Annals of Microbiology, 54 (3), 289-297 (2004)
Gene cloning and biochemical characterization
of chitinase CH from Bacillus cereus 28-9
C.-J. HUANG, C.-Y. CHEN*
Department of Plant Pathology and Microbiology, National Taiwan University, No. 1, Sec. 4, Roosevelt Rd., Taipei, Taiwan 106, Republic of China
Abstract - Bacillus cereus 28-9 is a chitinolytic bacterium showing antagonistic activity
against several fungi. One chitinase of 37 kDa, named chitinase CH (ChiCH), was purified by ammonium sulphate fractionation and anion exchange chromatography. The N-termi-nal sequence of purified ChiCH was determined as ANNLGSKLLVGYWHNFD. The
chiCH (1,083 bp), cloned from the genomic DNA of B. cereus 28-9, encodes a
polypep-tide of 360 amino acids containing the N-terminal signal peppolypep-tide and a catalytic domain. ChiCH, partially purified from an Escherichia coli transformant harbouring chiCH, exhibit-ed chitinase activity with an optimal pH of 6.0 and an optimal temperature of 40 °C. This ChiCH was slightly inhibitory to conidial germination of Botrytis elliptica. It was suggested that ChiCH is one of the factor involved in the antagonism of B. cereus 28-9 toward fungi.
Key words: chitinase, ChiCH, glycosyl hydrolase family 18, gene cloning.
INTRODUCTION
Bacillus cereus is a large, Gram-positive, endospore-forming bacterium that is
very common in soils and plants (Brunel et al., 1994; Martinez et al., 2002). For plant disease control, B. cereus UW85 has been proven as a reliable biocon-trol agent of Phytophthora damping off and root rot of soybean (Emmert and Handelsman, 1999), and capable of producing two antibiotics responsible for disease suppression (Silo-Suh et al., 1994). In addition, an endophytic B.
cereus strain 65 producing a chitobiosidase is effective against Rhizoctonia solani in cotton (Pleban et al., 1997). However, the role of chitobiosidase in the
antagonism of B. cereus strain 65 toward fungal plant pathogens is not clearly understood.
In this study, we analysed the chitinases produced by a chitinolytic strain of
B. cereus and found that this B. cereus strain excreted two chitinases. One of
them was partially purified and its encoding gene was cloned. In addition, this chitinase was characterized and investigated on its antifungal activity toward
Botrytis elliptica, a fungal pathogen of lily leaf and blossom blight.
MATERIALS AND METHODS
A chitinolytic strain 28-9 was classified as Bacillus cereus / Bacillus
thuringien-sis according to carbon source utilization ability by using BIOLOG plate
(Bacte-ria & Yeast Identification System, Biolog, Inc., Hayward, CA) and identified as a strain of B. cereus by PCR analysis of a gyrase gene (Yamada et al., 1999).
Chitinase activity was determined using a fluorometric substrate, 4-methy-lumbelliferyl β-D-N, N’-diacetylchitobioside (Sigma, St. Louis, MO, USA), fol-lowing the method of Morimoto et al. (1997). One unit of chitinase activity was defined as the amount of enzyme required to release 1 µmol of 4-methylum-belliferone per min. In addition, protein concentration was measured using Bradford’s method (1976) and bovine serum albumin was used as a standard. For the purification of ChiCH, all steps were carried out at 4 °C. Bacillus
cereus 28-9 was cultured in 500 mL of M9 broth that contained 0.4% GlcNAc at
37 °C on a rotary shaker at 175 rpm for three days. The culture supernatant was collected by centrifugation at 10,000 × g for 15 min, and proteins in the su-pernatant were precipitated with ammonium sulphate at 40-70% saturation. The precipitate was dissolved in 0.1 M of Tris-HCl buffer (pH 8.0) and dialyzed overnight in the same buffer. The dialysate was loaded onto a Hyper-D anion exchange column (Sigma) and proteins were eluted with 0.1-0.5 M NaCl gradi-ent in 0.1 M of Tris-HCl buffer (pH 8.0). ChiCH was eluted with 0.1 M NaCl and fractions that exhibited chitinase activity were pooled, concentrated by ultrafil-tration through a Centriplus YM-10 membrane (10 kDa MW cut-off, Millipore, Bedford, MA, USA), and finally stored at –20 °C.
