Other uses, including reproduction and distribution, or selling or
licensing copies, or posting to personal, institutional or third party
websites are prohibited.
In most cases authors are permitted to post their version of the
article (e.g. in Word or Tex form) to their personal website or
institutional repository. Authors requiring further information
regarding Elsevier’s archiving and manuscript policies are
encouraged to visit:
Association of polymorphisms in the genes of the urokinase plasminogen activation
system with susceptibility to and severity of non-small cell lung cancer
Chuen-Ming Shih
a, Wu-Hsien Kuo
b,c, Chiao-Wen Lin
d, Wei Chen
e, Wei-Erh Cheng
a,
Shuo-Chueh Chen
a, Yao-Ling Lee
f,g,⁎
a
Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, China Medical University Hospital, Taiwan, ROC
b
Department of Medicine, Armed-Force Taichung General Hospital, Taichung, Taiwan, ROC
c
General Education Center, Central Taiwan University of Science and Technology, Taiwan, ROC
d
Institute of Biochemistry and Biotechnology, Chung Shan Medical University, Taiwan, ROC
eDivision of Pulmonary and Critical Care Medicine, Department of Internal Medicine, Chia-Yi Christian Hospital, Taiwan, ROC f
School of Medical Laboratory and Biotechnology, Chung Shan Medical University, Taiwan, ROC
g
Clinical Laboratory, Chung Shan Medical University Hospital, Taiwan, ROC
a b s t r a c t
a r t i c l e i n f o
Article history: Received 20 June 2010
Received in revised form 30 September 2010 Accepted 4 October 2010
Available online 16 October 2010 Keywords:
Lung cancer
Urokinase plasminogen activator (uPA) Urokinase plasminogen activator receptor (uPAR)
Single nucleotide polymorphisms (SNPs) Polymerase chain reaction- restriction fragment-length polymorphism (PCR-RFLP)
Background: Urokinase plasminogen activating (uPA) system is implicated in neoplastic progression. High tissue levels of uPA system components correlate with a poor prognosis in lung cancer. The present study examined the single nucleotide polymorphisms (SNPs) of uPA and the corresponding receptor, uPAR, for exploring their roles in non-small cell lung cancer (NSCLC).
Methods: The allele frequencies and genotype distributions of uPA rs4065 C/T and uPAR rs344781 (−516 T/C) among 375 NSCLC cases and 380 healthy controls were examined using polymerase chain reaction-restriction fragment-length polymorphism (PCR-RFLP) analysis. Putative association between the above SNPs and clinicopathological characteristics of NSCLC were also analyzed.
Results: The genotype frequencies of the variant homozygotes of uPA and uPAR were significantly different between NSCLC and control subjects. Significant association was also observed between the examined genotypes and disease stage of NSCLC. Logistic regression analysis revealed that individuals with uPA rs4065 TT genotype have higher odds ratios (ORs) for lung cancer. Whereas, subjects with uPAR-344781 CC genotype have lower ORs for lung cancer. The patients carrying a homozygous TT genotype at uPA rs4065, or at least a T allele at uPAR-344781 (−516), had a tendency to develop advanced disease.
Conclusions: Our results revealed that genetic polymorphisms of the uPA rs4065 C/T and uPAR rs344781 (−516 T/C) were associated with the susceptibility and severity of NSCLC.
© 2010 Elsevier B.V. All rights reserved.
1. Introduction
Extracellular matrix (ECM) is a complex assembly of proteins and polysaccharides that are secreted, assembled and modeled by cells. The physiological function of ECM is to provide support and organization to tissues. However, alterations in the physical properties and composition of the ECM as well as in the expression or regulation of its corresponding receptors are implicated in many diseases[1,2]. Degradation of ECM and basement membrane is an essential mechanism for invasion and metastasis of cancer cells[3]. ECM proteinases are divided into three groups: metalloproteinases, cysteine proteinases and serine proteinases. Urokinase plasminogen activator (uPA) system is one of the serine proteinase systems involved in ECM degradation. Members of this system,
including uPA and the corresponding receptor uPAR, are overexpressed in several malignant tumors[4]. Clinical studies have shown that over-expression of the uPA/uPAR components correlates with increased proliferation, migration, and invasion of many cancers, including lung cancer[5–7].
