of total RNA, which was isolated from a tumor tissue of
NSCLC patient designated 8T in Fig. 1, 10 pmoles of adapter primer and
12 µl. The subsequent procedures were conducted entirely as those in the instruction
manual (3´ RACE system, Life Technologies). When amplifying the target cDNA,
20 pmoles of primer FP1-A76A, 10 pmoles of primer UAP, 1 µl of cDNA, 2 mM
MgCl2, 0.2 mM dNTP and 2.5 units of Ex. Taq DNA polymerase (TaKaRa Shuzo Co.,
Shiga, Japan) were included in 50 µl of reaction mixture. After preincubating the reaction at 95℃ for 5 min, thirty-five cycles of reaction were performed (95℃ for
30 s, 60℃ for 30 s and 72℃ for 30 s). The same conditions were also applied to
the nested amplification reaction, except using primers FP2-A76A and AUAP and 1
µl of primary PCR product as template. The subsequent protocols for subcloning the secondary PCR product and sequencing were equivalent with those mentioned above.
Prepar ations of Recombinant Proteins for Immunization. Three
recombinant proteins were prepared to immunize the mice in the study. Each primer
pair, used in RT-PCR, was designed to incorporate a restriction enzyme site (BamHI
or XhoI, Table 1) at their 5’-ends. For recombinant protein encompassing the
complete 370 residues of shorter transmembrane GTPase, TG370 (GenBank
accession number AK000700) (9), the primers used were B-AEPVSP (residues 2-7)
and RFHVQ-X (residues 366-370). The PCR product, treated with BamHI and XhoI,
was subcloned into the corresponding sites of the expression vector pET-29a+
lacking of exon 11, into the expression host, E. coli strain BL21(DE3), the
recombinant protein was introduced to over-express after 3-h induction in the
presence of 1 mM IPTG. The isolated inclusion body was washed twice with
distilled water and re-dissolved in 1% SDS overnight at room temperature. The
prepared recombinant protein showed near homogeneity as judged by SDS-PAGE and
was ready for immunization.
Similar methods were also applied to prepare the other two recombinant proteins
encompassing the C-terminal 100 residues and the internal 226 residues of
transmembrane GTPase, TG741 (10). In the former, primers B-FKQQFV (residues
642-647 of TG741) and SNEES-X (residues 737-741 of TG741) were used. In the
latter, primers B-YSVEER (residues 368-373 of TG741) and LASVTS-X (residues
588-593 of TG741) were employed to amplify the coding region followed by a
putative transmembrane region of residues 600-622 of TG741.
Prepar ations of Polyclonal Antibodies. The prepared recombinant proteins
were used to immunize the 6-weeks-old female Balb/c mice. First of all, each mouse
was initially injected with 0.5 ml of pristane. About 100 µg of antigen, mixed with
equal volume of complete Freund’s adjuvant, was applied subcutaneously after 10-15
days. Equal amounts of antigen, emulsified with incomplete Freund’s adjuvant, was
Finally, the serum-free myeloma cells (0.5-1x106) in PBS were injected
intraperitoneally into the mouse. The ascite fluids, normally accumulated after one
week, were collected daily for about 5-8 days.
Detection of Tr ansmembr ane GTPase by Flow Cytometr y. The expression
of transmembrane GTPase of the lung cancer cell line A549, at cell surface or
intracellularly, was illustrated by flow cytometry (FCM), using the three prepared
polyclonal antibodies mentioned above (11, 12). For detecting the surface antigen,
cells were washed twice, resuspended with the given antibody and incubated at 4℃
for 35-min staining. After being washed three times with 10% FBS in RPMI 1640
with 0.1 % sodium azide, cells were stained with FITC-labeled secondary antibody
(goat anti-mouse IgG, Jackson ImmunoResearch Laboratories, West Grove, PA, USA)
at 4℃ for 35 min. The cells were then washed three times, resuspended in PBS and
analyzed by flow cytometry.
For detecting the intracellular antigen, cells were initially washed twice,
resuspended in 100 µl of ice-cold 1% formaldehyde for 5 min, and mixed with 100 µl
of ice-cold 99% methanol for 30 min. Then the cells were washed three times with
0.1% BSA in PBS and mixed with 100 µl of 0.1% Triton X-100 in PBS with 0.1%
sodium citrate on ice for 45 min. After being washed three times with the same
and then washed three times with 0.1% BSA in FBS. The subsequent procedures
were equivalent to those for detection of surface antigen.
Immunohistochemistr y. Polyclonal antibody for the C-terminal 100 residues
of TG741 was used in the immunohistochemistry. Four-micrometer-thick
paraffin-embedded tissue sections on poly-L-lysine coated slides were deparaffinized.
