Developmental expression of Apnanos during oogenesis and
embryogenesis in the parthenogenetic pea aphid
Acyrthosiphon pisum
CHUN-CHE CHANG
1,2,*, TING-YU HUANG
1,3, CHARLES E. COOK
1, GEE-WAY LIN
1, CHUN-LIANG SHIH
1and RITA P.-Y. CHEN
3,41Laboratory for Genetics and Development, Department of Entomology, College of Bio-Resources and Agriculture, National Taiwan University, 2Institute of Biotechnology, College of Bio-Resources and Agriculture, National Taiwan University, 3Institute of Biochemical
Sciences, College of Life Science, National Taiwan University and 4Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan ABSTRACT Among genes that are preferentially expressed in germ cells, nanos and vasa are the
two most conserved germline markers in animals. Both genes are usually expressed in germ cells in the adult gonads, and often also during embryogenesis. Both nanos-first or vasa-first expres-sion patterns have been observed in embryos, implying that the molecular networks governing germline development vary among species. Previously we identified Apvasa, a vasa homologue expressed in germ cells throughout all developmental stages in the parthenogenetic and vivipa-rous pea aphid Acyrthosiphon pisum. In asexual A. pisum, oogenesis is followed by embryogen-esis, and both occur within the ovarioles. In order to understand the temporal and spatial distribution of nanos versus vasa during oogenesis and embryogenesis, we isolated a nanos homologue, Apnanos, and studied its expression. In adults, Apnanos is preferentially expressed in the ovaries. In early embryos, Apnanos transcripts are localized to the cytoplasm of cellularizing germ cells, and soon thereafter are restricted to the newly segregated germ cells in the posterior region of the cellularized blastoderm. These results strongly suggest that the Apnanos gene is a germline marker and is involved in germline specification in asexual A. pisum. However, during the middle stages of development, when germline migration occurs, Apnanos is not expressed in the migrating germ cells expressing Apvasa, suggesting that Apnanos is not directly associated with germline migration.
KEY WORDS: Acyrthosiphon pisum, germ cell, nanos, pea aphid
Introduction
The establishment of a germline lineage begins with the specification of germ cells and usually takes place during embryo-genesis. Two distinctive strategies for specifying germ cells, namely “preformation” and “epigenesis”, have been described. In the “preformation” mode, germline determinants are synthesized during oogenesis and then they are asymmetrically localized to a subcellular cytoplasm (germ plasm) or a perinuclear region (nuage) within the oocyte (Eddy, 1975; Strome and Lehmann, 2007). In model organisms such as Caenorhabditis elegans (nematode), Drosophila melanogaster (fruit fly), Danio rerio (zebrafish), and Xenopus laevis (frog), embryonic cells inheriting maternal
germ-BIOLOGY
www.intjdevbiol.com*Address correspondence to: Chun-che Chang. Laboratory for Genetics and Development, Department of Entomology, National Taiwan University, No. 27, Lane 113, Roosevelt Road, Sec. 4, Taipei 106, Taiwan. Fax: +886-2736-9366. e-mail: chunche@ntu.edu.tw
Electronic Supplementary Material for this article, consisting of an audio PodCast, is available online at: http://dx.doi.org/10.1387/ijdb.082570cc Accepted: 25 March 2008. Published online: 28 October 2008.
ISSN: Online 1696-3547, Print 0214-6282 © 2008 UBC Press
Printed in Spain
Abbreviations used in this paper: Apnanos, Acyrthosiphon pisum nanos; Apvasa,
Acyrthosiphon pisum vasa.
line determinants from these structures become primordial germ cells. In the “epigenesis” mode, specification of germ cells does not depend on a preformed subcellular structure containing germline determinants. Instead, germ cells are derived from pluripotent progenitors responding to extraembryonic molecules secreted from adjacent somatic cells (Saffman and Lasko, 1999; Wylie, 1999; Extavour and Akam, 2003). For example, in the mouse Mus musculus proximal epiblast cells induced by signaling
BMP8b from the extraembryonic ectoderm acquire germline competence at about embryonic day 6.25 (E6.25) (Hayashi et al.,
2007).
Although animals utilize different strategies to specify germ cells, they share some common factors to sustain germline survival and development. According to Extavour and Akam (2003), there are at least six gene homologues (boule/Dazl, germ-cell-less, nanos, pumilio, staufen, vasa) required for germline
development in D. melanogaster, a “preformation” organism, and
in M. musculus, an “epigenesis” organism. Among these
germ-line-related homologues, nanos and vasa are most conserved
because they have been identified in germ cells of the five model organisms mentioned above and many non-model species across invertebrates and vertebrates (Extavour and Akam, 2003). Ho-mologues of both nanos and vasa are specifically expressed in
germ cells within the adult gonads in most animals, yet embryonic distribution of nanos and vasa varies among species. For
ex-ample, mRNAs or proteins of nanos and vasa are components of
maternal germ plasm in C. elegans, D. melanogaster, and D. rerio
(Ikenishi, 1998; Subramaniam and Seydoux, 1999; Knaut et al.,
2000; Koprunner et al., 2001), whereas in X. laevis it is nanos
(Xcat2), rather than vasa (XVLG1), that is identified in the
mater-nal germ plasm; XVLG1 is first detectable in the germ cells of
hatching tadpoles (Forristall et al., 1995; Ikenishi and Tanaka,
2000).