Lung cancer is the leading cause of cancer-related mortality around the world. Among various factors, genetic variation is proved to contribute to the development and progression of lung cancer[8–11]. The median levels of uPA and uPAR expression are higher in lung tumor tissues than the adjacent lung parenchyma. Clinically, the association between highly expressed levels of uPA proteins and shorter survival of cancer patients supports the hypothesis that the over-expressed uPAR facilitates matrix degradation by promoting uPA activity in the microenvironment[12]. Particularly, uPAR is proved to be significantly correlated with the overall survival of patients with non-small cell lung cancer (NSCLC).
The uPA and uPAR gene is located at chromosome 10q24 and 19q13.2, respectively. Several single nucleotide polymorphisms (SNPs) located within the promoter or other regulatory regions of the genes in the uPA
Clinica Chimica Acta 412 (2011) 194–198
⁎ Corresponding author. School of Medical Laboratory and Biotechnology, Chung Shan Medical University. No.110, Sec. 1, Chien-Kuo N. Road, Taichung 402, Taiwan, ROC. Tel.: + 886 424 730 022; fax: + 886 423 248 171.
E-mail address:yllee@csmu.edu.tw(Y.-L. Lee).
0009-8981/$– see front matter © 2010 Elsevier B.V. All rights reserved. doi:10.1016/j.cca.2010.10.004
Contents lists available atScienceDirect
Clinica Chimica Acta
system may affect their expression and activity[13–17]. The functional impact of the SNPs of uPA rs4065 and uPAR rs344781 had been reported [18,19]. In addition, genotypes and frequencies of alleles in the uPA and uPAR genes are correlated with tumor progression[20–24], including NSCLC of Han Chinese in Taiwan[25–30]. Nevertheless, the effects of uPA/ uPAR genetic polymorphisms on NSCLC have not been explored[31–33] although the genetic polymorphisms in the uPA activation system had been studied in breast cancer, oral cancer, ovarian cancer and colorectal cancer[23,24,34]. In this context, we hypothesized that investigation of the SNPs located in uPA/uPAR genes may be a simple and efficient method to predict the risk and prognosis of lung cancer. To explore the influence of genetic polymorphisms of uPA/uPAR on the susceptibility and clinico-pathological development of NSCLC, the present study investigated the relationship between uPA/uPAR SNPs and the clinicopathological char-acteristics of NSCLC.
2. Materials and methods 2.1. Study population
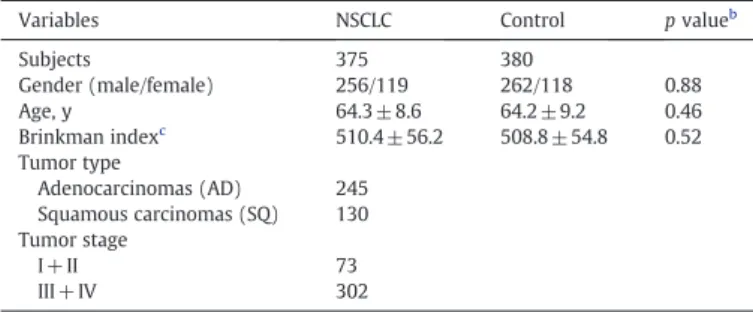
A total of 375 Han Chinese NSCLC patients (256 male and 119 female; median age 64.3) admitted to China Medical University Hospital between January 2005 and April 2010 were recruited. Among the patients, 245 had adenocarcinomas (AD) and 130 had squamous carcinomas (SQ). The histological determination, including tumor types and stages, was
performed according to the WHO classification (WHO, 1982) and the
TNM system (Mountain, 1986). In addition, 380 unrelated healthy control subjects (262 male and 118 female; median age 64.2) were randomly selected from a pool of healthy volunteers who visited the General Health Check-up Center of China Medical University Hospital during the same period. All the control subjects had no medical illness and hereditary disorders without taking any medications during the study period.
