Pseudomonas cichorii (Swingle) Stapp (10) (lettuce)(13) (cabbage)(27) (celery)(15) (tomato)(33) (chrysanthemum)(16) (geranium)(16) (dwarf schefflera)(11) (sunflower)(22) (magnolia)(19) (5, 10) (5) (3) (3) P. cichorii (10) (14) ( polymerase chain reaction PCR) (20)
RFLP (
Pseudomonas cichorii
1 1 1, 2 1 2 cylin@wufeng.tari.gov.tw 95 12 02 2006 Pseudomonas cichorii 15 275-285 60 (RAPD)Pseudomonas cichorii (Swingle) Stapp 1,100 bp Topo
pCR®
II-TOPO Pseudomonas
cichorii SfL1/SfR2 (polymerase chain
reaction, PCR) 379 bp 6 21 SfL1 / SfR2 P. cichorii DNA 5~10 pg 5.5~9 (P. cichorii) 106 cfu/ml 105 ~108 cfu/ml SfL1/ SfR2 P. cichorii 3 - 4 hr SfL1/SfR2 P. cichorii PCR
restriction fragment length polymorphism) AFLP (amplified fragment length polymorphism) RAPD (random amplified polymorphic DNA)(17, 18, 32)
Pseudomonas Pseudomonas syringae pv. cannabina efe gene DNA
ETH1 ETH2 ETH3 P. syringae pv.
cannabina P. syringae pv. glycinea P. syringae pv.
phaseolicola (24)
P. syringae pv. atropurpurea cfl gene DNA
Primer 1 Primer 2 P. syringae pv. glycinea P. syringae pv. maculicola P. syringae pv. tomato
(8) P. syringae pv. atropurpurea(30) P. syringae pv. pisi(7) P. syringae pv. phaseolicola(25) pel Erwinia carotovora (12) RFLP Erwinia (6, 12, 29) DNA PCR Seal (26)
RAPD Ralstonia solanacearum PCR RAPD
Pseudomonas cichorii Pseudomonas
cichorii PCR
35 Pseudomonas cichorii (Swingle) Stapp
Sf ( ) DNA 5 ml LB broth 30 16 hr 12,000 rpm 10 min 1 ml STE 12,000 rpm 10 min 0.5 ml STE 50 ml 10 % SDS 65 30 min proteinase K (100 g/ ml ) 30 min RNase A ( 50 g/ml ) 37 30 min phenol/chloroform
(phenol chloroform isoamylalcohol 25 24 1) 12,000 rpm 4 30 min 1/10 3 M NaOAc 2 95 % -20 1 hr DNA 70 % 12,000 rpm 4 30 min 50 15 min 100 l 4 DNA -30 (23) (plasmid)
Plasmid miniprep purification kit (Genemark) Kanamycin (50 g/ml) 5 ml LB 37 16 hr 2 ml 12,000 rpm 5 min 200 l solution I 200 l solution II 200 l solution III 12,000 rpm 4 10 min DNA 700 l washing solution 12,000 rpm RAPD PCR
Table 1. Bacterial isolates used in experiments of RAPD and PCR.