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed following the method of Laemmli (1970) using Mini-Protein II ap-paratus (Bio-Rad, Herculus, CA, USA). A separating gel (10%) containing 0.01% of glycol chitin was used for detection of chitinase activity. After elec-trophoresis, separated proteins were renatured by soaking the gel in 0.1 M ac-etate buffer (pH 5.0) containing 1% Triton X-100 at 37 °C with gentle shaking for 2 h. The gel was stained with 0.01% Calcofluor White M2R (Sigma) in 0.5 M Tris-HCl (pH 8.9). Protein bands exhibiting chitinolytic activities were visualized under a UV transilluminator (Trudel and Asselin, 1989). Proteins in the poly-acrylamide gel were stained with Coomassie Brilliant Blue G-250.
The protein was electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore), using a Mini-Electroblot apparatus (Bio-Rad). Proteins on the membrane were stained with 0.1% amido black. The protein band cor-responding to that exhibiting chitinase activity was cut out from the membrane and subjected to N-terminal amino acid sequencing by automated Edman degradation using the Applied Biosystems model 477A protein sequencer (Ap-plied Biosystems, Perkin Elmer, Foster City, Calif., USA).
The N-terminal amino acid sequence of ChiCH and the conserved se-quence of family 18 chitinases were used to design degenerated primers. Primer dchf (5’-TAITGGCAIAACTTTG-3’) corresponding to the amino acid se-quence YWHNF and primer dchr (5’-TTCITCITCIATITCTATTCC-3’) correspon-ding to the amino acid sequence G(L/I)D(L/I)DXE were used in polymerase
last cycle. Amplified DNA fragments were cloned into pGEMT-easy vector and sequenced. The insert of recombinant plasmid, encoding amino acid sequence of chitinase was used as a probe in Southern blot analysis and subsequent colony hybridisation. Probe was prepared using a PCR DIG Probe Synthesis Kit (Roche Molecular Biochemicals, Mannheim, Germany) following the method described by the manufacturer. A subgenomic library of B. cereus 28-9 was constructed in pBluescript II KS(-) and transformed into E. coli TOP10F’. After colony hybridisation, the insert DNA from a selected clone was sequenced using the ABI-310 autosequencer (Applied Biosystems).
The DNA fragment carrying the chiCH gene and 17-bp upstream region was amplified by PCR with primer chf, 5’-GTATAGGAGTGTTGATAATGTTAAA CAAG-3’, and primer chr, 5’-GTTATTTTTCGAAGGAAAGACCATC-3’. The am-plified chiCH-containing fragment was cloned into pGEMT-easy vector to cate recombinant plasmid, pGH51, and transformed into E. coli DH5α. The re-sulting E. coli DH5α(pGH51) was cultured in LB broth containing 50 µg/ml ampicillin under constant shaking at 37 °C for 20 h. The periplasmic protein of
E. coli DH5α(pGH51) was extracted following the method of Manoil and
Beck-with (1986). Purification of ChiCH from the periplasmic fraction of B. cereus 28-9 was performed by the same procedures used for purification of ChiCH from culture supernatant of B. cereus 28-9.