After quenching endogenous peroxidase with 3% H2O2 in methanol, the sections were
hydrated with gradient alcohol and PBS, then incubated with 10 mM citrate buffer and,
finally, heated at 100 ℃ for 20 minutes in PBS. After being exposed to
50-fold-diluted antibody for 30 minutes at room temperature, slides were incubated
with a HRP/Fab polymer conjugate (Zymed, PicTure Polymer Kit, South San
Francisco, CA, USA) for the same time. The sections were thoroughly washed with
PBS at each interval. The sites of peroxidase were visualized with
3,3’-diaminobenzidine tetrahydrochloride. Hematoxylin was used for
counterstaining. Appropriate positive and negative controls were also included.
Statistical Analysis. Statistical comparisons were carried out using the
Fisher’s exact test to determine the significance of the association between different
Results
Identification of Putative Tr ansmembr ane GTPase in NSCLC. After
making a trial to profile the difference of gene expression patterns using arbitrary
primer H-AP76 and reverse primer H-T11A, it was found that a DNA fragment of
347-bases length, termed A76A, is up-regulated in most of the tumor tissues
examined (Fig. 1). The DNA sequence of this fragment, except the primer
sequences at both ends, showed 100% similarity to that of nucleotides 2767-3084 of
human cDNA FLJ 20693 (GenBank accession number AK000700) (9). A76A was
undoubtedly amplified from the cDNA FLJ 20693, based upon the sequence analysis
of the nested PCR product of 3' RACE possessing near 200-bp extension at 3’ end.
The 3148-bp cDNA FLJ20693 encoded a 370-residue protein whose function was not
understood. A search for sequence homologies was subsequently performed based
upon the 370-residue sequence. Significant sequence similarities were observed in
the N-terminal 370 residues of human mitofusin 1 (98%, 362/370) (10) and mouse
putative transmembrane GTPase (90%, 333/370) (GenBank accession number
AK018181) (13). Therefore, it was speculated that the gene product encoded by
cDNA FLJ20693 represented a shorter form of transmembrane GTPase and is termed
TG370 herein to distinguish it from TG741 of 741-residue human mitofusin 1.
for TG370 and TG741. The difference is the presence of an extra 127-bp region in
nts 1098-1224 of cDNA coding region for TG741 (10) listed as:
GCATTATTCAGTGGAAGAGAGGGAAGACCAAATTGATAGACTGGACTTTAT
TCGAAACCAGATGAACCTTTTAACACTGGATGTTAAGAAAAAAATCAAGG
AGGTTACCGAGGAGGTGGCAAACAAA. It seemed likely that both cDNAs
resulted from the alternative splicing, i. e. such a 127-bp sequence constituted an exon.
Therefore, we analyzed the genome sequence and, as expected, both cDNAs were
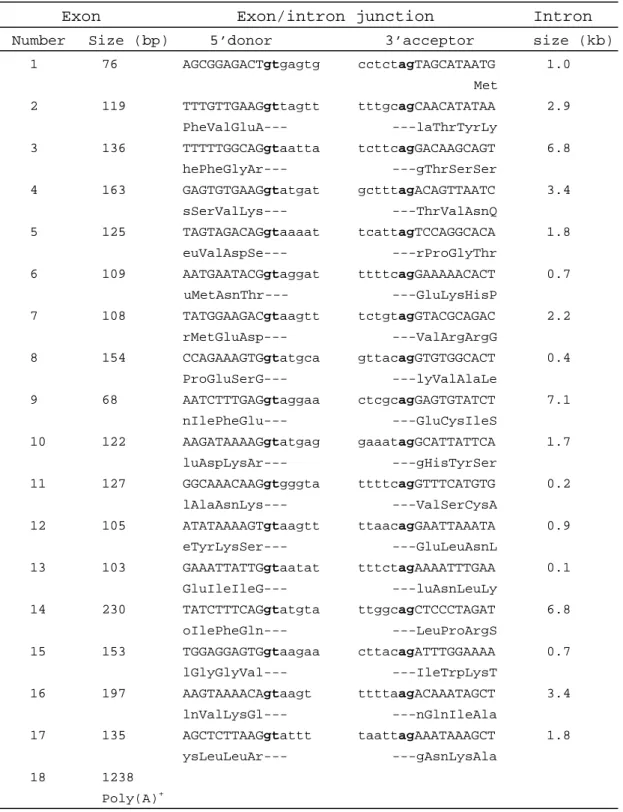
mapped to localize at the chromosome 3q25.1-25.33 (14). The genomic DNA
structure for TG370/TG741, spanning over 45 kb, was made up of 18 exons, in which
all intron excisions followed the GT-AG rules shown in bold and exon 2 encoded the
initiation codon (Table 2). In the case of cDNA for TG370, exon 11 was spliced out
and the reading frame was in-frame with the stop codon TGA in exon 12.