Bacterium Strain
Burkholderia andropogonis Pan1
B. caryophylli Tw7, Tw9
B. gladioli pv. gladioli Bg
Erwinia carotovora subsp. carotovora Zan01~15
Erwinia chrysanthemi Ech10~22
Sr53~56
Pantoea agglomerans Yx5, Yx7
Pseudomonas aeruginosa Pae
P. cichorii Sf P. fluorescens Pf P. putida Pu P. syringae pv. glycinea Psg P. syringae pv. lachrymans Psl P. syringae pv. phaseolicola Psph
P. syringae pv. pisi Pspi
P. syringae pv. syringae Pss
P. syringae pv. tabaci Psta
P. syringae pv. tomato Psto
Ralstonia solanacearum Ia57, 29
Xanthomonas axonopodis pv. dieffenbachiae Ao72 X. axonopodis pv. vesicatoria Xv15 X. campestris pv. mangiferaeindicae X58
4 30 sec 12,000 rpm 4 3 min DNA 1.5 ml 60 5~10 min 50 l 12,000 rpm 4 30 sec DNA RAPD Sf01 Sf02 Sf07 Sf09 Sf31 5 DNA OPERON
10-MER KITS OPX 01~20, OPY 01~20 OPZ 01~20
60 RAPD 20 l
100 ng DNA 200 M dNTPs 0.25 M
0.8 U DNA 1
GeneAmp PCR System 2400 94 1
min 94 1 min, 40 1 min, 72 2 min
40 72 20 min 8 l 1X TAE 1.5% agarose gel 100 V UV box DNA RAPD DNA DNA DNA RAPD 1X TAE buffer 1.2% agarose DNA Ultrafree®
-DA (MILLIPORE USA ) KIT Gel Nebulizer Ultrafree-MC Vial
6,000 rpm 10 min
DNA -30 DNA
Random Primer Fluorescein Labeling Kit with Antifluorescein-AP (NEN) (probe)
19 l DNA 5
min 5 min 5 l Random
primer and reaction buffer mix 5 l Fluorescein nucleotide mix 1 l Klenow fragment
30 l 37 1 hr 5 l 0.1 M EDTA (pH8.0) -30 (Southern hybridization) Southern (28) DNA Sf41 Sf80
DNA OPY-20 RAPD
0.25 N HCl 10 min denature buffer (0.5 M NaOH 1.5 M
NaCl) 30 min
neutralization buffer (0.1 M Tris-HCl, pH 7.5; 1.5 M NaCl) 30 min
10X SSC
10X SSC 16
hr DNA
UVC-515 Ultraviolet Crosslinker (ULTRA-LUM, Inc. U.S.A.) 254 nm
2X SSC salmon sperm
DNA 200 l ( hybridization
solution: 5X SSC, 0.5% SDS, 50% Formamide, 5X
Denhardts) 3 min 5 min
hybridization solution 65
16 hr 0.1 % SDS
2X SSC 5 min 2 0.1 %
SDS 0.2X SSC 65 15 min 2
buffer I (0.1 M Tris-HCl , pH 7.5 ; 0.15 M NaCl) 5 min buffer (0.5 % blocking reagent in buffer
II, 4 ) 1 hr Anti-fluorescein-AP
conjugate (NEN®
Life Science Products, Boston, MA) buffer II (1: 5000) 1 hr buffer I 4
5 min buffer (0.1 M Tris-HCl, pH 9.5 0.15 M NaCl) 2 5 min
CDP-Star 12.5 mM (Tropix, U.S.A.) buffer (1 200)
5 min X-ray 10 min
DNA TOPO TA Cloning® kit DNA 4 l DNA 1 l salt solution 1 l pCR® II-TOPO vector 5 min 2~6 l
(competent cell) 30 min 42
30 sec 2 min 250 l SOC 200 rpm 37 1 hr 40 l 100 mM IPTG 40 l 40 g/ml X-gal Kanamycin (50 ppm) LA 37 DNA EcoRI
NCBI (National Center for Biotechnology
Information) (GenBank)
Vector-NTI 8 (InforMax)
G+C hairpin
duplex structure (forward primer) ( reverse primer)
(dNTP) (primer) Taq polymerase (Dynazyme)
PCR PCR
PCR
DNA Sf DNA
Pseudomonas putida, P. aeruginosa, P. fluorescens, P. syringae pv. syringae, Agrobacterium tumefaciens, Erwinia carotovota pv. carotovota, E. chysanthemi, Ralstonia solanacearum, Xanthomonas axonopodis pv.