ChiCH purified from the periplasmic fraction of E. coli DH5α(pGH51) was used to determine the effects of pH and temperature on chitinase activity of ChiCH. Glycol chitin was used as a substrate. Chitinase activity was analysed by a procedure described by Imoto and Yogishita (1971). Hydrolysis reaction of ChiCH was performed at 37 °C for 25 min in the following buffers of 0.1 M: sodi-um citrate (pH 3-5), potassisodi-um phosphate (pH 6-7), Tris-HCl (pH 8), and glycine-NaOH (pH 9-11) buffers. Temperature effect on chitinase activity of ChiCH was measured in 0.1 M potassium phosphate buffer (pH 6.0) from 20 °C to 80 °C. In addition, substrate specificity of ChiCH was examined on soluble substrates, namely glycol chitin (Sigma), glycol chitosan (Sigma), car-boxymethylcellulose (Hayashi, Osaka, Japan), laminarin (Sigma), and soluble starch (Hayashi).
For antifungal assay, conidial suspension of Botrytis elliptica, at a final con-centration of 4 × 105conidia/ml, was mixed with purified ChiCH and incubated at room temperature for 12 h. After incubation, the percentage of germinated spores of Botrytis elliptica was calculated and the inhibition rate was used as an indication of antifungal activity. The experiment was repeated for three times and analysed statistically by Duncan’s Multiple Range Test.
The nucleotide sequence data of chiCH have been submitted to Gen-Bank/EMBL DNA Databases under accession numbers AF510723.
RESULTS AND DISCUSSION
Three chitinolytic B. cereus strains isolated from Israel (Pleban et al., 1997), United States (Wang et al., 2001), and Japan (Mabuchi et al., 2000), have been reported. These B. cereus strains produce chitinases, such as chitobiosidase of strain 65 (Pleban et al., 1997), Chi36 of strain 6E1 (Wang et al., 2001), and ChiA of strain CH (Mabuchi and Araki, 2001). In this report, we found that
ChiCH produced by the chitinolytic B. cereus 28-9 from Taiwan was similar to these chitinases. However, the biological function of this kind of chitinases from different B. cereus strains has not yet been studied. Therefore, we cloned the ChiCH-encoding gene and investigated the biological function of ChiCH of B.
cereus 28-9 herein.
B. cereus 28-9 produced at least two chitinases and secreted both enzymes
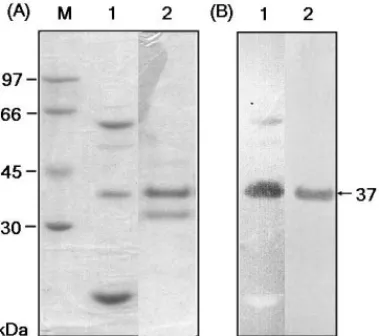
into culture medium. Fig. 1 shows the zymogram of partially purified chitinases produced by B. cereus 28-9. One chitinase with estimated molecular mass of 37 kDa was named ChiCH and its N-terminal amino acid sequence was deter-mined as ANNLGSKLLVGYWHNFD.
The N-terminal amino acid sequence of ChiCH and the conserved amino acid sequence of catalytic domains of family 18 chitinases were used to design degenerate primers, dchf and dchr. A DNA fragment of about 300 bp was am-plified and determined as the partial sequence of a chitinase gene based on se-quence analysis data. This fragment was subsequently used as a probe in Southern blot analysis and colony hybridisation.
Figure 2 shows the result of Southern blot analysis using the 300-bp DNA probe. EcoRI-digested genomic DNA of B. cereus 28-9 yielded a single band at the 1.7 kb-position. Single bands were also detected in the EcoRV- and PvuII-digested B. cereus 28-9 genomic DNA at the positions of about 4.0 kb and 8.0 kb, respectively. Therefore, EcoRI fragments (1.5-3 kb) of B. cereus 28-9 were used to construct a subgenomic library. One clone, screened from this subge-nomic library, harboured a recombinant plasmid carrying a 1.7-kb EcoRI insert. Sequence analysis revealed an open reading frame (ORF) in this region. The
FIG. 1 – SDS-PAGE and zymogram analysis. (A) Gel was stained with Coomassie Brilliant Blue G-250. (B) Chitinase activity was detected by staining the gel containing 0.01% glycol chitin with 0.01% Calcofluor. Lanes: 1, proteins tially purified from the culture supernatant of B. cereus 28-9; 2, ChiCH par-tially purified from E. coli DH5α(pGH51). ChiCH with estimated molecular mass of 37 kDa is indicated by an arrow.