Alter native Splicing in Lung Cancer. As the identified 347-bp DNA
fragment was mapped to the 3’ untranslated region of cDNA FLJ20693 for TG370 (9),
two specific primers FP0-A76A (residues 180-186 of TG370) and RFHVQ-X
(residues 366-370 of TG370) were designed to evaluate the occurrence of alternative
splicing in lung cancer. As shown in Fig. 2A, two PCR products of 714- and 587-bp
length, in which the former was the predominant, were simultaneously observed in all
the pair-wise normal lung tissue of patients, numbering 6 and 7 in Fig. 1, were not
included to evaluate. The alternative splicing was also observed and nearly occurred
in all tissues examined (Fig. 2B). Moreover, the up-regulated expression of
transmembrane GTPase was implicated in at least 5 out of 7 tumor tissues. Such
observation was also consistent with the result of differential display in Fig. 1.
Another distinct PCR product migrating between those two major ones was
significantly, however, observed in cell line H520 (Fig. 2A). Sequence analysis
showed that the striking feature of the 673-bp product was the presence of an extra
86-bp region lying between exons 9 and 10 (Fig. 2C). Based upon the genome
sequence, the additional 86-bp sequence was mapped to which was followed
immediately by exon 10. Primers RP5-A76A, in the 86-bp region, and FP0-A76A
were further used to interpret the retention of such a sequence in the splicing process.
The observation of the 486-bp product showed its universal occurrence in most tumor
tissues and cell lines (Fig. 2D).
Prior to the illustration of cDNA for TG741, another human cDNA for putative
transmembrane GTPase (GenBank accession number U95822) had been described by
Fuller and Hales (15). The partial 493-residues gene product exhibited the lack of
111 residues, which exactly constituted exons 13 and 14, between residues Ser443 and
in the formation of cDNA encoding at least three gene products with different residue
numbers. However, the retention of the 86-bp region preceding exon 10 complicated
the alternative splicing as illustrated in Fig. 2. Moreover, another type of exon
deletion was also observed. As shown in Fig. 2D, besides the predicted 486-bp
product, a distinct 332-bp product was detected in all tumor tissues and cell lines.
Sequence analysis identified the unique deletion of exon 8. Equivalent result was
also obtained using primers RP5-A76A and B-AEPVSP from which, besides the
predicted 1028-bp product, the 874-bp PCR product, lacking of exon 8, was also
derived (data not shown). Actually, there were still distinct splice variants to be
identified in the experiment. As an example, the splicing pattern of exons 1-18 was
illustrated using primers FP6-A76A, in exon 1, and RP3-A76A, in exon 18, from
which a splice variant, lacking of exons 12-14, was thus identified in tumor tissues
(data not shown).
Locating Tr ansmembr ane GTPase at Cell Sur face and Cytosol. The results
of locating transmembrane GTPase by flow cytometry are presented in Fig. 3. It
indicates that, using antibody raised against TG370, the proportion of cells expressing
antigens at the surface and intracellularly was 78% and 99% respectively, as
illustrated in Fig. 3A and 3B. About 73% of cells expressed the antigen at the
transmembrane region of TG741 (Fig. 3C). After increasing the antibody
concentration, the proportion of cells being stained increased only to about 80%. It
may be that some cells were not mature enough to produce the surface antigen. In
contrast, it was observed that low percentages of cells, comparable to the control
group, were stained for cytoplasmic antigen expression using the same antibody (Fig.
3D). On the other hand, the intracellular antigen expression was significantly
detected in 92% of cells, using antibody for the C-terminal 100 residues of TG741
(Fig. 3F). In this case, antigen was not detected at the surface (Fig. 3E). Combined
with these results, the following two facts were concluded. First, transmembrane
GTPase was destined to cell surface or cytosol. Depending on the length of
polypeptide chain, the shorter form(s) might retain at cytosol and that with
transmembrane region was translocated to plasma membrane. Second, the
full-length TG741 was anchored at cell surface in which its N-terminal domain was
exposed extracellularly.