dieffenbachiae 5 9 10 ng SfL1/SfR2 PCR SfL1/SfR2 P. cichorii DNA Sf NA 24 hr 108 cfu/ml 10 106 cfu/ml Pseudomonas syringae pv. syringae P. putida Agrobacterium tumefaciens 104 ~108 cfu/ml Sf 106 cfu/ml 50 l 200 ml 50 l 0.5 N NaOH 50 l 50 l 1 M Tris-HCl (pH 8.0) 2 l SfL1/SfR2 PCR DNA Sf02 Sf76 DNA 100, 10, 1 ng 100, 10, 1, 5 1 0.1 0.5 pg SfL1/SfR2 PCR 20 l PCR SfL1/SfR2 DNA Sf02 Sf058 NA (nutrient agar) 24 hr OD620 0.3 ( 10 8 cfu/ml) 10 101 ~107 cfu/ml 50 l 200 l 50 l 0.5 N NaOH 50 l 50 l 1 M Tris-HCl (pH 8.0) 2 l SfL1/SfR2 PCR spiral plater (model D, Spiral systems, Inc. Bethesde, Maryland)
NA 30 24 hr Wang(31) 50 l 0.5 N NaOH 25 l 25 l 1 M Tris-HCl (pH 8.0) 2 l SfL1/SfR2 PCR PCR (Pseudomonas cichorii) Sf06 Sf09 Sf26 Sf28 Sf30 Sf62 NA 24 hr Spectrophotometer A620 nm O.D 0.3 ( 1 108 cfu/ml) 30 24 48 72 96 120 hr 1 10 1 30 1 50 50 l
500 l PBS-Tween-20 ( phosphate-buffer saline
0.5% Tween-20) 10,000 rpm 20 min 100 l PBS-Tween-20 (19) 2 l SfL1/SfR2 PCR SfL1/SfR2 RAPD Sf01 Sf02 Sf07 Sf09 Sf31 5 DNA OPX 01~20 OPY 01~20 OPZ 01~20 60 RAPD 10
OPX-17 1,100 bp ( ) OPX-17 DNA ( ) RAPD 1,100 bp DNA Sf 1,100 bp 1,100 bp 1,100 bp 1,100 bp DNA D N A Sf RAPD 1,100 bp DNA pCR® II-TOPO vector DNA DNA 99% (identity) NCBI DNA 1,100 bp 8~242 bp 272~304 bp 480~537 bp 765~1138 bp 4 P. syringae pv. tomato DC3000 84~93% ( ) P. syringae pv. tomato 304~765 (
DNA 15%) Vector NTI
suite 8 (InforMax) 2 SfL1 SfL2
2 SfR1 SfR2
PCR SfL1/SfR2
(Data not shown) Sf 379 bp
P. cichorii
SfL1/SfR2 PCR
PCR 20 l
50 ng DNA 1X Taq buffer 200 M dNTP 0.25 M Primer 0.4 Unit Dynazyme
PCR (a) 94 5 min 1
(b) 94 1 min 65 30 sec 72 30 sec 30 (c) 72 10 min 1
2 % agarose
SfL1/SfR2
Sf02 Sf10 Sf28 Sf33 Sf41 Sf68 Sf74
Sf85 DNA Pseudomonas putida, P.
aeruginosa, P. fluorescens, P. syringae pv. syringae, Agrobacterium tumefaciens, Erwinia carotovota pv. carotovota, E. chysanthemi, Ralstonia solanacearum, Xanthomonas axonopodis pv. dieffenbachiae 5 9
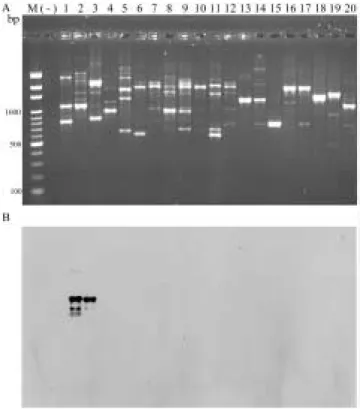
DNA Sf DNA ( ) P. cichorii Sf41 Sf41 106 cfu/ml 2 3 (108 ~104 cfu/ml) PCR Sf 379 bp P. cichorii OPX-17 RAPD (A), Sf X17-Sf1100 (B). Lane 1 lane 2 Sf M Bio 100 marker (-) Lane 3-20
Fig 1. Agarose gel electrophoresis shows RAPD patterns of strains of Pseudomonas cichorii and other bacteria using OPX-17 random primer (A) and southern hybridization of the RAPD products with X17-Sf1100 probe cloned from Pseudomonas cichorii (B). Lanes 1 and 2, Pseudomonas cichorii Sf isolates; M, Bio100 marker; lanes 3 - 20, other bacteria; (-), negative control.