FIG. 2 – Southern blot analysis of the genomic DNA of B. cereus 28-9. The genomic DNA was digested with EcoRI (lane 1), EcoRV (lane 2) and PvuII (lane 3). The Southern blot showed signals of hybridization with the 300-bp DNA probe corresponding to chiCH sequence. The estimated DNA fragment size of each signal is indicated beside the arrow.
FIG. 3 – Nucleotide and the deduced amino acid sequences of chiCH. Amino acid residues corresponding to those determined by N-terminal sequencing are in bold type. The putative active site of ChiCH is underlined. The stop codon of
chiCH is indicated by an asterisk. The putative Shine-Dalgarno sequence,
Sequence analysis indicated that chiCH gene is 1,083 bp in length with an ATG start codon and a TAA stop codon (Fig. 3). The putative Shine-Dalgarno sequence, AGGAG, was located 8 nucleotides upstream of the start codon. The deduced protein (ChiCH precursor) consisted of 360 amino acid residues with a calculated molecular weight of 39,372 and isoelectric point of 6.21.
Alignment of the deduced amino acid sequence of ChiCH precursor with the N-terminal amino acid sequence of excreted ChiCH of B. cereus 28-9 showed that the deduced amino acid sequence of ChiCH precursor contained a signal peptide which was cleaved off between Ala-27 and Ala-28 by the signal pepti-dase. The signal peptide of ChiCH produced by B. cereus 28-9 had 27 amino acid residues with the common characteristics of a signal peptide, including a positive-charged region, a hydrophobic central core and a signal peptidase recognition site, Ala-X-Ala (Perlman and Halvorson, 1983).
In addition to the N-terminal signal peptide, the deduced ChiCH contained a catalytic domain as that shown in the conserved domain database of National Center for Biotechnology (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The conserved amino acid sequence of the catalytic domain, from Gly-139 to Leu-150, was homologous to a number of family 18 chitinases (Henrissat and Bairoch, 1993) (Fig. 3). This region included two aspartate residues, Asp-141 and Asp-143, and one glutamate residue, Glu-145. These residues have also been found in ChiA1 of Bacillus circulans WL-12 (Watanabe et al., 1993) and ChiA of Serratia marcescens (Perrakis et al., 1994). Furthermore, Glu-145 in the deduced ChiCH seemed to correspond to Glu-315 of S. marcescens ChiA, which has been reported to be involved in the catalysis of chitinase (Perrakis et
al., 1994).