In D. melanogaster, posterior localization of nanos mRNA to
the pole plasm (germ plasm) requires the RNA helicase Vasa, which promotes the translation of nanos mRNA (Gavis et al.,
1996). The CCHC zinc finger motifs in Drosophila Nanos protein
then guide abdomen formation and germline migration (Arrizabalaga and Lehmann, 1999). In other dipteran insects such
as D. virilis, Musca domestica (housefly), and Chironomus
samoensis (midge), homologues of nanos mRNA are also
local-ized to the pole plasm, and they can functionally substitute for
nanos in D. melanogaster (Curtis et al., 1995). This suggests that
posterior localization and translation of nanos depends on Vasa
and is conserved in dipterans. In other non-dipteran insects such as the orthopteran Schistocerca americana (grasshopper) and
the hymenopteran Apis mellifera (honeybee), nanos is expressed
in the posterior region of early embryos and nanos is regarded as
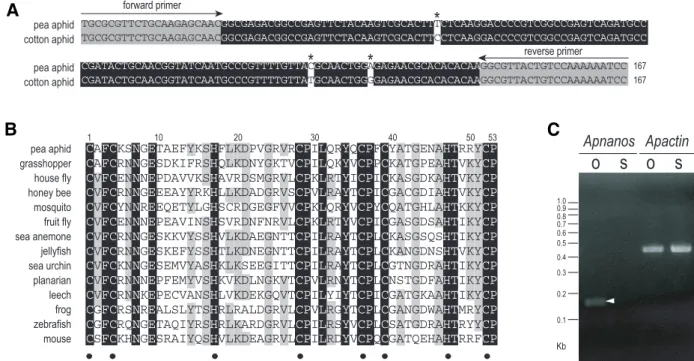
Fig. 1. Characterization of Apnanos. (A) Alignment of Apnanos and a nanos homologue from the cotton aphid Aphis gossypii (accession number:
DR389642). DNA sequences of these two nanos homologues encode zinc-finger motifs conserved in Nanos-related proteins in metazoans. Nucleic acids identical in these two species are highlighted in black. Grey area indicates primer sequences adopted from the Aphis nanos sequences. Asterisks indicate different residues in the aligned region. Both of these two nanos fragments encode identical amino acid sequences. (B) Alignment of amino acid sequences in zinc-finger motifs of ApNanos protein and thirteen other Nanos homologues across invertebrates and vertebrates. Residues identical in all taxa are highlighted in black; those identical in at least 50% of taxa are highlighted in grey. Amino acid residues constituting the conserved double CCHC zinc-finger motifs are indicated with dots beneath sequences. Sequences are labelled with common names of species. Full names and GenBank accession numbers are as follows: pea aphid ( Acyrthosiphon pisum, EU180023); grasshopper ( Schistocerca americana, AAO38523) (Lall et al., 2003); house fly ( Musca domestica, AAA87461) (Curtis et al., 1995); honey bee ( Apis mellifera, ABC41342) (Dearden, 2006); mosquito ( Aedes aegypti, EAT35750) (Calvo et al., 2005); fruit fly ( Drosophila melanogaster, AAA28715) (Wang and Lehmann, 1991); sea anemone ( Nematostella vectensis, AAW29070) (Extavour et al., 2005); jellyfish ( Podocoryne carnea, AAU11513) (Torras et al., 2004); sea urchin ( Hemicentrotus pulcherrimus, BAE53723) (Fujii et al., 2006); planarian ( Schmidtea mediterranea, ABO52809) (Wang et al., 2007); leech ( Helobdella robusta, AAB63111) (Pilon and Weisblat, 1997); frog ( Xenopus laevis, CAA51067) (Mosquera et al., 1993); zebrafish ( Danio rerio, AAL15474) (Köprunner et al., 2001); mouse ( Mus musculus, BAC82558) (Tsuda et al., 2003). (C) Detection of the expression of Apnanos mRNA in ovaries (O) and somatic (S) tissues with semi-quantitative RT-PCR. A PCR fragment (arrowhead) corresponding to the expected size of the 167-bp Apnanos amplicon was identified in ovaries dissected from adult pea aphids. In remaining tissues, which were somatic, transcripts of mRNA were almost undetectable. Expression of Apactin, an actin homologue in pea aphids, served as an internal control.
C
Apnanos Apactin
o
s
o
s
1.0 0.9 0.8 0.7 0.6 0.5 0.4 0.3 0.2 0.1 KbCAFCKSNGETAEFYKSHFLKDPVGRVRCPILQRYQCPFCYATGENAHTRRYCP CAFCRNNGESDKIFRSHQLKDNYGKTVCPILQKYVCPPCKATGPEAHTVKYCP CVFCENNNEPDAVVKSHAVRDSMGRVLCPKLRTYICPICKASGDKAHTVKYCP CVFCRNNGEEEAYYRKHLLKDADGRVSCPVLRAYTCPICGACGDIAHTVKYCP CVFCYNNREEQETYLGHSCRDGEGFVVCPKLQRYVCPYCQATGHLAHTKKYCP CVFCENNNEPEAVINSHSVRDNFNRVLCPKLRTYVCPICGASGDSAHTIKYCP CVFCRNNGESKKVYSSHVLKDAEGNTTCPILRAYTCPLCKASGSQSHTIKYCP CVFCRNNGESKEFYSSHTLKDNEGNTTCPILRAYTCPLCKANGDNSHTVKYCP CVFCKNNGESEMVYASHKLKSEEGITTCPILRAYTCPLCGTNGDRAHTIKYCP CVFCRNNNEPFEMYVSHKVKDLNGKVTCPVLRNYTCPLCNSTGDFAHTIKYCP CVFCRNNKEPECVANSHLVKDEKGQVTCPILYIYTCPICGATGKAAHTIKYCP CGFCRSNREALSLYTSHRLRALDGRVLCPVLRGYTCPLCGANGDWAHTMRYCP CGFCRQNGETAQIYRSHRLKARDGRVLCPILRSYVCPLCSATGDRAHTRYYCP CSFCKHNGESRAIYQSHVLKDEAGRVLCPILRDYVCPQCGATQEHAHTRRFCP
1 10 20 30 40 50 53 pea aphid grasshopper house fly honey bee mosquito fruit fly sea anemone jellyfish sea urchin planarian leech frog zebrafish mouse
TGCGCGTTCTGCAAGAGCAACGGCGAGACGGCCGAGTTCTACAAGTCGCACTTTCTCAAGGACCCCGTCGGCCGAGTCAGATGCC
TGCGCGTTCTGCAAGAGCAACGGCGAGACGGCCGAGTTCTACAAGTCGCACTTCCTCAAGGACCCCGTCGGCCGAGTCAGATGCC
CGATACTGCAACGGTATCAATGCCCGTTTTGTTACGCAACTGGAGAGAACGCACACACAAGGCGTTACTGTCCAAAAAATCC 167
CGATACTGCAACGGTATCAATGCCCGTTTTGTTATGCAACTGGGGAGAACGCACACACAAGGCGTTACTGTCCAAAAAATCC 167
pea aphid cotton aphid pea aphid cotton aphid forward primer reverse primer
*
*
*
B
A
a conserved posterior determinant in these insects. However, colocalization of nanos and vasa mRNA/protein occur neither to
the posterior region of mature oocytes nor in newly-laid eggs in A. mellifera, S. americana, or the closely related S. gregaria, where
a preformed germ plasm has not been identifiable (Chang et al.,
2002; Lall et al., 2003; Dearden, 2006). In our previous studies we
used cross-reacting antibodies against Nanos and Vasa to iden-tify a preformed germ plasm in the posterior region of the syncy-tium in the parthenogenetic pea aphid Acyrthosiphon pisum.
Additionally, we found that posterior localization of Nanos signals preceded that of Vasa signals during early embryogenesis in this species, which is different from the localization order of Nanos and Vasa in D. melanogaster (Chang et al., 2006). Taken
to-gether, this indicates that the hierarchical relationship between
nanos and vasa in the molecular network governing germline
development is not conserved, even within species relying on a preformed germ plasm to drive germ-cell formation. In effect, why animals need both nanos and vasa to sustain germline
develop-ment and how they interact with other germline-specific compo-nents are still not clear.
We have cloned Apvasa, a Drosophila vasa homologue in
parthenogenetic A. pisum, using it as a germline marker to
monitor the migration of germ cells during embryogenesis. Apvasa
is specifically expressed in germ cells throughout all developmen-tal stages except for a short interval before the invasion of endosymbiotic bacteria (Chang et al., 2007). In order to
under-stand how germline development was regulated by nanos and the
regulatory relationship between nanos and vasa in A. pisum, we
cloned a pea-aphid nanos homologue and made riboprobes for in situ hybridization to detect its developmental distribution. In our
previous immunostaining experiments the cross-reacting anti-body against Drosophila Nanos only marked aphid germ cells
until the blastoderm is formed, but after that germ cells become devoid of staining (Chang et al., 2006). We infer that either germ
cells do not express Nanos or that the antibody cannot penetrate into the embryos after blastulation. We expect that under stringent
in situ hybridization conditions the nanos riboprobes can have
better penetration than that of Nanos antibody. In this report we analyze the developmental expression of nanos and discuss its
potential roles in germline development.