A detailed questionnaire, including information regarding the average number of cigarettes smoked daily and the number of years the subjects had been smoking, was completed by each recruited subject under the instruction of a trained interviewer. This study was approved by the Research Ethics Committee of China Medical University, and informed consent was obtained from each participant prior to the commencement. No significant differences in the demographic information between case and control participants were observed (Table 1).
2.2. Samples collection and genomic DNA extraction
Venous blood (5–10 mL) from each subject was drawn into Vacutainer tubes containing EDTA and stored at 4 °C. Genomic DNA was extracted by QIAamp DNA blood mini kits (Qiagen, Valencia, USA) according to the manufacturer's instructions. DNA was dissolved in TE buffer [10 mM Tris (pH 7.8), 1 mM EDTA] and quantitated by spectrophotometer. The
extracted DNA were stored at −20 °C and used as templates for
polymerase chain reaction (PCR).
2.3. Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP)
Genotypes of the uPA (rs4065, C/T SNP) and uPAR (rs344781, T/C SNP) among the study subjects were determined by PCR-RFLP. The sequences of forward and reverse primers were as follows: 5
′-AGTCACACCAAGGAA-GAGAA-3′ and AGACAAGTTGCTGGTCAGTA-3′ for uPA(291 bps);
5′-AATCG CTCTCCACTGCTGTA-3′ and 5′-CAATGCCTGGAATAGCTG CT-3′ for
uPAR (308 bps). The PCR was performed in a 10μL volume containing
100 ng DNA template, 1.0μL of 10× PCR buffer (Invitrogen, Carslbad, CA, USA), 0.25 U of Taq DNA polymerase (Invitrogen, Carslbad, CA, USA), 0.2 mM dNTPs (Promega, Madison, WI, USA), and 200 nM of each primer (MDBio Inc, Taipei, Taiwan). The PCR cycling conditions were 5 min at 94 °C followed by 35 cycles of 1 min at 94 °C, 1 min at 60 °C, and 2 min at 72 °C, with afinal step at 72 °C for 20 min to allow a complete extension of all PCR fragments. Then the genotypes of uPA and uPAR genes were determined by HphI and MspA1I digestion, respectively. In brief, a 10μL aliquot of PCR product was subjected to digestion at 37 °C for 4 h in a 15μL reaction containing 5 U of restriction enzyme (New England Biolabs,
Beverly, MA) and 1.5μL 10× buffer (New England Biolabs). Digested
products were separated on a 3% agarose gel and then stained with ethidium bromide. As a result, the PCR fragment containing T allele yielded 187- and 104- bp products for SNP of uPA, while the that containing C alleles yielded a 291-bp product; the PCR fragment containing C allele yielded 200- and 108- bp products for SNP of uPAR, while that containing T alleles yielded a 308-bp product. Representative results were shown inFig. 1.
2.4. Statistical analysis
Differences in clinical data between the NSCLC patients and the control subjects were examined. All continuous data were expressed as mean± standard deviation and compared using a two-tailed Student's t-test. Categorical variables were reported as a percentage and compared using Chi-square (χ2) or Fisher's exact test. Hardy–Weinberg equilibrium was assessed using a goodness-of-fit χ2
test for biallelic markers. The genotype distribution of uPA and uPAR between healthy subjects and patients was examined by theχ2test. Significance was
Table 1
Clinical features of the study populations.a
Variables NSCLC Control p valueb
Subjects 375 380 Gender (male/female) 256/119 262/118 0.88 Age, y 64.3 ± 8.6 64.2 ± 9.2 0.46 Brinkman indexc 510.4 ± 56.2 508.8 ± 54.8 0.52 Tumor type Adenocarcinomas (AD) 245 Squamous carcinomas (SQ) 130 Tumor stage I + II 73 III + IV 302 a
Data are presented as no. or mean ± SEM.
b p values were calculated using the Mann–Whitney U test.
c Brinkman index = daily cigarette numbers multiplied by smoking years.