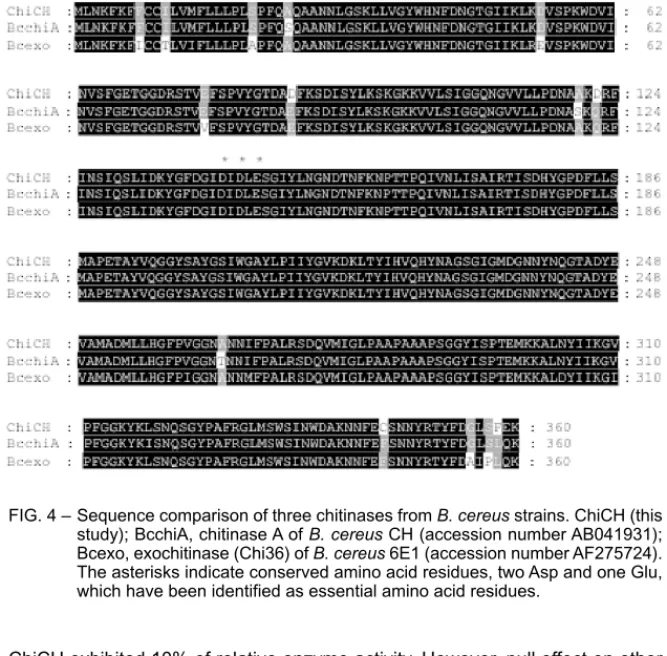
The amino acid sequence of the deduced ChiCH showed 97.5% homology to that of the ChiA of B. cereus CH (Mabuchi and Araki, 2001) and 94.7% ho-mology to that of the Chi36 of B. cereus 6E1 (Wang et al., 2001) as analysed by the comparison program in the GCG package (Fig. 4). In addition, Wang et al. (2001) have speculated that the chitobiosidase of B. cereus strain 65 is similar to Chi36 of strain 6E1 (Pleban et al., 1997; Wang et al., 2001). Although ho-mologous chitinases are distributed in different B. cereus strains from different countries, it is unclear whether this kind of family 18 chitinase gene is species-specific in B. cereus. However, a species-species-specific family 19 chitinase gene has been found in Burkholderia gladioli (Kong et al., 2001). Therefore, the distribu-tion of chiCH-like genes in B. cereus strains and other Bacillus species be-comes a subject to study.
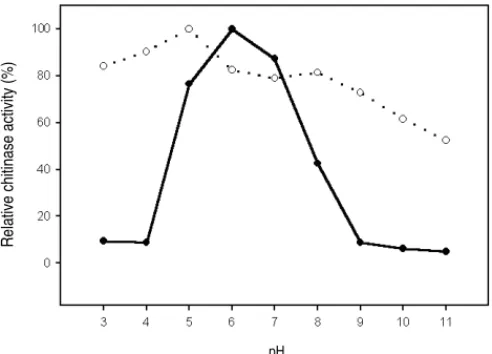
ChiCH was expressed and purified from the periplasmic fraction of E. coli DH5α(pGH51) as shown by SDS-PAGE and in-gel activity assay (Fig. 1, lane 2). A 33-kDa protein without chitinase activity as shown by in-gel activity assay was consistently co-purified with ChiCH in our procedure. Therefore, the par-tially purified ChiCH was used for further characterization. The results indicated that ChiCH had an optimal pH of 6 (Fig. 5) and an optimal temperature of 40 °C (Fig. 6). It retained over 75% of optimal activity between pH 5.0–7.0. Further-more, pH and temperature stabilities of ChiCH could be maintained in the ranges of pH 3-8 (Fig. 5) and 40-50 °C (Fig. 6), respectively.
FIG. 4 – Sequence comparison of three chitinases from B. cereus strains. ChiCH (this study); BcchiA, chitinase A of B. cereus CH (accession number AB041931); Bcexo, exochitinase (Chi36) of B. cereus 6E1 (accession number AF275724). The asterisks indicate conserved amino acid residues, two Asp and one Glu, which have been identified as essential amino acid residues.
ChiCH exhibited 10% of relative enzyme activity. However, null effect on other substrates, including laminarin (β-1,3-glucan), carboxymethyl cellulose (β-1,4-glucan), and soluble starch (β-1,4/1,6-(β-1,4-glucan), was observed.
ChiCH of 17 µunit inhibited about 10% of the conidial germination of
Botry-tis elliptica and the inhibition level was not augmented by increasing chitinase
activity from 17 to 66 µunit. On the other hand, inhibition of the conidial germi-nation of Botrytis elliptica was much stronger (55.2% of inhibition) by the cul-ture supernatant of B. cereus 28-9 at 20 µunits than by the partially purified ChiCH from E. coli DH5α(pGH51).
According to the study of Pleban et al. (1997), a chitobiosidase is present in the endophytic B. cereus strain 65. They have suggested that chitobiosidase activity is important for antifungal activity of strain 65. Our present work indicat-ed that ChiCH possibly has antifungal activity of mild strength. Since this anti-fungal level was much lower than that exhibited by the culture supernatant of
B. cereus 28-9, we presume that not only ChiCH but also other antifungal
fac-tors were produced by B. cereus 28-9 and exhibited synergistic or combined ef-fect against target fungi.