SfL1/SfR2 SfL1/SfR2 Sf02 Sf76 DNA PCR Sf76 DNA 5 pg 10 pg ( ) Sf02 Sf58 NaOH DNA PCR 5.5 ~ 9 ( ) Sf02 Sf07 Sf09 Sf27 Sf31 Sf35 Sf41 Sf58 Sf68 Sf70 Sf71 Sf75 Sf77 Sf80 Sf83 Sf85 0.5 N NaOH DNA SfL1/SfR2 PCR 379 bp P. cichorii PCR pX17-Sf1100 bp DNA SfL1 SfL2 SfR1 SfR2
Fig.2. The sequence of the inserted DNA fragment in recombinant plasmid pX17-Sf1100 bp. The sequence of the primers SfL1, SfL2, SfR1 and SfR2 designed are underlined, and the sequence of random primers OPX 17 are indicated red bold-faced.
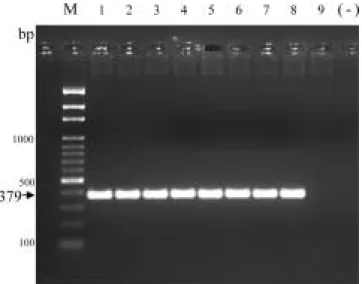
1 10 1 30 1 50 PBST 2 l SfL1/SfR2 PCR 24 hr 48 hr 72 hr (1 10 1 30 1 50) 96 hr 120 hr ( ) Pseudomonas spp. (7, 8, 25, 30) P. cichorii Sf41 DNA SfL1/SfR2 1-8 Pseudomonas
putida, P. fluorescens, P. syringae pv. syringae, Erwinia carotovora pv. carotovora, E. chrysanthemi, Xanthomonas axonopodis pv. dieffenbachiae, Agrobacterium trmefaciens, Ralstonia solanacerum 9 Pseudomonas aeruginosa DNA; M 100 bp marker (-) Fig 3. Polymerase chain reaction amplification products of total DNA from strains of Pseudomonas cichorii Sf41 mixed with other bacterial with primer pair SfL1/SfR2. Lanes 1-8, Pseudomonas putida, P. fluorescens, P. syringae pv. syringae, Erwinia carotovora pv. carotovora, E. chrysanthemi, Xanthomonas axonopodis pv. dieffenbachine, Agrobacterium trmefaciens, Ralstonia solanacerum, respectively; lane 9, only Pseudomonas aeruginosa DNA; M, 100 bp marker; (-), negative control.
SfL1/SfR2
Sf76 DNA
M 100 bp marker (-) 1-9 100
ng 10 ng 1 ng 100 pg 10 pg 5 pg 1 pg 0.5 pg 0.1 pg
Fig 4. Sensitivity of polymerase chain reaction using primer pair SfL1 / SfR2 to detect total DNA of Pseudomonas cichorii Sf76. Lanes 1-9, 100 ng 10 ng 1 ng 100 pg 10 pg 5 pg 1 pg 0.5 pg and 0.1 pg of DNA, respectively. M, Bio 100 DNA marker; (-), negative control. SfL1/SfR2 Sf58 1-3 5.5~9.0 103 cfu 4-6 5.5~9.0 102 cfu 7-9 5.5~9.0 101 cfu 10-12 5.5~9.0 cfu 13-15 5.5~9.0 10-1 cfu 16-18 5.5~9.0 10-2 cfu 19-21 5.5~9.0 10-3 cfu M 100 bp marker (-)
Fig 5. Sensitivity of PCR for detection of cells of Pseudomonas cichorii Sf58 with primer pair SfL1/SfR2. Lanes 1-3, 5.5~9.0 103
cfu; lanes 4-6, 5.5~9.0 102
cfu; lanes 7-9, 5.5~9.0 101
cfu; lanes 10-12, 5.5~9.0 cfu; lanes 13-15, 5.5~9.0 10-1
cfu; lanes 16-18, 5.5~9.0 10-2
cfu; lanes 19-21, 5.5~9.0 10-3
cfu; M, 100 bp marker; (-), negative control.