Immunohistochemical studies. The expression of transmembrane GTPase in
NSCLC was further illustrated immunohistochemically using antibody raised for the
C-terminal 100 residues of TG741. The results showed that differential expression
of transmembrane GTPase in tumor tissues was revealed in 13 out of 18 cases of
staining results of representative case of each tumor type, accompanied with adjacent
normal lung tissue from the same patient, were shown in Fig. 4. As observed in
most cases of adenocarcinoma and SCC examined in the experiment, the expression
of transmembrane GTPase was detected to a significant lower extent in type 2
Discussion
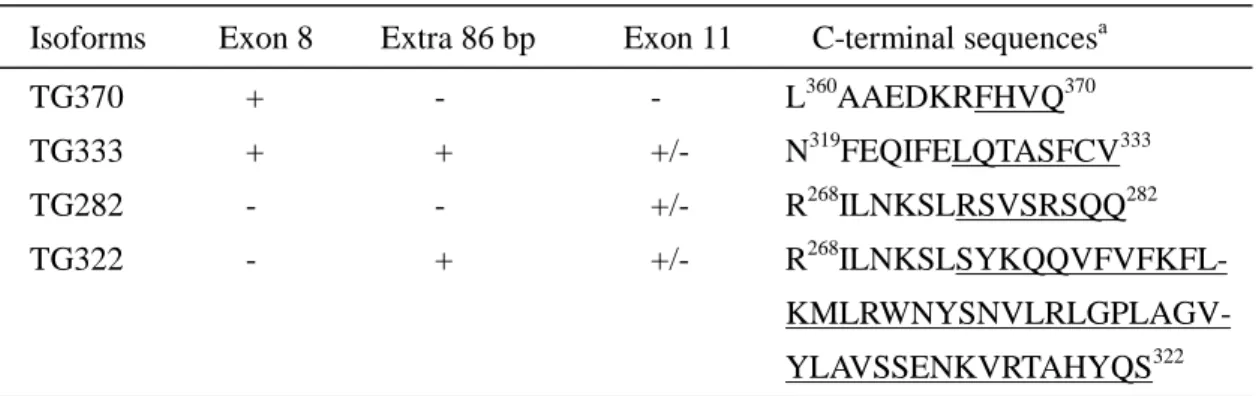
In our experiment, the alternative splicing obviously resulted from the
retention/deletion of exon 8, the 86-bp region and exon 11. The putative shorter
isoforms and deduced C-terminal unique sequences are listed at Table 3. If exon 8
was spliced out, a stop codon TGA or TAA would be encountered in exon 10 in the
absence or presence of the 86-bp region, i.e., the predicted gene products of 282 and
322 residues were not relevant to the exon 11. In the case of retention of exon 8 and
the 86-bp region, an in-frame stop codon TAA in the 86-bp region caused the
predicted protein to have the C-terminal unique sequence LQTASFCV333..
Therefore, four putative shorter isoforms of transmembrane GTPase, including
TG370, theoretically existed. Also, the multiplicity of shorter isoforms may be
appropriate to account for the detection of intracellular antigen using antibodies for
TG370 (Fig. 3B). In the future, synthetic peptide, its design based upon the
C-terminal unique sequence, will be used to prepare the antibody to identify the
existence of the individual shorter isoform by flow cytometry.
Protein sequence analysis of TG741 revealed the presence of a transmembrane
region of I600IVGGVIWKTIGWKLLSVSLTMY622 but no putative signal peptide or
mitochondrial prosequence. From the results of flow cytometry in Fig. 3, TG741
domain exposed extracellularly. However, Fuller et al. demonstrated that human
mitofusin 1, equivalent with TG741, was destined to mitochondrial membrane and
implicated in mitochondrial fusion (10) and the homologues of fruit fly (Fzo) (15) and
yeast (Fzo1p) (16) played the same roles. There is yet no reasonable explanation for
the controversy of locating TG741 destination. Actually, increasing members of
putative transmembrane GTPase family displayed similar structures of polypeptide
chain length, P-loop near the N-terminus and transmembrane region near the
C-terminus (13, 15-19). There were no reports to reveal the presence of the shorter
isoforms in other species. Distinct kinds of transmembrane GTPase, which showed
no sequence similarity to TG741, were also previously described, including
interleukin-1 receptor (20) and beta subunit of SRP (signal recognition particle)
receptor in ER membrane (21). This provided an insight that TG741, whose precise
functions were still under-established, served as a membrane-anchored receptor to
participate in the signal transduction pathway.