Results
Isolation and characterization of a nanos homologue in parthenogenetic pea aphids
For animals lacking completely sequenced genomes the most common method used for isolating a nanos gene is via PCR
cloning with degenerate primers designed from the conserved zinc-finger motifs. For example, Nvnos1 and Nvnos2, two nanos
homologues in the sea anemone Nematostella vectensis, were
cloned with this strategy (Extavour et al., 2005). In the pea aphid Acyrthosiphon pisum we attempted to amplify nanos-related
genes using the same approach, but degenerate primers encod-ing amino acid sequences at both N and C termini of the zinc-finger motifs did not work for annealing temperatures of 40-60°C. As an alternative, we searched for nanos homologues in
se-quences from an Aphis gossypii EST (expressed sequence tag)
library created at USDA-ARS (United States Dept. Agriculture-Agricultural Research Service) and submitted to GenBank and
from an A. pisum EST database (http://urgi.versailles.inra.fr/).
The A. pisum EST database contained no nanos-like sequences,
but we did identify a single nanos-like sequence in A. gossypii that
includes the same zinc-finger motifs used for the degenerate
nanos primers. We used this sequence to design aphid-specific nanos primers that we successfully used to amplify and clone a
167 base-pair sequence from A. pisum. This fragment differed
from the A. gossypii sequence at only three nucleotides (Fig. 1A),
suggesting that DNA sequences encoding Nanos zinc-finger motifs in aphids are highly conserved. BLASTX searches clearly demonstrate that both Agnanos and Apnanos are nanos
homo-logues rather than genes encoding some other zinc-finger pro-teins. Additionally, comparison of the putative ApNanos with other Nanos proteins shows that ApNanos also contain amino acids constituting the highly conserved CCHC zinc-finger domain of Nanos (Fig. 1B). We then investigated gene expression using semi-quantitative RT (reverse transcription)–PCR in asexual adults
of A. pisum. This revealed that Apnanos mRNA is preferentially
expressed in ovarioles accommodating germ cells, while in so-matic tissues it was expressed at a much lower level (Fig. 1C).
Expression of Apnanos mRNA in germaria, oocytes and early embryos before gastrulation
We synthesized a DIG-labelled antisense Apnanos riboprobe
to detect the temporal and spatial distribution of Apnanos mRNA
in parthenogenetic A. pisum during oogenesis and
embryogen-esis. Detection of Apnanos expression was carried out with
whole-mount in situ hybridization in this study. In the germarial
lumen, a central space within the germarium, we identified an enrichment of Apnanos transcripts (Fig. 2A). Preferential
expres-sion of Apnanos mRNA also took place in the trophic cord (Fig.
2B), a structure known to transport nutrients from the germarium to the developing oocytes (Blackman, 1987), suggesting that
Apnanos transcripts synthesized by the nurse cells (trophocytes)
were transported to developing oocytes via this channel. Apnanos
transcripts aggregated to granular material in the cytoplasm of developing oocytes and in these oocytes they were also identified in the periphery of nuclei (stage 0-2; Fig. 2 A-D). A posterior accumulation of Apnanos mRNA particularly appeared in the
oocyte undergoing maturation division (stage 2; Fig. 2D). How-ever, when the oocyte nucleus began to cleave synchronously, a cellular status categorized as the beginning of embryogenesis according to Miura et al. (2003), the posterior gathering of Apnanos
transcripts was not visible and the distribution patterns of Apnanos
mRNA became uniform (stage 3; Fig. 2E). A similar distribution pattern was identified in older embryos where the cleaved nuclei migrated toward the inner periphery of the syncytium (stage 4; Fig. 2F).
During formation of the blastoderm, expression of Apnanos
mRNA was down regulated in the anteriormost two thirds of the egg chamber, while in the posterior some Apnanos transcripts
aggregated to granules in the cytoplasm of the presumptive germ cells (stage 5; Fig. 2G). After blastoderm formation, expression of
Apnanos mRNA was restricted to the morphologically identifiable
germ cells in the posterior region, whereas in other areas of the egg chamber Apnanos mRNA was almost undetectable (early
stage 6; Fig. 2H). Specific expression of Apnanos mRNA
continu-ously occurred within multiplying germ cells invaginated into the embryonic inner cavity (stage 6; Fig. 2I). Thereafter, expression
of Apnanos mRNA in germ cells became weaker just before the
incorporation of the maternal endosymbiotic bacteria (late stage 6; Fig. 2J). When bacteria had entered into the egg chamber, in the stage just before gastrulation, Apnanos mRNA was
unidenti-fiable in germ cells, bacteria, or other places in the embryo (stage 7; Fig. 2K).
Identification of Apnanos mRNA in migrating germ cells during gastrulation, katatrepsis and germ band retraction
Germ cells start migrating out of the posterior egg chamber after gastrulation (stage 8). They stay at the dorsal region while the germ band is folding and elongating (stage 9-14); after that, when katatrepsis is initiated (stage 15), they start migrating from
the dorsal to the anteriormost region of the egg chamber (Chang
et al., 2007). During these developmental periods, we did not
detect Apnanos expression in migrating germ cells with the
antisense Apnanos riboprobe. The single in situ hybridization
results were confirmed by the aid of double-labelled embryos with both antisense riboprobes of Apnanos as well as Apvasa, a
germline marker expressed throughout whole embryogenesis in asexual A. pisum (Chang et al., 2007). Figure 3A shows that
co-localization of Apnanos and Apvasa in situ signals occurs in the
germarium and the stage 5 embryo; however, Apvasa, rather than Apnanos, occurs in germ cells in the late blastula (late stage 6),
the gastrulating embryo (stage 9) and the embryo undergoing germ band extension (stage 14). The absence of the preferential
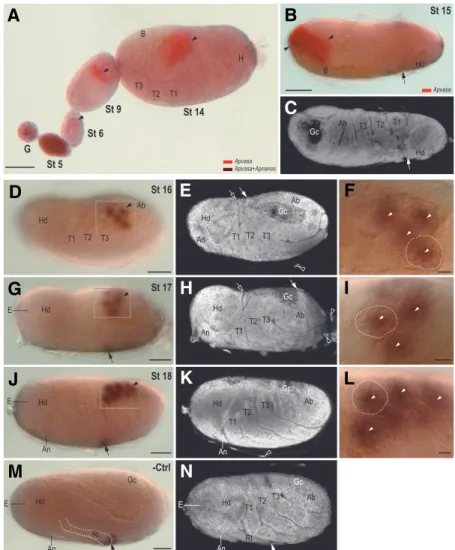
Fig. 2. Temporal and spatial distribution of Apnanos mRNA in germaria, developing oocytes and embryos by stage 7 of development. Samples were whole mount hybridized with a DIG-labelled Apnanos antisense riboprobe. Anterior regions of germaria, oocytes and egg chambers (accommodat-ing embryos) are to the left and posterior regions are to the right. (A,B) Germaria and developing oocytes (stage (St) 0). (A) Apnanos transcripts accumulate in the germarial lumen (Gl; arrowhead). Area of the germarium (G) is as indicated. Apnanos transcripts occur in the cytoplasm of nurse cells (Nc; arrow), and more weakly within the nuclei of the nurse cells (Nn). In oocytes not fully segregated from the germarium, Apnanos transcripts occur in the oocyte cytoplasm (Oc; arrowhead), and in lesser quantities in the oocyte nucleus (On). Follicle cells (Fc) are devoid of staining.