RAPD P. cichorii RAPD P. cichorii RAPD Arnold et al.(7) pX17-sf-1100 OPX-17 1.1 kb NCBI 8~242 bp 272~304 bp 480~537 bp 765~1,138 bp 4 P.
syringae pv. tomato DC3000 (AE016853)
84~93% ( ) 813-956 bp P.
aeruginosa PAO1 (AE004091) 88% P. cichorii
P. syringae pv. tomato P.
aeruginosa PAO1 304~765 bp
( DNA
15%) Vector NTI suite 8 (InforMax)
2 SfL1 SfL2 2 SfR1 SfR2 PCR SfL1 SfR2 SfL1/SfR2 P. cichorii P. cichorii (No. 12682)
Agrobacterium, Burkholderia, Erwinia, Xanthomonas, Ralstonia Pseudomonas spp.
PCR P. cichorii
P. cichorii 379 bp
Sato et al.(24)
efe gene ETH1
ETH2 ETH3 P. syringae pv. cannabina, pv. glycinea, pv. phaseolicola, pv. sesame
Bereswill et al.(8)
efl gene Primer 1 Primer 2
P. syringae pv. atropurpurea, pv. glycinea, pv. maculicola, pv. tomato DNA P. cichorii SfL1/SfR2 ( P. cichorii P. syringae ) DNA RAPD P. cichorii SfL1/SfR2 5 pg~10 pg DNA Bereswill (9) Ewinia amylovora 10 ng (2) SL1/SR1
Acidovorax avenae subsp. citrulli 100
pg SfL1/SfR2
Sf02 Sf58
5.5 ~ 9 (4)
5A/5B, Ec1/Ec2 Erwinia carotovora pv. carotovora E. chrysanthemi Ecc 1.3
102
Ech 5.2 101 (2)
SL1/SR1 Acidovorax avenae subsp. citrulli
1.2 102 (1)
nL/nR Burkholderia caryophylli 20 Prosen et al.(21)
P. syringae pv. phaseolicola 103 cfu SfL1/SfR2 P. cichorii Prosen et al.(21) P. syringae pv. phaseolicola (probe hybridization) 101 cfu/ml SfL1/SfR2 5.5 101 cfu SfL1/SfR2 P. cichorii DNA P. cichorii PCR SfL1/SfR2 PCR
Pseudomonas cichorii M 100 bp marker (-)
1-3 24 hr 4-6 48 hr 7-8 72 hr 9-12 96 hr 13-15 120 hr 1-15 1 10 1 30 50 16 Pseudomonas cichorii DNA
Fig 6. Detection of Pseudomonas cichorii in artificially infested leaf tissures of sunflower by polymerase chain reaction using primer pair SfL1/SfR2. Lanes 1-3, 24hr; lanes 4-6, 48 hr; lanes 7-9, 72 hr; lanes 10-12, 96 hr; lanes 13-15, 120 hr; lanes 1-15, symbol 1 10 1 30 and 1 50 of sample, respectively. 16, Pseudomonas cichorii ; M, Bio 100 DNA marker; (-), negative control.
379 bp P. cichorii P. cichorii P. cichorii Wang et al.(31) 0.5 N NaOH
genomic DNA
Tris-HCl buffer genomic DNA 0.5 N NaOH PCR DNA (2) (1) P. cichorii SfL1/SfR2 P. cichorii DNA P. cichorii PCR 24 hr 48 hr 72 hr 96 hr 120 hr SfL1/SfR2 SfL1/SfR2 P. cichorii P. cichorii 1. 1999 Burkholderia caryophylli 2. 1999 3. 2004 13 329-334 4. 1998 Erwinia 5. 1989 31 346-357
6. Anna, O. A., Hyman, L. J., Toth, R. L. and Toth, I. K. 2002. Application of amplified fragment length polymorphism fingerprinting for taxonomy and identification of the soft rot bacteria Erwinia carotovora and Erwinia chysanthemi. Appl. Environ. Microbiol. 68 : 1499-1508.