On the other hand, the shorter isoforms, all having no transmembrane regions,
were experimentally concluded to be soluble in cytoplasm. It may be suitable to
refer to the shorter isoforms of transmembrane GTPase as TG-related GTPase, which
still retained the P-loop of GR83TSSGK88S, like that of TG741, and showed no
superfamily. These shorter isoforms might be postulated to be functional based upon
the fact that the GTP-binding domain of TG-related GTPase was conserved in several
nucleotide-binding proteins. These candidates were widely distributed in human
(MMR_HSR1 and NPG1_HUMAN) (22, 23), yeast Schizosaccharomyces pombe
(YAWG_SCHPO and T39037) (24, 25), Mycoplasma pneumoniae (Y442_MYCPN)
(26), Aquifex aeolicus (ENGA_AQUAE) (27), Synechocystis PCC6803 (ENGA_SYNY3) (28), Buchnera aphidicola (ENGA_BUCAP) (29) and Chlamydia trachomatis (ENGA_CHLTR) (30). The aligned conserved domains are shown
below: TG RHMKVAFFGRTSSGKSSVINAMLWDKVLP-SGIGHITNCFLSVEGT-DGDKAYLM-TEGSDEKKSVK137 MMR_HSR1 KSIRVGVIGYPNVGKSSLINALVKKKRAIVSNRPGTTRDIQEVKLV-KDKKIYLIDTPGIRFPSSVD174 YAWG_SCHPO TKMTFGLVGYPNVGKSSTINALVGSKKVSVSSTPGKTKHFQTINL---SEKVSLLDCPGLVFPSFAT363 NGP1_HUMAN KQISVGFIGYPNVGKSSVINTLRSKKVCNVAPIAGETKVWQYITL---MRRIFLIDCPGVVYPS-ED371 T39037 KQISVGLIGFPNAGKSSIINTLRKKKVCNVAPIPGETKVWQYVAL---MKRIFLIDCPGIVPPSSND372 Y442_MYCPN HQFRLAVIGMPNVGKSSLINLLLNKNHLQVANRAGVTKSMSWNQI---SSEFYLSDTPGVFFKRIDE179 ENGA_AQUAE IKVAFIGRPNVGKSSLVNAILKDERVIVSPIAGTTRDAIEIPFRWKDKNFILIDTAGVRRPSNVE239 ENGA_SYNY3 IKVAIVGRPNVGKSSLLNALTGEQRAIVSPISGTTRDAIDMVVERNGQKYRLIDTAGIRRKKNVD241 ENGA_BUCAP NSVKIACIGKPNVGKSTLINSLLMKKRMITSNKAGTTLDTVLVPIKYNYKNYIFIDTAGMSKKKSKT252 ENGA_CHLTR RPLKVALIGHPNVGKSSIINALLKEERCITDNSPGTTRDNIDVAYTHNNKEYVFIDTAGLRKTKSIK291
Additionally, the similar sequences of the P-loop have also been found in cystic
fibrosis transmembrane conductance regulator (GRTGSGKS1221) (31), sulfonylurea
receptor 2B (GRTGSGKS1349) (32), ATP-binding cassette transporter abc1
(GRTGSGKS2134) (35). Several reports have revealed the identification and
characterization of various GTPase-interacting or -activating proteins (36-39). In
perspective, we need to investigate the target proteins interacting with or activating
TG741/TG-related GTPase for further illustrations of their physiological functions.
The up-regulated expression of transmembrane GTPase might be involved with
the tumorigenesis process. As shown in Fig. 4, transmembrane GTPase was
differentially expressed in tumor tissues, especially in adenocarcinoma. Most cases
of adenocarcinoma were clinically destined to peripheral lung. It had been
established that this tumor type was derived from type 2 pneumocytes of alveolar sac
and Clara cells of the bronchiole. Therefore, transmembrane GTPase may have the
References
1. Health and Vital Statistics. Department of Health, the Executive Yuan, Taiwan
ROC 1972-1989.
2. Moore, D. F. and Lee, J. S. Staging and prognostic factor: non-small cell lung
cancer. In: H. I. Pass, J. B. Mitchell, D. H. Johnson, and A. T. Turrisi (eds.),
Lung Cancer: Principle and Practice, pp. 481-494. Philadelphia: Lippincott-Raven
Publishers, 1996.
3. Liang, P. and Pardee, A. B. Differential display of eukaryotic messenger RNA by
means of the polymerase chain reaction. Science, 257: 967-970, 1992.
4. Tran, Y. K., Bogler, O., Gorse. K. M., Wieland, I., Green, M. R., and Newsham, I.
F. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing
properties in lung cancer. Cancer Res., 59: 35-43, 1999.
5. Manda, R., Kohno, T., Niki, T., Yamada, T., Takenoshita, S., Kuwano, H., and
Yokota, J. Differential expression of the LAMB3 and LAMC2 genes between
small cell and non-small cell lung carcinomas. Biochem. Biophys. Res.
Commun., 275: 440-445, 2000.
6. Martin-Satue, M. and Blanco, J. Identification of semaphorin E gene expression
in metastatic human lung adenocarcinoma cells by mRNA differential display. J.
7. Yu, L., Hui-chen, F., Chen, Y., Zou, R., Yan, S., Chun-xiang, L., Wu-ru, W., and Li,
P. Differential expression of RAB5A in human lung adenocarcinoma cells with
different metastasis potential. Clin. Exp. Metastas., 17: 213-219, 1999.
8. Hsu, N. Y., Ho, H. C., Chow, K. C., Lin, T. Y., Shih, C. S., Wang, L. S., and Tsai. C.
M. Overexpression of dihydrodiol dehydrogenase as a prognostic factor of
non-small cell lung cancer. Cancer Res., 61: 2727-2731, 2001.
9. Sugano, S., Suzuki, Y., Ota, T., Obayashi, M., Nishi, T., Isogai, T., Shibahara, T.,
Tanaka, T., and Nakamura, Y. EMBL/GenBank/DDBJ data banks accession
number AK000700, 2000.
10. Santel, A and Fuller, M. Control of mitochondrial morphology by a human
mitofusin. J Cell Sci., 114: 867-874, 2001.