(B) Another focal plane of (A), showing Apnanos expression in the trophic cord (Tc; highlighted with a dashed line). (C) Stage (St) 1, segregated oocyte. Preferential expression of Apnanos occurs in gran-ules mostly in the anterior (arrowhead), as well as some that are posteriorly associated with the oocyte nucleus (On; arrowhead). In the peripheral region of the nucleus Apnanos transcripts also accumulate (arrow), and weak expression occurs in the oocyte cytoplasm (Oc). Follicle cells (Fc) have almost unde-tectable signals. The trophic cord (Tc; highlighted with a dashed line) is not clear in this focal plane. (D) Stage (St) 2, the oocyte nucleus (On) undergoing maturation division moves anteriorly. Apnanos tran-scripts occur primarily in the cytoplasmic granules (arrowhead), in the periphery (arrow) of the oocyte nucleus (On), and in the posterior region (double arrowhead). (E,F) Stages (St) 3 and 4, embryos under-going nuclear division. In the stage 4 embryo, dividing nuclei (Dn) migrate to the inner periphery of the embryo. Apnanos transcripts are uniformly
distrib-uted in the cytoplasm. (G) Stage (St) 5, cellularization and blastoderm formation. Localization of Apnanos transcripts occurs in the cytoplasmic granules at the posterior (arrowhead), where formation of the primordial germ cells (PGCs) occurs. In situ signals are weaker from posterior to anterior.
(H-J) Stage (St) 6, morphogenesis of germ cells. (H) An early St-6 embryo shortly after cellularization of blastoderm. Apnanos transcripts specifically occur in the cytoplasm (arrowhead) of primordial germ cells invaginating into the center of the embryo. In (I), Apnanos mRNA is expressed in primordial germ cells (arrowhead), whereas in (J) expression of Apnanos is weaker. The embryo in (J) has more nuclei in the blastoderm (arrow) than (I), and is therefore slightly older. Double arrowhead in (I) indicates posterior localization of Apnanos in a stage-2 embryo (upper left), similar to that in (D). The anterior region of this embryo is not shown in (I). (K) Stage (St) 7, invasion of the maternal endosymbiotic bacteria (B; outlined with black dashed line). Expression of Apnanos is almost unidentifiable in primordial germ cells (PGCs, outlined with white dashed line) above the bacteria. (L) Negative control (-Ctrl), ovariole containing germarium (G), stage (St)-3 and 6 embryos were hybridized with a DIG-labelled Apnanos sense riboprobe. Only weak background signals are detected. (M) A schematic illustration of the expression of Apnanos shown in (A)-(K). Cellularized blastoderm in embryos at stages 6 and 7 of development is marked with grey color. Cs, central syncytium; Ps, posterior syncytium. Scale bars, 10 µm.
Gl Nn Nc G G G G Gl Gl Gl Fc Oc On Tc Nc Fc On Oc Fc Nn Tc Nn Fc On Nn Nc St 0 St 0 St 1 St 2 St 3 St 4 St 5 Dn Fc Dn Fc St 7 B PGCs -Ctrl G St 3 St 6 Gl Nn Dn Cs Ps Cs Ps Cs Ps St 6 Early St 6 Late St 6 Nurse-cell nucleus (Nn) Nurse-cell cytoplasm (Nc) Follicle cell (Fc) Oocyte nucleus (On) Oocyte cytoplasm (Oc) Trophic cord (Tc) On Oc Germarial lumen (Gl) Germarium (G) Fc On Oc Fc Dividing nucleus (Dn) Fc Dividing nucleus (Dn) St 0 St 1 St 2 St 3 St 4
Primordial germ cell (PGC; under cellularization)
Fc
Primordial germ cell (PGC; cellularized) Fc Cellularized blastoderm Central syncytium (Cs) Posterior syncytium (Ps) Cellularized blastoderm PGC Fc Bacteria (B) St 5 St 6 St 7 Fc Apnanos
-
+
Cs PsG
B
C
D
E
F
I
J
K
L
A
M
H
expression of Apnanos mRNA in germ cells continues in embryos
undergoing katatrepsis. Figure 3B is an example showing that germ cells reaching the anteriormost region of the egg chamber only express Apvasa: co-localization of Apvasa and Apnanos
mRNAs is not visible.
We again identified specific expression of Apnanos mRNA in
germ cells of stage-16 embryos, after katatrepsis was complete and the germ band was about to retract (Fig. 3 D,E). From this stage onward, germ cells expressed Apnanos mRNA (Fig. 3 G,J).
Compared with germ cells in the stage-15 embryo (Fig. 3B), germ cells in the stage-16 embryo were separated into subclusters due to the formation of germaria (Fig. 3F). As with the distribution pattern in germaria dissected from adult A. pisum (Fig. 2 A-D),
accumulation of Apnanos transcripts occurred in the lumina of
these newly formed germaria (Fig. 3F). While the germ band was retracting from dorsal anterior to dorsal posterior (stage 17; Fig. 3 G,H), and finally merging into the abdomen (stage 18; Fig. 3 J,K), germ cells expressing Apnanos were located within the
germaria and they migrated dorsally toward the midline of the abdominal cavity (Fig. 3 I,L).