Fig. 1. Polymerase chain reaction–restriction fragment length polymorphism of uPA and uPAR gene. PCR products of uPA and uPAR gene were subjected to enzymatic digestion by incubation with HphI and MspA1 at 37 °C for 4 h and then submitted to electrophoresis in 3% agarose gels. For C/T SNP of uPA, T allele yielded 187- and 104-bp products, while C alleles yielded a 291-bp product; for T/C SNP of uPAR, C allele yielded 200- and 108-bp products, while T alleles yielded a 308-bp product.
accepted at pb0.05. Odds ratios (ORs) and 95% confidence intervals (CI) for lung cancer of each specific genotype were calculated with logistic regression to quantitatively assess the degree of association observed. 3. Results
The association of the SNPs in uPA and uPAR genes with clinicopath-ological parameters of lung cancer patients was analyzed and shown in Tables 2 and 3. Overall, distributions of the uPA and uPAR genotypes were significantly different between non-cancer controls and lung cancer patients (pb0.0001). The frequency of uPA variant polymorphic (T/T) homozygote was 26% and 5% in the case and control subgroup, respectively, while that of the wild-type allele was higher (58% for case and 74% for control). The frequency of uPAR variant polymorphic C/C homozygote was 15% and 25% in the case and control subgroup, respectively, while that of the wild-type allele was higher (60% for case and 0.52% for control). Results of theχ2goodness-of-fit test showed that
genotype frequencies of uPA and uPAR were consistent with Hardy–
Weinberg equilibrium in the population.
Logistic regression analysis revealed that individuals carrying the homozygous uPA variant allele (T/T) had higher ORs for NSCLC (6.28, 95% CI 3.79–10.4, pb0.0001), adenocarcinoma (7.20, 95% CI 4.24–12.2;
pb0.0001) and squamous cell carcinoma (4.72, 95% CI 2.54– 8.76;
pb0.0001), compared with the subjects carrying wild-type allele (C/C or C/T genoytpe). On the contrary, individuals with homozygous uPAR variant allele (C/C) had lower ORs for NSCLC (0.51, 95% CI 0.36–0.74,
pb0.0001), adenocarcinoma (0.55, 95% CI 0.37–0.84; p=0.005) and
squamous cell carcinoma (0.44, 95% CI 0.25–0.77; p=0.003), compared with those carrying wild-type allele (T/T or T/C genotype). In addition, subjects carrying homozygous TT genotype at uPA rs4065 (p = 0.001) or
at least a T allele genotype at uPAR-344781–516 (p=0.001) had a
tendency to develop advanced disease. 4. Discussion
The uPA-mediated ECM degradation is an important mechanism in physiological and pathological tissue remodeling[35,36]. The plasmino-gen pathway plays an important role in the behavior of many tumors
including lung cancer[20,24,33,34,37,38]. The roles of uPA expression in tumor occurrence, invasion and prognosis have been established. Hence genetic variants encoding (variants in the genes encoding uPA, uPAR and uPA inhibitor plasminogen activator inhibitor (PAI)) may contribute to cancer prognosis. Our previous study revealed that high plasma uPA levels might be involved in tumor cell invasion and play an important role in NSCLC metastasis[39]. In this context, we hypothesize that genotyping of the SNPs located within the promoter or regulatory regions of the genes in the uPA system may be a simple method to predict the risk and prognosis of cancer because the abovementioned SNPs may affect the expression of the gene or the activities of their corresponding proteins. The present study provides novel information regarding the effects of genetic polymorphisms of uPA and uPAR on the susceptibility and clinicopath-ologic characteristics of NSCLC.