11. Jacobberger, J. W., Fogelman, D., and Lehman, J. M. Analysis of intracellular
antigens by flow cytometry. Cytometry, 7: 356-364, 1986.
12. Francis, C. and Connelly, M. C. Rapid single step method for flow cytometric
detection of surface and intracellular antigens using whole blood. Cytometry, 25:
58-70, 1996.
13. The RIKEN Genome Exploration Research Group Phase II Team and FANTOM
Consortium. Functional annotation of a full-length mouse cDNA collection.
14. Worley, K. C. EMBL/GenBank/DDBJ data banks accession number
AC007823, 2000.
15. Hales, K. G. and Fuller, M. T. Developmentally regulated mitochondrial fusion
mediated by a conserved, novel, predicted GTPase. Cell, 90: 121-129, 1997.
16. Hermann, G. J., Thatcher, J. W., Mills, J. P., Hales, K. G., Fuller, M. T., Nunnari,
J., and Shaw, J. M. Mitochondrial fusion in yeast requires the transmembrane
GTPase Fzo1p. J. Cell Biol., 143: 359-373, 1998.
17. Nagase, T., Seki, N., Ishikawa, K., Ohira, M., Kawarabayasi, Y., Ohara, O.,
Tanaka, A., Kotani, H., Miyajima, N., and Nomura, N. Prediction of the coding
sequences of unidentified human genes. VI. The coding sequences of 80 new genes
(KIAA0201-KIAA0280) deduced by analysis of cDNA clones from cell line KG-1
and brain. DNA Res., 3: 321-329, 1996.
18. Adams, M. D., Celniker, S. E., Gibbs, R. A., Rubin, G. M., and Venter, C. J.
EMBL/GenBank/DDBJ data banks accession number AAF46161, 2000.
19. Latreille, P. EMBL/GenBank/DDBJ data banks accession number T34496,
1995.
20. Hopp, T. P. Evidence from sequence information that the interleukin-1 receptor
is a transmembrane GTPase. Protein Sci., 4: 1851-1859, 1995.
signal recognition particle receptor is a transmembrane GTPase that anchors the
alpha subunit, a peripheral membrane GTPase, to the endoplasmic reticulum
membrane. J. Cell Biol., 128: 273-282, 1995.
22. Vernet, C., Ribouchon, M. T., Chimini, G., and Pontarotti, P. Structure and
evolution of a member of a new subfamily of GTP-binding proteins mapping to the
human MHC class I region. Mamm. Genome, 5: 100-105, 1994.
23. Racevskis, J., Dill, A., Stockert, R., and Fineberg, S. A. Cloning of a novel
nucleolar guanosine 5'-triphosphate binding protein autoantigen from a breast
tumor. Cell Growth Differ., 7: 271-280, 1996.
24. Murphy, L., Harris, D., Barrell, B. G., Rajandream, M. A., and Walsh, S. V.
EMBL/GenBank/DDBJ data banks accession number Q10190, 1996.
25. Gentles, S., Churcher, C. M., Barrell, B. G., Rajandream, M. A., and Wood, V.
EMBL/GenBank/DDBJ data banks accession number T39037, 1997.
26. Himmelreich, R., Hilbert, H., Plagens, H., Pirkl, E., Li, B. C., and Herrmann, R.
Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res., 24: 4420-4449, 1996.
27. Deckert, G., Warren, P. V., Gaasterland, T., Young, W. G., Lenox, A. L., Graham,
D. E., Overbeek, R., Snead, M. A., Keller, M., Aujay, M., Huber, R., Feldman, R.
hyperthermophilic bacterium Aquifex aeolicus. Nature, 392: 353-358, 1998.
28. Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y.,
Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., Kimura, T., Hosouchi, T.,
Matsuno, A., Muraki, A., Nakazaki, N., Naruo, K., Okumura, S., Shimpo, S.,
Takeuchi, C., Wada, T., Watanabe, A., Yamada, M., Yasuda, M., and Tabata, S.
Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis
sp. strain PCC6803. II. Sequence determination of the entire genome and
assignment of potential protein-coding regions. DNA Res., 3: 109-136, 1996.
29. Clark, M. A., Baumann, L., and Baumann, P. Sequence analysis of a 34.7-kb
DNA segment from the genome of Buchnera aphidicola (endosymbiont of aphids)
containing groEL, dnaA, the atp operon, gidA, and rho. Curr. Microbiol., 36:
158-163, 1998.
30. Stephens, R. S., Kalman, S., Lammel, C. J., Fan, J., Marathe, R., Aravind, L.,
Mitchell, W. P., Olinger, L., Tatusov, R. L., Zhao, Q., Koonin, E. V., and Davis, R.
W. Genome sequence of an obligate intracellular pathogen of humans:
Chlamydia trachomatis. Science, 282: 754-759, 1998.