Acknowledgements
This work was financially supported by National Science Council, Taiwan, Re-public of China.
REFERENCES
Bradford M.M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., FIG. 5 - Effect of pH on the chitinase activity and stability of ChiCH. ChiCH partially pu-rified from E. coli DH5α(pGH51) was assayed for the effect of pH on its en-zyme activity (solid circle) and stability (open circle).
FIG. 6 - Effect of temperature on the chitinase activity and stability of ChiCH. ChiCH partially purified from E. coli DH5α(pGH51) was assayed for the effect of tem-perature on its enzyme activity (solid circle) and stability (open circle).
pH
Temperature (°C)
Relative chitinase activity (%)
Emmert E.A.B., Handelsman J. (1999). Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett., 171: 1-9.
Henrissat B., Bairoch A. (1993). New families in the classification of glycosyl hydro-lases based on amino acid sequence similarities. Biochem. J., 293: 781-788. Imoto T., Yogishita K. (1971). A simple activity measurement of lysozyme. Agric. Biol.
Chem., 35: 1154-1156.
Kong H., Shimosaka M., Ando Y., Nishiyama K., Fujii T., Miyashita K. (2001). Species-specific distribution of a modular family 19 chitinase gene in Burkholderia gladioli. FEMS Microbiol. Ecol., 37: 135-141.
Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227: 680-685.
Mabuchi N., Araki Y. (2001). Cloning and sequencing of two genes encoding chitinas-es A and B from Bacillus cereus CH. Can. J. Microbiol., 47: 895-902.
Mabuchi N., Hashizume I., Araki Y. (2000). Characterization of chitinases excreted by
Bacillus cereus CH. Can. J. Microbiol., 46: 370-375.
Martinez C., Michaud M., Belanger R.R., Tweddell R.J. (2002). Identification of soils suppressive against Helminthosporium solani, the causal agent of potato silver scurf. Soil Biol. Biochem., 34: 1861-1868.
Manoil C., Beckwith J. (1986). A genetic approach to analyzing membrane protein topology. Science, 233: 1403-1408.
Morimoto K., Korita S., Kimura T., Sakka K., Ohmiya K. (1997). Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J. Bacteriol., 179: 7306-7314.
Perlman D., Halvorson H.O. (1983). A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J. Mol. Biol., 167: 391-409.
Perrakis A., Tews I., Dauter Z., Oppenheim A.B., Chet I., Wilson K.S., Vorgias C.E. (1994). Crystal structure of a bacterial chitinase at 2.3Å resolution. Structure, 2: 1169-1180.
Pleban S., Chernin L., Chet I. (1997). Chitinolytic activity of an endophytic strain of
Bacillus cereus. Lett. Appl. Microbiol., 25: 284-288.
Silo-Suh L.A., Lethbridge B.J., Raffel S.J., He H., Clardy J., Handelsman J. (1994). Bi-ological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol., 60: 2023-2030.
Trudel J., Asselin A. (1989). Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal. Biochem., 178: 362-366.
Watanabe T., Kobori K., Miyashita K., Fujii T., Sakai H., Uchida M., Tanaka H. (1993). Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus
circulans WL-12 as essential residues for chitinase activity. J. Biol. Chem., 268:
18567-18572.
Wang S.Y., Moyne A.L., Thottappilly G., Wu S.J., Locy R.D., Singh N.K. (2001). Purifi-cation and characterization of a Bacillus cereus exochitinase. Enzyme Microbial Technol., 28: 492-498.
Yamada S., Ohashi E., Agata N., Venkateswaran K. (1999). Cloning and nucleotide se-quence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B.
anthracis and their application to the detection of B. cereus in rice. Appl. Environ.