Expression of Apnanos mRNA in ovarioles dissected from mature embryos
Parthenogenetic A. pisum embryos are in fact pregnant before
they are born. We also investigated the distribution of Apnanos in
developing ovarioles within mature embryos still in the mother. These mature embryos are already fully cuticularized so the
Apnanos riboprobe did not penetrate the embryonic cavity during
our studies of earlier stage embryos in the mother. We therefore cut open the abdomen and exposed the ovarioles directly to the hybridization solution. These young ovarioles were not dissected out of the mature embryos until in situ signals were developed to
prevent them being washed off. We found that oocytes and embryos developed asynchronously between ovarioles and that embryos after stage 7 of development were not identifiable (Fig. 4). Distribution patterns of Apnanos mRNA appeared similar to
G St 5 St 6 St 9 St 14 St 15 Apvasa Apvasa+Apnanos B H T3 T2 T1 Hd Hd B Ab Gc T3 T2 T1 Apvasa Hd T3 T2 T1 Gc Ab An Hd T3 T2 T1 Ab Hd An T3 T2 T1 Ab Gc Hd E E E Hd Hd T3 T2 T1 Ab Gc An Gc Rt Hd Hd T2T3 T1 Ab E Rt An An An St 16 St 17 St 18 -Ctrl Gc
G
B
C
D
E
F
H
I
J
K
L
A
M
N
Fig. 3. Whole-mount identification of Apnanos mRNA in
embryos during mid and late embryogenesis. Unless
otherwise noted, anterior is to the left, dorsal is uppermost and all views are lateral. (A-C) Ovariole double probed with antisense Apvasa and Apnanos riboprobes. Color features of single and double in situ signals are indicated on the figure. (A) Colocalized signals of Apvasa and Apnanos appear in the germarium (G) and stage (St) 5 embryo. In embryos at St-6, 9 and 14 of development only Apvasa transcripts are detected in germ cells (arrowhead). Expression of Apnanos is not identifiable in embryos older than stage 5 of development in this preparation. (B) Stage 15, embryos undergoing katatrepsis. Arrow indicates the tip of the head migrating toward the anterior region of the egg chamber. Expression of Apvasa, rather than Apnanos, preferentially occurs in migrating germ cells (arrowhead) in the anterior region. (C) Nuclear staining of embryo in (B). (D-L) Embryos hybridized only with an antisense Apnanos riboprobe. (D-F) Stage 16, embryo after katatrepsis. Head (Hd) flips to the anterior region of the egg chamber. (D) In situ signals of Apnanos mRNA occur in germ cells (arrow-head) in the dorsolateral region. (E) Nuclear staining of (D). Posterior tip (arrow) of the abdomen (Ab) is close to the posteriormost part of the head (hollow arrow), indicating that germ band retraction has just initiated. Large cells of the serosal membrane are indicated with hollow arrows. (F) Magnification of the inset shown in (D). Arrowheads indicate the location of the presumptive germarial lumen, where the preferential expression of Apnanos takes place. Dashed line marks the boundary of a germarium visible in the presented focal section. (G-I) Stage 17, embryo undergoing germ band retraction. (G) Germ cells (arrowhead) labelled with Apnanos probes are migrating to the uppermost region of the embryo.
(H) Nuclear staining of (G). Posterior tip (arrow) of the abdo-men (Ab) is retracting to the posterior region of the embryo.
(I) Magnification of the inset shown in (G). The expression pattern of Apnanos mRNA is similar to that shown in (F).
(J-L) Stage 18, germ band retraction completed. (J) Germ cells (arrowhead) labelled with Apnanos probes are closer to the
dorsal midline, but not visible from a lateral view. (K) Nuclear staining of (J). (L) Magnification of the inset shown in (J). Expression pattern of Apnanos is similar to that described in (F) and (I). (M,N) Negative control (-Ctrl), embryos slightly older than stage 18, but not yet reaching the average size of St-19 embryos, were hybridized with a DIG-labelled Apnanos sense riboprobe. In situ signals are not preferentially identified in germ cells (Gc; with visible germarium shape located dorsally) but background signals are detectable. In most embryos after germ band retraction, non-specific in situ signals can be identified in the terminal region of the rostrum (Rt; arrows in (G), (J), (M)) with either antisense or sense Apnanos riboprobes. Other abbreviations: An, antenna; B, bacteria; E, eyes. T1-T3, the three thoracic segments. Scale bars: (F,I, L), 10 µm; others, 50 µm.
those identified in ovarioles dissected from the adult (Fig. 2 A-I). For example, we identified an aggregation of Apnanos transcripts
in the germarial lumina (Fig. 4 A-C), a uniform distribution in the syncytium (stage 4; Fig. 4C), and a down regulation of Apnanos
expression in germ cells from stage 5 (Fig. 4A) to stage 6 of development (Fig. 4 B,C).
Discussion
We cloned Apnanos, a Drosophila nanos homologue in the
parthenogenetic and viviparous pea aphid Acyrthosiphon pisum
(Fig. 1 A,B). In adults, Apnanos mRNA (Apnanos) is preferentially
expressed in the ovaries (Fig. 1C). In embryos, Apnanos
tran-scripts are localized to the cytoplasm of cellularizing germ cells (stage 5; Fig. 2G), and thereafter Apnanos is restricted to the
newly-segregated germ cells in the posterior region of the cellularized blastoderm (stage 6; Fig. 2 H,I). These results strongly suggest that Apnanos is a germline marker and is involved in
germline specification in asexual A. pisum. However, from stage
7 to 15, the developmental period during which germline migration occurs (Chang et al., 2007), Apnanos is not identifiable in the
migrating germ cells (Fig. 2K; Fig. 3 A,B), suggesting that Apnanos
is not directly associated with germline migration.
In our previous studies, we used a cross-reacting antibody against the Drosophila Nanos protein to identify presumptive
germ plasm first localized to the posterior region of the oocyte undergoing maturation division (stage 2) (Hanyu-Nakamura et al., 2004; Chang et al., 2006). The Nanos signals remain localized
in the posterior region during early embryogenesis and are finally incorporated into the morphologically identifiable germ cells at the posterior. This suggests that the specification of germ cells in asexual A. pisum depends on germ plasm expressing Nanos
protein (Nanos) (Chang et al., 2006). Accumulation of Apnanos
occurs in the posterior during stage 2 of development (Fig. 2D), which corresponds with the Nanos expression mentioned above. Nevertheless, unlike the distribution pattern of the Nanos protein, we did not detect the posterior localization of the Apnanos signal
continuously in embryos from stage 3 to stage 4 of development, which is a period of nuclear division (Fig. 2 E,F). Accordingly, we infer that the posterior localization of Nanos protein is via the translational control of Apnanos, or depends on a “posterior
molecular anchor”, localized prior to Nanos, that can localize
Nanos circulating to the posterior region. However, we cannot exclude the possibility that the Nanos signals detected by the cross-reacting antibody is not the protein transcribed from Apnanos.
If this is the case, then there may be more than one nanos
homologue in A. pisum.
In asexual A. pisum, migrating germ cells expressing Apvasa
remain an integrated group from gastrulation to katatrepsis (Fig. 3 A,B), and they first appear as subclusters within germaria after the completion of katatrepsis (Fig. 3D) (Chang et al., 2007).
Accordingly, this suggests that the coalescence between migrat-ing germ cells and the somatic gonadal tissue takes place while germ cells are migrating from the anteriormost region of the egg chamber into the body cavity (Fig. 3 B,D), and that the formation of gonads, which will differentiate into the germarial primordia, is accomplished after the embryo flips (Fig. 3D) (Chang et al., 2007).
Given that the re-expression of Apnanos occurs to germ cells
within the newly formed germaria, we hypothesize that the syn-thesis of Apnanos mRNA may be induced by germarial somatic
cells surrounding the germ cells. According to Blackman (1978) and Büning (1985), undifferentiated germ cells within the germarium give rise to nurse cells and, under environmental conditions appropriate to asexual oogenesis, nurse cells at the posterior germarium further differentiate into oocytes. We thus surmise that Apnanos within the germ cells freshly incorporated
into the germarium is involved in the production of oocytes (Fig. 3 D,G,J) and that Apnanos in the oocytes and embryos by stage
4 of development is associated with the maintenance of oogen-esis and early embryogenoogen-esis (Fig. 2 C-F (ovarioles in adults); Fig. 4 A-C (ovarioles in embryos)). However, at present we do not have direct evidence via functional assay of Apnanos to support
the above inference. In Drosophila melanogaster (fly) and Danio rerio (zebrafish), it has been demonstrated that nanos is required
for production of oocytes (Forbes and Lehmann, 1998; Draper et al., 2007), suggesting that this feature may be conserved in A. pisum as well.