31. Hart, P., Warth, J. D., Levesque, P. C., Collier, M. L., Geary, Y., Horowitz, B.,
and Hume, J. R. Cystic fibrosis gene encodes a cAMP-dependent chloride
32. Tanemoto, M., Abe, T., and Hebert, S. C. EMBL/GenBank/DDBJ data banks
accession number T46645, 1997.
33. Christensen, P. U., Davis, K., Nielsen, O., and Davey, J. Abc1: a new ABC
transporter from the fission yeast Schizosaccharomyces pombe. FEMS Microbiol.
Lett., 147: 97-102, 1997.
34. Ouellette, M., Fase-Fowler, F., and Borst, P. The amplified H circle of
methotrexate-resistant Leishmania tarentolae contains a novel P-glycoprotein gene.
EMBO J., 9: 1027-1033, 1990.
35. Perrone, C. A., Myster, S. H., Bower, R., O'Toole, E. T., and Porter, M. E.
Insights into the structural organization of the I1 inner arm dynein from a domain
analysis of the 1beta dynein heavy chain. Mol. Biol. Cell, 11: 2297-2313, 2000.
36. Brondyk, W. H., McKiernan, C. J., Fortner, K. A., Stabila, P., Holz, R. W., and
Macara, I. G. Interaction cloning of Rabin3, a novel protein that associates with
the Ras-like GTPase Rab3A. Mol. Cell. Biol., 15: 1137-1143, 1995.
37. Haataja, L., Groffen, J., and Heisterkamp, N. Identification of a novel
Rac3-interacting protein C1D. Int. J. Mol. Med., 1: 665-670, 1998.
38. Cuif, M. H., Possmayer, F., Zander, H., Bordes, N., Jollivet, F.,
Couedel-Courteille, A., Janoueix-Lerosey, I., Langsley, G., Bornens, M., and Goud,
which associates with the centrosome. EMBO J., 18: 1772-1782, 1999.
39. Hoffenberg, S., Liu, X., Nikolova, L., Hall, H. S., Dai, W., Baughn, R. E., Dickey,
B. F.., Barbieri, M. A., Aballay, A., Stahl, P. D., and Knoll, B. J. A novel
membrane-anchored Rab5 interacting protein required for homotypic endosome
Table 1. Sequences of the primers used in the study
Primer Sequence (5´→3´)a Orientationb
B-AEPVSP CACCCGGATCCGCAGAACCTGTTTCTCCA S RFHVQ-X GGTCCCTCGAGTCATTGCACATGAAACCT A B-FKQQFV CACCCGGATCCTTTAAACAGCAGTTTGTA S SNEES-X GGTCCCTCGAGTTAGGATTCTTCATTGCT A B-YSVEER CACCCGGATCCTATTCAGTGGAAGAGAGG S LASVTS-X GGTCCCTCGAGTCAAGATGTAACGGACGCCAA A FP0-A76A CCAGGCACAGATGTCACTAC S FP1-A76A CAGCTTTGCTCCCATT S FP2-A76A GGAACGCTTTCCTAGTGCA S FP4-A76A TGTTTAAATTCCTGAAAATGTTGCG S FP6-A76A GTTGCCGGGTGATAGTTGGAG S RP3-A76A ACAGCCCCACCCCTCAGGGG A RP5-A76A CATGGCAACATTTTCAGGAATTTAA A
Adapter primer GGCCACGCGTCGACTAGTACT17 A
UAP CUACUACUACUAGGCCACGCGTCGACTAGTAC A
AUAP GGCCACGCGTCGACTAGTAC A
H-AP76 AAGCTTGTTATAG S
H-T11A AAGCTTTTTTTTTTTA A
a
The underlined sequences GGATCC, CTCGAG and AAGCTT represented the sites recognized by restriction enzymes BamHI, XhoI and HindIII, respectively.