Przybylowska et al. reported the relation between protein levels and gene polymorphisms of uPA in colorectal cancer[40]. The uPAR in proved to play a central role in sustaining the malignant phenotype
and promoting tumor metastasis[41]. The interaction between uPA
and uPAR mediates various tumor cell activities , including tissue remodeling, chemotaxis, tumor invasion, dissemination, proliferation, and angiogenesis[42].
A significant difference in genotypic frequencies of uPA and uPAR genes between controls and NSCLC patients is demonstrated in this study. Individuals with uPA T/T homozygotes had a 6.28-fold higher risk of having NSCLC, compared with individuals carrying C/C homozygotes or C/ T heterozygotes. Whereas individuals with uPAR C/C homozygotes had a 0.51-fold risk of developing NSCLC, compared with those carrying T/T homozygotes or T/C heterozygotes. To the best of our knowledge, the relationship between PAI gene polymorphisms and lung cancer had never been studied except for one report from Di Bernardo et al.[33]. In this study, we focus upon NSCLC patients. Our data revealed that genetic polymorphisms of the uPA and uPAR are associated with the susceptibility of NSCLC. Significant differences were found in the genotype distribution of uPA and uPAR between NSCLC patients and controls. Individuals carrying a homologous uPA rs4065 TT genotype had a higher risk, whereas that carrying an uPAR rs344781 CC genotype had a lower risk for developing NSCLC. In addition, the uPA and uPAR genotypes are associated with the clinicopathologic status of NSCLC patients. Significant association between the genotypes of uPA and uPAR genes with advanced stages of
Table 2
The association between the uPA polymorphism and the clinicopathologic parameters of the studied subjects.
Characteristics Genotypes Total p value Odds ratio (95% CI) p valuea CC (%) CT (%) TT(%) Non-cancer control 202 (53) 158 (42) 20 (5) 380 1.00 Lung cancer 160 (43) 118 (31) 97 (26) 375 b0.0001b 6.28 (3.79–10.4) 0.0001 Tumor type AD 101 (41) 74 (30) 70 (29) 245 b0.0001b 7.20 (4.24–12.2) 0.0001 SQ 59 (45) 44 (34) 27 (21) 130 b0.0001b 4.72 (2.54– 8.76) 0.0001 Tumor stage I + II 44 (60) 21 (29) 8 (11) 73 0.001c 3.40 (1.56–7.37) 0.001 III + IV 116 (38) 97 (32) 89 (30) 302
AD: adenocarcinoma, SQ: squamous cell carcinoma.
a Odds ratios and p value were calculated using logistic regression to measure
the association of the variant genotypes TT with lung cancer risk, with that of the CC/CT genotype being referred to as 1.
b
The frequencies of the genotypes between the cancer and non-cancer control groups were compared with chi-square analysis.
c
The frequencies of the genotypes between lung cancers with different tumor stages were compared with chi-square analysis.
Table 3
The association between the uPAR polymorphism and the clinicopathologic parameters of the studied subjects.
Characteristics Genotypes Total p value Odds ratio (95% CI) p valuea TT (%) TC (%) CC (%) Non-cancer control 109 (29) 174 (46) 97 (25) 380 1.00 Lung cancer 130 (35) 189 (50) 56 (15) 375 0.001b 0.51(0.36–0.74) 0.0001 Tumor type AD 80 (33) 126 (51) 39(16) 245 0.018b 0.55 (0.37–0.84) 0.005 SQ 50 (38) 63(49) 17 (13) 130 0.007b 0.44 (0.25–0.77) 0.003 Tumor stage I + II 16 (22) 36 (49) 21 (29) 73 0.0001c 0.33 (0.18–0.60) 0.001 III + IV 114 (38) 153(51) 35(11) 302
AD: adenocarcinoma, SQ: squamous cell carcinoma.
a
Odds ratios and p value were calculated using logistic regression to measure the association of the variant genotypes CC with lung cancer risk, with that of the TT/TC genotype being referred to as 1.
b
The frequencies of the genotypes between the cancer and non-cancer control groups were compared with chi-square analysis.