Besides the conserved character of being a germline marker,
nanos genes in insects have been regarded as determinants that
regulate posterior development (Curtis et al., 1995; Lall et al.,
2003). Functional assays show that abdominal development is largely prevented in D. melanogaster mutants with nanos loss of
function alleles (Gavis and Lehmann, 1992). In the grasshopper
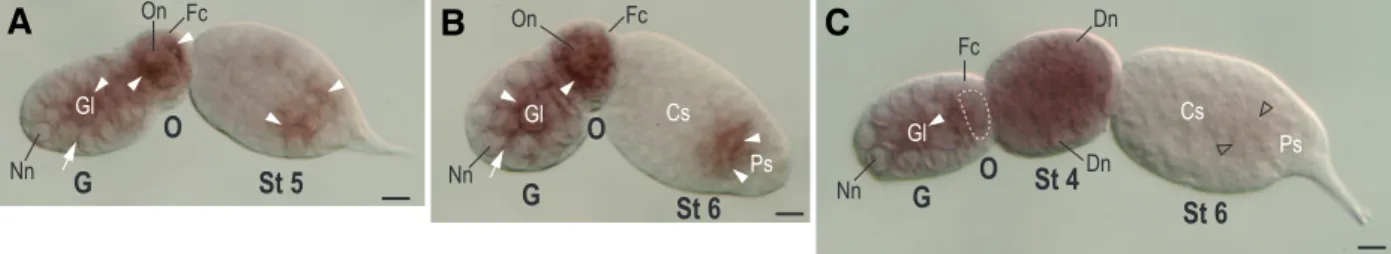
Schistocerca americana tools for genetic manipulation and knock-Fig. 4. Expression of Apnanos mRNA in ovarioles of mature embryos. Ovarioles were dissected from fully cuticularized embryos at stage 20 of
development, a stage just before larviposition. These ovarian tubules contained germaria, developing oocytes and embryos up to stage 6 of development. Anterior is to the left but note that the above structures in an ovariole cannot be aligned horizontally and straight in all preparations. For the exact orientation of each respective structure please refer to Fig. 2. All abbreviations and labels are also identical to those in Fig. 2. In all panels, distribution of Apnanos mRNA in the germaria (G) and developing oocytes (O) is similar to Fig. 2 A-C. (A,B) Expression of Apnanos occurs in the cytoplasm of germ cells (arrowhead) in stage (St)-5 and St-6 embryos respectively. (C) In an older St-6 embryo, expression of Apnanos is almost undetectable in germ cells (hollow arrowhead). The oocyte is not fully segregated from the germarium, and is not clear in this focal plane: its boundary is highlighted with a dashed line. Distribution of Apnanos in St-4 embryo is similar to that shown in Fig. 2F. Scale bars, 10 µm.
Gl Gl Gl
G
St 5
O
On Nn FcG
St 6
O
On Fc Nn NnG
O
St 4
St 6
Dn Dn Fc Cs Ps Cs PsB
C
A
down experiments have not yet been developed; nevertheless,
nanos has been implicated as a posterior regulator by an empty
patch of hunchback expression in the nanos-positive area (Lall et al., 2003). In addition, it has been proposed that nanos genes
expressed in the posterior region of oocytes and early embryos in honeybees (Dearden, 2006), mosquitoes (Calvo et al., 2005), and
wasps (Olesnicky and Desplan, 2007) are associated with poste-rior development. In asexual A. pisum, asymmetric localization of Apnanos mRNA occurs in the posterior region of the oocyte
undergoing maturation division (stage 2; Fig. 2D), suggesting that
Apnanos is also conserved in regulating posterior development.
From stage 3 to stage 4 of development Apnanos is not localized
to the posterior and there is no posterior to anterior gradient of
Apnanos (Fig. 2 E,F). During the same developmental period
localized Nanos signals are detectable in the posterior region (Chang et al., 2006), suggesting that it is the Nanos protein, rather
than the nanos mRNA, that maintains the posterior development.
In D. melanogaster (Wang and Lehmann, 1991; Curtis et al.,
1995), S. americana (Lall et al., 2003), Apis mellifera (Dearden,
2006), and A. pisum, where nanos expression has been studied
throughout developmental stages, we find a common feature that
nanos transcripts are again detectable within the germ cells that
have just been incorporated into the presumptive gonads. How-ever, the point at which nanos become undetectable varies in
these four insect species. In S. americana and Ac. pisum, nanos
becomes undetectable after germ cells are specified (Lall et al.,
2003) (Fig. 2K; Fig. 3 A,B). In Ap. mellifera, this occurs after the
formation of a posterior gradient of nanos during early
embryo-genesis, although whether germ cells are specified in early embryos is not clear (Dearden, 2006). In D. melanogaster, the
breakdown of maternally-inherited nanos occurs in migrating
germ cells in the midgut prior to germ band retraction (D. melano-gaster developmental stage 10) (Wang and Lehmann, 1991;
Curtis et al., 1995). Thus, of these four insects D. melanogaster
is the only species in which nanos is detected in migrating germ
cells (D. melanogaster developmental stage 6-10): pole cells
lacking nanos activity fail to migrate to the gonads (Kobayashi et al., 1996). Whether nanos regulation of germline migration only
takes place in Drosophila or is common to other insects requires
further investigation. In our target insect, the asexual Ac. pisum,
we propose experiments for knocking down Apnanos with tools
such as RNA interference or antisense morpholino oligonucle-otides in order to investigate how Apnanos affects germline
specification and migration, and how Apvasa is expressed under
the knock-down of Apnanos and vice versa.
Materials and Methods
Pea aphid culture
We reared parthenogenetic pea aphids, Acyrthosiphon pisum, on
garden pea plants Pisum sativum at 15°C in a growth chamber with a long-day period (16 hours light/8 hours dark). Staging of aphid development follows the scheme established by Miura et al. (2003).