7. Arnold, D. L., Atheypollard, A., Gibbon, M. J., Taylor, J. D. and Vivian, A. 1996. Specific oligonucleotide primers for the identification of Pseudomonas syringae pv pisi yield one of two possible DNA fragments by PCR amplification : evidence for phylogenetic divergence. Physiol. Mol. Plant Pathol. 49 : 233-245. 8. Bereswill, S., Pshl, A., Bellemann, P., Zeller, W. and
Geider, K. 1992. Sensitive and species-specific detection of Erwinia amylovora by polymerase chain detection analysis. Appl. Environ. Microbiol. 58 : 3522-3526.
9. Bereswill, S., Bugert, P., Volksch, B., Ullrich, M., Bender, C. L., et al. 1994. Identification and relatedness of coronatine-producing pseudomonas syringae pathovars by PCR analysis and sequence determination of the amplification products. Appl. Environ. Microbiol 60:2924-2930.
10. Chase, A. R. 1988. Compendium of ornamental foliage plant diseases. APS PRESS 92pp.
11. Chase, A. R. and Jones, J. B. 1986. Effects of host nutrition, leaf age, and Preinoculation light levels on severity of leaf spot of dwarf schefflera caused by Pseudomonas cichorii. Plant Dis. 70 : 561-563. 12. Derrasse, A., Prious, S., Kotoujansky, A. and
Bertheau, Y. 1994. PCR and restriction fragment length polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato disease. Appl. Environ. Microbiol. 60: 1437-1443.
13. Grogan, R. G., Misaghi, I. J., Kimble, K. A., Greathead, A. S., Ririe, D. and Bardin, R. 1977. Varnish spot, destructive disease of lettuce in California caused by Pseudomonas cichorii. Phytopathology. 67 : 957-960.
14. Gulya, T. J., Woods, D. M., Bell, R. and Mancl, M. K. 1991. Disease of sunflower in calfornia. Plant Dis. 75 : 572-574.
15. Jagger, I. C. 1914. Bacterial leaf spot disease of celery. Phytopathology. 4 : 395(Abstr.).
16. Jones, J. B., B. C. Raju., and A. W. Engelhard. 1984. Effects if temperature and leaf wetness on development of bacterial spot of geranium and chrysanthemum incited by Pseudomonas cichorii. Plant Dis. 68 : 248-251.
17. Louws, F. J., Rademarker, J. L. W. and de Bruijn, F. J. 1999. The three Ds of PCR-based genomic analysis of
phytobacteria: diversity, detection, and disease diagnosis. Annu. Rev. Phytopathol. 37: 81-125. 18. Manulis, S., Valinsky, L., Lichter, A. and Gabriel, D.
W. 1994. Sensitive and specific detection of Xanthomonas campestris pv. pelargonii with DNA primers and probes identified by random amplified polymorphic DNA analysis. Appl. Environ. Microbiol. 60 : 4094-4099.
19. Mullen, J. M. and G. S. Cobb. 1984. Leaf spot of southern magnolia caused by Pseudomonas cichorii. Plant Dis. 68 : 1013-1015.
20. Mullid, K. B. and Faloona, F. A. 1987. Specific synthesis of DNA in virto via a polymerase-catalysed chain reaction. Methods in Enzymology. 155 : 335-350.
21. Prosen, D., Hatziloukas, E., Schaad, N. W. and Panopoulos, N. J. 1993. Specific detection of pseudomonas syringae pv. phaseolicola DNA in bean seed by polymerase chain reaction-based amplification of a phaseolotoxin gene region. Phytopathology 83 : 965-970.
22. Robbs, C. F. and Almeida, A. M. R. 1982. Bacterial leaf blight of sunflower caused by Pseudomonas cichorii (Swingle) Stapp. First record in Brazil. Rev. Plant Pathol. 61 : 356.