c The frequencies of the genotypes between lung cancers with different tumor stages
were compared with chi-square analysis. C.-M. Shih et al. / Clinica Chimica Acta 412 (2011) 194–198
NSCLC is also observed. Patients carrying a homozygous TT genotype at uPA rs4065 or at least 1 T allele at uPAR rs344781 had a tendency to develop advanced disease. Nevertheless, the possibility that the relation-ship between the uPA/uPAR genotypes with NSCLC susceptibility in the present study is an ethnic-dependent observation cannot be entirely excluded because multiple risk factors and etiology contribute to the pathophysiology of NSCLC development. Besides, uPA and uPAR may have differential expression, functions and regulatory mechanisms in various tissues and tumors[43–47]. Nevertheless, our data demonstrate that the polymorphisms of uPA and uPAR gene are significantly asso-ciated with the susceptibility and severity of NSCLC in Taiwanese popu-lation. The possible selection bias has been taken into consideration and reduced to as low as possible (lowest level). Given that all lung cancer patients are diagnosed and treated at our hospital, the demographics and clinical characteristics of the cancer patients included in the current study were compatible with those of lung cancer patients in Taiwan in general, and it is reasonable to assume that the case group is repre-sentative of the lung cancer patients in our community. In addition, all cases and controls were ethnically Han Chinese in Taiwan with a relatively homogenous genetic background[48]. Therefore, the poten-tial confounding effect of population stratification for genotyping data should not be a major concern.
However, the small sample size may be a limitation of the present study. To enlarge the sample size and to analyze the relationship between NSCLC and uPA haplotypes as well as the relationship between NSCLC and other genotypes are required to further explore and understand the association of the genetic factors with lung cancer and the development of metastases.
In conclusion, our study demonstrates a significant association
between uPA/uPAR genotypes and NSCLC. The results of this study uncover the significant relationship between genetic polymorphisms of uPA and uPAR with the susceptibility and severity of lung cancer. Conflict of interest statement
All authors have no declared conflict of interest. Acknowledgements
This study was supported in part by grant DMR-98-017 from China Medical University and grant DOH 99-TD-C-111-005 from the Department of Health (The Executive Yuan, Republic of China). References
[1] Shi Z., Stack M.S. Molecules of cell adhesion and extracellular matrix proteolysis in oral squamous cell carcinoma. Histol Histopathol 2010;25:917–932.
[2] Xu S, Grande-Allen KJ. The role of cell biology and leaflet remodeling in the progression of heart valve disease. Methodist Debakey Cardiovasc J 2010;6: 2–7.
[3] Mekkawy AH, Morris DL, Pourgholami MH. Urokinase plasminogen activator system as a potential target for cancer therapy. Future Oncol 2009;5:1487–99. [4] Blasi F, Sidenius N. The urokinase receptor: focused cell surface proteolysis, cell
adhesion and signaling. FEBS Lett 2010;584:1923–30.
[5] Hildenbrand R., Allgayer H., Marx A., Stroebel P. Modulators of the urokinase-type plasminogen activation system for cancer. Expert Opin Investig Drugs 2010;19: 641–652.
[6] Ho M.L., Chen P.N., Chu S.C., et al. Peonidin 3-glucoside inhibits lung cancer metastasis by downregulation of proteinases activities and MAPK pathway. Nutr Cancer 2010;62:505–516.
[7] Altundag O, Altundag K, Morandi P, Gunduz M. Cytokines and chemokines as predictive markers in non-small cell lung cancer patients with brain metastases. Lung Cancer 2005;47:291–2.
[8] Huang YT, Heist RS, Chirieac LR, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol 2009;27:2660–7.
[9] Mostertz W., Stevenson M., Acharya C., et al. Age- and sex-specific genomic profiles in non-small cell lung cancer. Jama 2010;303:535–543.
[10] Lee E.B., Jeon H.S., Yoo S.S., et al. Polymorphisms in apoptosis-related genes and survival of patients with early-stage non-small-cell lung cancer. Ann Surg Oncol 2010;17:2608–2618.