Cloning and reverse transcription (RT)-PCR of Apnanos
A partial nanos fragment of A. pisum was amplified from
complemen-tary DNA (cDNA) reverse transcribed from total RNA of A. pisum. cDNA
was synthesized using StrataScript reverse transcriptase (Stratagene), and RNA was purified with RNeasy Mini Kit (Qiagen). Experiments were carried out according to the manufacturer’s instructions. Primers were
designed using the Aphis gossypii nanos sequence (Fig. 1) as follows: (1)
forward: 5'- TGCGCGTTCTGCAAGAGCAAC-3' (CAFCKSN); (2) reverse: 5'- GGATTTTTTGGACAGTAACGCC-3' (RYCPKN). Amplification pa-rameters were: 95°C for 5 minutes, followed by 40 cycles at 94°C for 30 seconds, 50°C for 30 seconds, 70°C for 30 seconds and, finally, 70°C for 5 minutes. PCR products were cloned into the pGEM-T Easy Vector (Promega) for sequencing and in vitro transcription. Sequences were
aligned using MacVector 8.0 (Accelrys). PCR cloning of an actin
homo-logue in A. pisum (Apactin) was carried out under the same conditions as
above, except that the annealing temperature was 45°C. Degenerate primers used for cloning Apactin were: (1) forward:
5’-GCATCATCACCAACTGGGAYGAYATGGA-3' (FITNWDDME); (2) re-verse: 5'-CTTCCGGATGTCCACGTCRCAYTTCAT-3' (MKCDVDIRK). GenBank accession number of Apactin: EU346758.
Semi-quantitative RT-PCR was performed using cDNA from ovaries and somatic tissues dissected from adult A. pisum. The concentration of
cDNA was normalized to 250 ng for each PCR amplification. We tested various numbers of amplification cycles for Apnanos and found that: (1)
After 40 cycles, PCR amplification was saturated–it was no longer possible to discriminate product intensity for the ovary or somatic groups on the agarose gel; (2) Under 22 cycles, PCR products became almost undetectable. The best detection of differential expression of Apnanos in
both ovary and somatic tissues was obtained from 25 cycles of amplifica-tion. PCR conditions were the same as those used for cloning Apnanos
except that the annealing temperature was 58°C. Primers used for semi-quantification of Apnanos mRNA were identical to those used in cloning Apnanos; primers used for semi-quantification of Apactin were: (1)
forward: 5'-AATCCTGTTGACCGAAGCCC-3' (ILLTEA); (2) reverse: 5’-TTCCGATGGTGATGACCTG TCC-3’ (GQVITIG).
Whole-mount in situ hybridization and microscopy
Apnanos riboprobes for in situ hybridization were synthesized from
linearized plasmids containing the 167 base pairs encoding the zinc-finger domain. For single in situ hybridization to detect the expression of Apnanos mRNA, Digoxigenin (DIG)-labelled UTP was incorporated into
the probe with a DIG RNA Labeling Kit (SP6/T7) (Roche). Apvasa
riboprobes for double in situ hybridization experiments were synthesized
with Fluorescein Labeling Mix (Roche). Ovarioles were dissected from apteriform adults or mature embryos in 1x phosphate buffered saline (PBS), then fixed in 3.8% formaldehyde in 1x PBS at 4°C overnight. Other steps for single in situ hybridization were based upon the protocol
described in Chang et al. (2007). For double in situs we first developed Apnanos signals with NBT/BCIP (20 µl stock solution (Roche) in 1 ml 1x detection buffer containing 1 mM levamisole). Before applying anti-Fluorescein antibody (Roche; Alkaline phosphatase (AP) conjugated), activity of the AP conjugated to the anti-DIG antibody was blocked with 0.1 M Glycine-HCl (pH 2.2) in 0.1% Tween 20 for 10 minutes. Apvasa signals
were developed with a Fast Red tablet (Roche). Ovarioles, whether dissected from adults or mature embryos, were cleared in 70% glycerol in 1x PBS at room temperature for 6 hours or at 4°C overnight. Embryos older than stage 11 of development were mounted under a bridged coverslip. Samples were photographed with a Leica DMR connected to a Fuji FinePix S2 Pro digital camera. Nuclear staining was carried out with DAPI (2 ng/µl; Sigma) and pictures were taken with a Zeiss LSM510 META laser-scanning microscope.
Acknowledgements
We are grateful to Sue-Ping Lee for technical support on confocal microscopy; Te-pin Chang, Hsiao-Ling Lu, Hui Chiu and Jou-Han Chen for careful manuscript proofreading; Wen-Che Wu and How-Jing Lee for reagent support. C.C. would like to thank Akira Nakamura (Center for Developmental Biology, RIKEN) and Sophia Wang (NYU Medical Center) for providing information about the expression of Drosophila nanos. This work was supported by the National Science Council of Taiwan (95-2313-B-002-097-MY2), BAPHIQ of the Agricultural Council and the Program for
Academic Comprehensive Promotion of the College of Bio-Resources and Agriculture at the National Taiwan University.
References
ARRIZABALAGA, G. and LEHMANN, R. (1999). A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics 153: 1825-1838.
BLACKMAN, R.L. (1978). Early development of parthenogenetic egg in three species of aphids (Homoptera Aphididae). Int. J. Insect Morphol. 7: 33-44.
BLACKMAN, R.L. (1987). Chapter 3. Reproduction, Cytogenetics and Develop-ment. In Aphids: their biology, natural enemies and control, vol. A (ed. MINKS,
A. K. and HARREWIJN, P.). Elsevier, pp.163-195.
BÜNING, J. (1985). Morphology, ultrastructure, and germ-cell cluster formation in ovarioles of aphids. J. Morphol. 186: 209-221.
CALVO, E., WALTER, M., ADELMAN, Z.N., JIMENEZ, A., ONAL, S., MARINOTTI, O. and JAMES, A.A. (2005). Nanos (nos) genes of the vector mosquitoes, Anopheles gambiae, Anopheles stephensi and Aedes aegypti. Insect Biochem. Mol. Biol. 35: 789-798.
CHANG, C.C., DEARDEN, P. and AKAM, M. (2002). Germ line development in the grasshopper Schistocerca gregaria: vasa as a marker. Dev. Biol. 252: 100-118.
CHANG, C.C., LEE, W.C., COOK, C.E., LIN, G.W. and CHANG, T. (2006). Germ-plasm specification and germline development in the parthenogenetic pea aphid Acyrthosiphon pisum: Vasa and Nanos as markers. Int. J. Dev. Biol. 50:
413-421.
CHANG, C.C., LIN, G.W., COOK, C.E., HORNG, S.B., LEE, H.J. and HUANG, T.Y. (2007). Apvasa marks germ-cell migration in the parthenogenetic pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Dev. Genes Evol. 217: 275-87.
CURTIS, D., APFELD, J. and LEHMANN, R. (1995). nanos is an evolutionarily conserved organizer of anterior-posterior polarity. Development 121:
1899-1910.
DEARDEN, P.K. (2006). Germ cell development in the Honeybee (Apis mellifera); vasa and nanos expression. BMC Dev. Biol. 6: 6.
DRAPER, B.W., MCCALLUM, C.M. and MOENS, C.B. (2007). nanos1 is required to maintain oocyte production in adult zebrafish. Dev. Biol. 305: 589-598.
EDDY, E.M. (1975). Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 43: 229-280.
EXTAVOUR, C.G. and AKAM, M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130:
5869-5884.
EXTAVOUR, C.G., PANG, K., MATUS, D.Q. and MARTINDALE, M.Q. (2005). vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol. Dev. 7: 201-215.
FORBES, A. and LEHMANN, R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125:
679-690.
FORRISTALL, C., PONDEL, M., CHEN, L. and KING, M.L. (1995). Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development 121: 201-208.