b
Table 2. Genomic organization of TG (transmembrane GTPase) gene
Exon Exon/intron junction Intron
Number Size (bp) 5’donor 3’acceptor size (kb)
1 76 AGCGGAGACTgtgagtg cctctagTAGCATAATG 1.0 Met 2 119 TTTGTTGAAGgttagtt tttgcagCAACATATAA 2.9 PheValGluA--- ---laThrTyrLy 3 136 TTTTTGGCAGgtaatta tcttcagGACAAGCAGT 6.8 hePheGlyAr--- ---gThrSerSer 4 163 GAGTGTGAAGgtatgat gctttagACAGTTAATC 3.4 sSerValLys--- ---ThrValAsnQ 5 125 TAGTAGACAGgtaaaat tcattagTCCAGGCACA 1.8 euValAspSe--- ---rProGlyThr 6 109 AATGAATACGgtaggat ttttcagGAAAAACACT 0.7 uMetAsnThr--- ---GluLysHisP 7 108 TATGGAAGACgtaagtt tctgtagGTACGCAGAC 2.2 rMetGluAsp--- ---ValArgArgG 8 154 CCAGAAAGTGgtatgca gttacagGTGTGGCACT 0.4 ProGluSerG--- ---lyValAlaLe 9 68 AATCTTTGAGgtaggaa ctcgcagGAGTGTATCT 7.1 nIlePheGlu--- ---GluCysIleS 10 122 AAGATAAAAGgtatgag gaaatagGCATTATTCA 1.7 luAspLysAr--- ---gHisTyrSer 11 127 GGCAAACAAGgtgggta ttttcagGTTTCATGTG 0.2 lAlaAsnLys--- ---ValSerCysA 12 105 ATATAAAAGTgtaagtt ttaacagGAATTAAATA 0.9 eTyrLysSer--- ---GluLeuAsnL 13 103 GAAATTATTGgtaatat tttctagAAAATTTGAA 0.1 GluIleIleG--- ---luAsnLeuLy 14 230 TATCTTTCAGgtatgta ttggcagCTCCCTAGAT 6.8 oIlePheGln--- ---LeuProArgS 15 153 TGGAGGAGTGgtaagaa cttacagATTTGGAAAA 0.7 lGlyGlyVal--- ---IleTrpLysT 16 197 AAGTAAAACAgtaagt ttttaagACAAATAGCT 3.4 lnValLysGl--- ---nGlnIleAla 17 135 AGCTCTTAAGgtattt taattagAAATAAAGCT 1.8 ysLeuLeuAr--- ---gAsnLysAla 18 1238 Poly(A)+
Table 3. Putative shorter isoforms of transmembrane GTPase
Isoforms Exon 8 Extra 86 bp Exon 11 C-terminal sequencesa
TG370 + - - L360AAEDKRFHVQ370 TG333 + + +/- N319FEQIFELQTASFCV333 TG282 - - +/- R268ILNKSLRSVSRSQQ282 TG322 - + +/- R268 ILNKSLSYKQQVFVFKFL- KMLRWNYSNVLRLGPLAGV-YLAVSSENKVRTAHYQS322 a
FIGURE LEGENDS
Fig.1. The gene expression patterns of NSCLC are profiled by differential display
using the arbitrary primer H-AP76 and reverse primer H-T11A. The
up-regulated DNA fragment of 347-bp length termed A76A in most tumor
tissues is indicated by arrowhead. The symbols ‘N’ and ‘T’ represent
non-tumor and pair-wise tumor fractions of surgical resections of 9 NSCLC
patients.
Fig. 2. Occurrence of alternative splicing and up-regulated expression of cDNA for
transmembrane GTPase in lung cancer cell lines and NSCLC patients. A.
Presence or absence of exon 11 is determined using primers FP0-A76A and
RFHVQ-X. The two predicted PCR products of 714- and 587-bp length, in A
and B, and an additional distinct PCR product of 673-bp length are shown at
right. Lung cancer cell lines used in the study are indicated at the top of each
lane. The left lane is 100-bp DNA ladder. B. To evaluate the alternative
splicing and up-regulated expression, 7 out of 9 NSCLC patients in Fig. 1,
which are indicated at top of each lane, are used in the experiment. The
symbols ‘N’ and ‘L’ following the patient number are normal and lung cancer
tissues. The left lane is 100-bp DNA ladder. C. DNA sequence of the 673-bp
boldface. The exon/intron boundaries are denoted under the DNA sequence.
D. Universal presence of the extra 86-bp region is illustrated using primers
FP0-A76A and RP5-A76A and the predicted 486-bp product is indicated. The
unexpected 332-bp product is derived from the deletion of exon 8 and described
in the Results.
Fig. 3. Flow cytometry (FCM) histograms of transmembrane GTPase expression in
lung cancer cell line A549 using antibodies for surface and cytoplasmic staining.
The X-axis and Y-axis in each histogram represent fluorescence intensity and
cell number. After being stained by a primary antibody, the cells were then
stained by a fluorescence-labeled secondary antibody and analyzed by FCM as
described in Materials and Methods. The left (A, C and E) and right (B, D and
F) panels indicate the detection of surface and cytoplasmic antigens, respectively.
The antibodies used were raised for TG370 (A and B), for the internal 226
residues of TG741 (C and D) and for the C-terminal 100 residues of TG741 (E
and F).
Fig. 4. Immunohistochemical staining of transmembrane GTPase in representative
examples of adenocarcinoma and SCC of NSCLC. A and B. Tumor tissues of
adenocarcinoma and SCC. C and D. Adjacent normal lung tissues of
expression of transmembrane GTPase is detected in tumor tissues (A and B),
whereas type 2 pneumocytes and alveolar macrophages show weak staining and
type 1 pneumocytes appear negative in the pair-wise normal lung tissues (C and
Fig. 2
Fig. 3