[11] Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE 2010;5:e10312.
[12] Smith H.W., Marshall C.J. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol 2010;11:23–36.
[13] Arcaroli J, Sankoff J, Liu N, Allison DB, Maloney J, Abraham E. Association between urokinase haplotypes and outcome from infection-associated acute lung injury. Intensive Care Med 2008;34:300–7.
[14] Chou HT, Chen YT, Wu JY, Tsai FJ. Association between urokinase-plasminogen activator gene T4065C polymorphism and risk of mitral valve prolapse. Int J Cardiol 2004;96:165–70.
[15] Begin P, Tremblay K, Daley D, et al. Association of urokinase-type plasmin-ogen activator with asthma and atopy. Am J Respir Crit Care Med 2007;175: 1109–16.
[16] Thornton-Wells TA, Moore JH, Martin ER, Pericak-Vance MA, Haines JL. Confronting complexity in late-onset Alzheimer disease: application of two-stage analysis approach addressing heterogeneity and epistasis. Genet Epidemiol 2008;32:187–203.
[17] Stewart CE, Hall IP, Parker SG, et al. PLAUR polymorphisms and lung function in UK smokers. BMC Med Genet 2009;10:112.
[18] Nanbu R, Menoud PA, Nagamine Y. Multiple instability-regulating sites in the 3′ untranslated region of the urokinase-type plasminogen activator mRNA. Mol Cell Biol 1994;14:4920–8.
[19] Tran H, Maurer F, Nagamine Y. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol 2003;23:7177–88.
[20] Przybylowska K, Szemraj J, Kulig A, Dziki A, Ulanska J, Blasiak J. Antigen levels of urokinase-type plasminogen activator receptor and its gene polymorphism related to microvessel density in colorectal cancer. Acta Biochim Pol 2008;55: 357–63.
[21] Kohonen-Corish MR, Wang Y, Doe WF. A highly polymorphic CA/GT repeat in intron 3 of the human urokinase receptor gene (PLAUR). Hum Genet 1996;97: 124–5.
[22] Wu CY, Wu MS, Chen YJ, et al. Clinicopathological significance of urokinase-type plasminogen activator genotypes in gastric cancer. Hepatogastroenterology 2008;55:1890–4.
[23] Forsti A, Lei H, Tavelin B, et al. Polymorphisms in the genes of the urokinase plasminogen activation system in relation to colorectal cancer. Ann Oncol 2007;18:1990–4.
[24] Minisini AM, Fabbro D, Di Loreto C, et al. Markers of the uPA system and common prognostic factors in breast cancer. Am J Clin Pathol 2007;128:112–7. [25] Hsieh YS, Lee YL, Yang SF, et al. Association of EcoRI polymorphism of the
metastasis-suppressor gene NME1 with susceptibility to and severity of non-small cell lung cancer. Lung Cancer 2007;58:191–5.
[26] Hsu HS, Lee IH, Hsu WH, Kao WT, Wang YC. Polymorphism in the hMSH2 gene (gISV12-6 TNC) is a prognostic factor in non-small cell lung cancer. Lung Cancer 2007;58:123–30.
[27] Shih CM, Lee YL, Chiou HL, et al. Association of TNF-alpha polymorphism with susceptibility to and severity of non-small cell lung cancer. Lung Cancer 2006;52: 15–20.
[28] Shih CM, Lee YL, Chiou HL, et al. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer 2005;50:291–7. [29] Tseng RC, Hsieh FJ, Shih CM, Hsu HS, Chen CY, Wang YC. Lung cancer susceptibility
and prognosis associated with polymorphisms in the nonhomologous end-joining pathway genes: a multiple genotype–phenotype study. Cancer 2009;115: 2939–48.
[30] Wang YC, Chen CY, Chen SK, Chang YY, Lin P. p53 codon 72 polymorphism in Taiwanese lung cancer patients: association with lung cancer susceptibility and prognosis. Clin Cancer Res 1999;5:129–34.