FUJII, T., MITSUNAGA-NAKATSUBO, K., SAITO, I., IIDA, H., SAKAMOTO, N., AKASAKA, K. and YAMAMOTO, T. (2006). Developmental expression of HpNanos, the Hemicentrotus pulcherrimus homologue of nanos. Gene Expr. Patterns 6: 572-577.
GAVIS, E.R. and LEHMANN, R. (1992). Localization of nanos RNA controls embryonic polarity. Cell 71: 301-313.
GAVIS, E.R., LUNSFORD, L., BERGSTEN, S.E. and LEHMANN, R. (1996). A
conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122: 2791-2800.
HANYU-NAKAMURA, K., KOBAYASHI, S. and NAKAMURA, A. (2004). Germ cell-autonomous Wunen2 is required for germline development in Drosophila embryos. Development 131: 4545-4553.
HAYASHI, K., DE SOUSA LOPES, S.M. and SURANI, M.A. (2007). Germ cell specification in mice. Science 316: 394-396.
IKENISHI, K. (1998). Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Dev. Growth Differ. 40: 1-10.
IKENISHI, K. and TANAKA, T.S. (2000). Spatio-temporal expression of Xenopus vasa homolog, XVLG1, in oocytes and embryos: the presence of XVLG1 RNA in somatic cells as well as germline cells. Dev. Growth Differ. 42: 95-103.
KNAUT, H., PELEGRI, F., BOHMANN, K., SCHWARZ, H. and NUSSLEIN-VOLHARD, C. (2000). Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification.
J. Cell Biol. 149: 875-88.
KOBAYASHI, S., YAMADA, M., ASAOKA, M. and KITAMURA, T. (1996). Essential role of the posterior morphogen nanos for germline development in Drosophila.
Nature 380: 708-711.
KÖPRUNNER, M., THISSE, C., THISSE, B. and RAZ, E. (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15: 2877-2885.
LALL, S., LUDWIG, M.Z. and PATEL, N.H. (2003). Nanos plays a conserved role in axial patterning outside of the Diptera. Curr. Biol. 13: 224-229.
MIURA, T., BRAENDLE, C., SHINGLETON, A., SISK, G., KAMBHAMPATI, S. AND STERN, D. L. (2003). A comparison of parthenogenetic and sexual embryogen-esis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). J. Exp. Zool. 295B: 59-81.
MOSQUERA, L., FORRISTALL, C., ZHOU, Y. and KING, M.L. (1993). A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 117: 377-386.
OLESNICKY, E.C. and DESPLAN, C. (2007). Distinct mechanisms for mRNA localization during embryonic axis specification in the wasp Nasonia. Dev. Biol.
306: 134-142.
PILON, M. and WEISBLAT, D.A. (1997). A nanos homolog in leech. Development
124: 1771-1780.
SAFFMAN, E.E. and LASKO, P. (1999). Germline development in vertebrates and invertebrates. Cell. Mol. Life Sci. 55: 1141-1163.
STROME, S. and LEHMANN, R. (2007). Germ versus soma decisions: lessons from flies and worms. Science 316: 392-393.
SUBRAMANIAM, K. and SEYDOUX, G. (1999). nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861-4871.
TORRAS, R., YANZE, N., SCHMID, V. and GONZALEZ-CRESPO, S. (2004). nanos expression at the embryonic posterior pole and the medusa phase in the hydrozoan Podocoryne carnea. Evol. Dev. 6: 362-371.
TSUDA, M., SASAOKA, Y., KISO, M., ABE, K., HARAGUCHI, S., KOBAYASHI, S. and SAGA, Y. (2003). Conserved role of Nanos proteins in germ cell develop-ment. Science 301: 1239-1241.
WANG, C. and LEHMANN, R. (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66: 637-647.
WANG, Y., ZAYAS, R.M., GUO, T. and NEWMARK, P.A. (2007). nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. U.S.A. 104: 5901-5906.
Further Related Reading, published previously in the Int. J. Dev. Biol.
See our recent Special Issue Fertilization, in honor of David L. Garbers and edited by Paul M. Wassarman and Victor D. Vacquier at: http://www.ijdb.ehu.es/web/contents.php?vol=52&issue=5-6
See our recent Special Issue Plant Development edited by José Luis Micol and Miguel-Angel Blázquez at: http://www.ijdb.ehu.es/web/contents.php?vol=49&issue=5-6
Germ cell cluster formation and ovariole structure in viviparous and oviparous generations of the aphid Stomaphis quercus
Grazyna Pyka-Fosciak and Teresa Szklarzewicz Int. J. Dev. Biol. (2008) 52: 259-265
The distribution and behavior of extragonadal primordial germ cells in Bax mutant mice suggest a novel origin for sacrococcygeal germ cell tumors
Christopher Runyan, Ying Gu, Amanda Shoemaker, Leendert Looijenga and Christopher Wylie Int. J. Dev. Biol. (2008) 52: 2486-2486
Germ cell cluster formation and ovariole structure in viviparous and oviparous generations of the aphid Stomaphis quercus
Grazyna Pyka-Fosciak and Teresa Szklarzewicz Int. J. Dev. Biol. (2008) 52: 259-265
Interleukin-2 induces the proliferation of mouse primordial germ cells in vitro
Cristina Eguizabal, Maria D. Boyano, Alejandro Díez-Torre, Ricardo Andrade, Noelia Andollo, Massimo De Felici and Juan Aréchaga Int. J. Dev. Biol. (2007) 51: 731-738
Germ-plasm specification and germline development in the parthenogenetic pea aphid Acyrthosiphon pisum: Vasa and Nanos as markers
Chun-che Chang, Wen-chih Lee, Charles E. Cook, Gee-way Lin and Tschining Chang Int. J. Dev. Biol. (2006) 50: 413-421
2006 ISI **Impact Factor = 3.577**
Visualization of primordial germ cells in vivo using GFP-nos1 3’UTR mRNA
Taiju Saito, Takafumi Fujimoto, Shingo Maegawa, Kunio Inoue, Minoru Tanaka, Katsutoshi Arai and Etsuro Yamaha
Int. J. Dev. Biol. (2006) 50: 691-700
Posterior expression of nanos orthologs during embryonic and larval development of the anthozoan Nematostella vectensis
Raquel Torras and Sergio González-Crespo Int. J. Dev. Biol. (2005) 49: 895-899
The formation of primordial germ cells from germline cells in spherical embryos derived from the blastodisc of 2-cell embryos in goldfish, Carassius auratus
Satoshi Otani, Tomoe Kitauchi, Taiju Saito, Suzu Sakao, Shingo Maegawa, Kunio Inoue, Katsutoshi Arai and Etsuro Yamaha
Int. J. Dev. Biol. (2005) 49: 843-850
Role of cell division in branching morphogenesis and differentiation of the embryonic pancreas.
L D Horb and J M Slack
Int. J. Dev. Biol. (2000) 44: 791-796
Cell-cycle-dependent nuclear translocation of HSP70 in amphibian embryonic cells.
N Moreau, C Prudhomme and N Angelier Int. J. Dev. Biol. (1998) 42: 633-636