23. Sambrook, j., Mantis, T. I. and Fritsch, E. F. 1989. Molecular cloning: a laboratory manual. 2nd
ed. Cold Spring Harbor Laboratory, Press, N. Y.
24. Sato, M., Watanabe, K., Yazawa, M., Takikawa, Y. and Nishiyama, K. 1997. Detection of new ethylene-producing bacteria Pseudomonas syringae pvs. canncbina and sesame, by PCR amplification of genes for the ethylene-forming enzyme. Phytopathology 87 : 1192-1196.
25. Schaad, N. W., Azad, H., Peet, R. C. and Panopoulos,
N. J. 1989. Identification of Pseudomonas syringae pv. phaseolicola by a DNA hybridization probe. Phytopathology 79 : 903-907.
26. Seal, S. E., Jackson, L. A. and Daniels, M. J. 1992. Isolation of a Pseudomonas solanacearum specific DNA probe by hybridization and construction of species-specific oligonucleotide primers for sensitive detection by the polymerase chain-reaction. Appl. Environ. Microbiol. 58 : 3759-3761.
27. Smith, M. A. and Ramsey, G. B. 1956. Bacterial zonate spot of cabbage. Phytopathology 46 : 210-213. 28. Southern, E. M. 1975. Detection of specific sequences
among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98 : 503-517.
29. Toth, I. K., Hyman, L. J. and Wood, J. R. 1999. A one-step PCR-based method for the detection of economically important soft rot Erwinia species on micropropagated potato plants. J. Appl. Microbiol. 87 : 158-166.
30. Tskahashi, Y., Omura, T., Hibino, H. and Sato, M. 1996. Detection and identification of Pseudomonas syringae pv atropurpurea by PCR amplification of specific fragments from an indigenous plasmid. Plant Dis. 80 : 783-788.
31. Wang, H., Qi., M. and Cutler, A. J. 1993. A simple method of preparing plants samples for PCR. Nucleic Acids Res. 21: 4153-4154.
32. Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. and Tingey, S. V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18: 6531-6535.
33. Wilk, J. D. and Dye, D. W. 1974. Pseudomonas cichorii causing tomato and celery diseases in New Zealand. N. Z. J. Agr. Res. 17 : 123-130.
ABSTRACT
Hseu, S. H.,1
Shentue, H.1
and Lin, C. Y.1, 2
2006. Development of specific PCR primers for identification of Pseudomonas cichorii. Plant Pathol. Bull. 15: 275-285 (1
Department of Plant Pathology, Agricultural Research Institute, Council of Agriculture, Wufeng, Taichung, Taiwan, ROC.;
2
Corresponding author, E-mail: cylin@wufeng.tari.gov,tw)
A specific PCR (polymerase chain reaction) primer pair have been developed using RAPD ( random amplified polymorphic DNA ) to detect Pseudomonas cichorii. Totally sixty random primers were used to find specific DNA fragments of P. cichorii, and a specific DNA fragment of 1,100 bp amplified by the primer OPX17 was cloned into the pCR®
II-TOPO vector and further sequenced to design a specific primer pair SfL1 / SfR2. The primer pair could amplify a distinct band of 379 bp that was specific to P. cichorii, and no DNA fragment amplified by the same primer pair from the other tested 21 bacterial species in 6 genera. Sensitivity of PCR for detection of P. cichorii with primer pair SfL1 / SfR2 was between 5 ~ 10 pg for purified DNA and 5.5 ~9 cfu for cultured cells. Non-target bacteria did not affect the efficiency of specific amplification of P. cichorii in PCR assay with primer pair SfL1 / SfR2. PCR technique using primer pair SfL1 / SfR2 identifies the culture of P. cichorii within 3 - 4 hours and detected the bacterium in artificially inoculated sunflower leaf tissue. Based on the data provided, we conclude that the primer pair SfL1 / SfR2 might be a useful tool for rapid identification and diagnosis of P. cichorii.