[31] Xu J., Li W., Bao X., et al. Association of putative functional variants in the PLAU gene and the PLAUR gene with myocardial infarction. Clin Sci (Lond) 2010; 119:353–359.
[32] Gosselink J.V., Hayashi S., Elliott W.M., et al. Differential expression of tissue repair genes in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2010;181:1329–1335.
[33] Di Bernardo MC, Matakidou A, Eisen T, Houlston RS. Plasminogen activator inhibitor variants PAI-1 A15T and PAI-2 S413C influence lung cancer prognosis. Lung Cancer 2009;65:237–41.
[34] Tsai MH, Chen WC, Chen HY, Tsai FJ. Urokinase gene 3′-UTR T/C polymorphism is associated with oral cancer. J Clin Lab Anal 2004;18:276–9.
[35] Madsen CD, Sidenius N. The interaction between urokinase receptor and vitronectin in cell adhesion and signalling. Eur J Cell Biol 2008;87:617–29. [36] Sidenius N, Blasi F. The urokinase plasminogen activator system in cancer: recent
advances and implication for prognosis and therapy. Cancer Metastasis Rev 2003;22:205–22.
[37] Bentov Y, Brown TJ, Akbari MR, et al. Polymorphic variation of genes in thefibrinolytic system and the risk of ovarian cancer. PLoS ONE 2009;4:e5918.
[38] Lei H, Hemminki K, Johansson R, et al. PAI-1–675 4 G/5 G polymorphism as a prognostic biomarker in breast cancer. Breast Cancer Res Treat 2008;109:165–75. [39] Yang SF, Hsieh YS, Lin CL, et al. Increased plasma levels of urokinase plasminogen activator and matrix metalloproteinase-9 in nonsmall cell lung cancer patients. Clin Chim Acta 2005;354:91–9.
[40] Przybylowska K, Smolarczyk K, Kulig A, et al. Antigen levels of the urokinase-type plasminogen activator and its gene polymorphisms in colorectal cancer. Cancer Lett 2002;181:23–30.
[41] Carriero MV, Longanesi-Cattani I, Bifulco K, et al. Structure-based design of an urokinase-type plasminogen activator receptor-derived peptide inhibiting cell migration and lung metastasis. Mol Cancer Ther 2009;8:2708–17.
[42] Weidle UH, Konig B. Urokinase receptor antagonists: novel agents for the treatment of cancer. Expert Opin Investig Drugs 1998;7:391–403.
[43] Almasi CE, Hoyer-Hansen G, Christensen IJ, Pappot H. Prognostic significance of urokinase plasminogen activator receptor and its cleaved forms in blood from patients with non-small cell lung cancer. APMIS 2009;117:755–61.
[44] Ahmad A, Kong D, Wang Z, Sarkar SH, Banerjee S, Sarkar FH. Down-regulation of uPA and uPAR by 3,3′-diindolylmethane contributes to the inhibition of cell growth and migration of breast cancer cells. J Cell Biochem 2009;108:916–25. [45] Thomas C, Wiesner C, Melchior SW, et al. Urokinase-plasminogen-activator
receptor expression in disseminated tumour cells in the bone marrow and
peripheral blood of patients with clinically localized prostate cancer. BJU Int 2009;104:29–34.
[46] Kogianni G, Walker MM, Waxman J, Sturge J. Endo180 expression with cofunctional partners MT1-MMP and uPAR-uPA is correlated with prostate cancer progression. Eur J Cancer 2009;45:685–93.
[47] Illemann M, Bird N, Majeed A, et al. Two distinct expression patterns of urokinase, urokinase receptor and plasminogen activator inhibitor-1 in colon cancer liver metastases. Int J Cancer 2009;124:1860–70.
[48] Yang HC, Lin CH, Hsu CL, et al. A comparison of major histocompatibility complex SNPs in Han Chinese residing in Taiwan and Caucasians. J Biomed Sci 2006;13:489–98. C.-M. Shih et al. / Clinica Chimica Acta 412 (2011) 194–198