ORIGINAL PAPER
Qualitative analysis of the fluorophosphonate-based
chemical probes using the serine hydrolases from mouse
liver and poly-3-hydroxybutyrate depolymerase (PhaZ)
from Bacillus thuringiensis
Yi-Long Huang&Tsai-Wen Chung&Chia-Mao Chang&
Chih-Hau Chen&Chen-Chung Liao&Yeou-Guang Tsay&
Gwo-Chyuan Shaw&Shwu-Huey Liaw&Chung-Ming Sun&
Chao-Hsiung Lin
Received: 7 June 2012 / Revised: 3 August 2012 / Accepted: 9 August 2012 / Published online: 1 September 2012 # Springer-Verlag 2012
Abstract The serine hydrolase family consists of more than 200 members and is one of the largest enzyme families in the human genome. Although up to 50 % of this family remains unannotated, there are increasing evidences that activities of certain serine hydrolases are associated with diseases like cancer neoplasia, invasiveness, etc. By now, several activity-based chemical probes have been developed and are applied to profile the global activity of serine hydrolases in diverse pro-teomes. In this study, two fluorophosphonate (FP)-based chemical probes were synthesized. Further examination of
their abilities to label and pull down serine hydrolases was conducted. In addition, the poly-3-hydroxybutyrate depoly-merase (PhaZ) from Bacillus thuringiensis was demonstrated as an appropriate standard serine hydrolase, which can be applied to measure the labeling ability and pull-down efficien-cy of FP-based probes. Furthermore, mass spectrometry (MS) was used to identify the serine residue that covalently bonded to the active probes. Finally, these FP-based probes were shown capable of establishing the serine hydrolase profiles in diverse mouse tissues; the serine hydrolases pulled down from
Yi-Long Huang and Tsai-Wen Chung contributed equally to this work. Electronic supplementary material The online version of this article (doi:10.1007/s00216-012-6349-0) contains supplementary material, which is available to authorized users.
Y.-L. Huang
:
S.-H. Liaw:
C.-H. Lin (*)Department of Life Sciences and Institute of Genome Sciences, National Yang Ming University,
155 Linong Street, Sec. 2, 11221, Taipei, Taiwan e-mail: chlin2@ym.edu.tw
Y.-L. Huang
:
C.-H. LinInstitute of Biopharmaceutical Sciences, National Yang Ming University, 155 Linong Street, Sec. 2, 11221, Taipei, Taiwan
T.-W. Chung
:
C.-M. Chang:
C.-H. Chen:
C.-M. Sun (*)Department of Applied Chemistry, National Chiao Tung University, Science Building 2, 1001 Ta Hsueh Road, Hsinchu 300, Taiwan
e-mail: cmsun@mail.nctu.edu.tw
C.-C. Liao
:
Y.-G. Tsay:
C.-H. LinProteomics Research Center, National Yang Ming University, 155 Linong Street, Sec. 2, 11221, Taipei, Taiwan
Y.-G. Tsay
:
G.-C. ShawInstitute of Biochemistry and Molecular Biology, National Yang Ming University,
155 Linong Street, Sec. 2, 11221, Taipei, Taiwan DOI 10.1007/s00216-012-6349-0
mouse liver organ were further identified by MS. In summary, our study provides an adequate method to evaluate the reac-tivity of FP-based probes targeting serine hydrolases.
Keywords Activity-based probe . Serine hydrolase . PhaZ . Esterase . Fluorophosphonate
Abbreviations
ABP Activity-based probe
ABPP Activity-based protein profiling DPP7 Dipeptidylpeptidase 7
edo Ethylenedioxy FP Fluorophosphonate
PhaZ Poly-3-hydroxybutyrate depolymerase peg Polyethylene glycol
Introduction
Activity-based protein profiling (ABPP) has been widely used as an important tool to detect global enzyme activity in bio-logical samples [1–4]. ABPP is based on the development of activity-based probes (ABPs) capable of labeling a group of activated/active enzymes by stable covalent linkage. Detailed classifications and potential applications of ABPs in drug discovery have been extensively reviewed [1,5–10]. Among
them, members of the serine hydrolase family, including ser-ine proteases, esterases, lipases, and amidases, are specific targets of fluorophosphonate (FP)-based chemical probes [11]. Since the development of the first biotin-tagged FP probe against serine hydrolases in 1999 [12], several structurally modified FP-based probes have been proposed either with [13–17] or without [18–20] a polyethylene glycol linker (Fig.1a, b). Many targets of FP probes have been identified as serine hydrolases across proteomes of mammalian tissues [21, 22], cancer cells [23], Arabidopsis thaliana [24], and Saccharomyces cerevisiae [25] (Electronic supplementary material (ESM) Table S1). The resulted global profile identi-fied by FP probes provides an opportunity to investigate the specific or conserved serine hydrolases from prokaryotes to eukaryotes. Up to this date, only a few serine hydrolases have been studied for their in vitro reactivity with FP-based probes [26]. In addition, the kinetic study of FP probes using a single protein was only reported on human sera AChE and BChE [27], but systematic kinetics analysis of serine hydrolase probe using a standard enzyme remains unexplored. In the present study, we reported that a recently identified poly-3-hydroxybutyrate depolymerase (PhaZ) from Bacillus thurin-giensis [28] reacts well with FP probes in vitro. In addition, we have prepared two hydrophilic FP-based probes, FP-edo-biotin and FP-edo-fluorescein, with an ethylenedioxy (edo) group in the linker region (Fig. 1c). The kinetic studies of
Fig. 1 Chemical structures of FP-based activity probes in the previous and present studies. a FP probes with R substituent as
biotin [12], fluorescein [12], or
rhodamine [22]. b FP-peg
probes with R substituent as
biotin [15], rhodamine [33], or
fluorescein [33]. c FP-edo
probes with R substituent as biotin or fluorescein
these probes using PhaZ were carried out to illustrate their characteristics. Furthermore, 30 serine hydrolases could be identified in mouse liver homogenate by FP-edo-biotin and LC-MS/MS. Most of these proteins were also reported by an original FP–biotin probe [29], indicating that FP-edo-probes are suitable reagents for targeting serine hydrolases.
Materials and methods
Preparation of probes
Syntheses of FP-edo-biotin and FP-edo-fluorescein were performed following a previous procedure [12], with a slight modification, and described in ESM Fig. S1 and ESM
Method. FP-edo-biotin and FP-edo-fluorescein were dis-solved in chloroform to make a stock solution and then stored at−20 °C. Prior to the reaction, the stock solution was added to a clean Eppendorf tube and chloroform was removed under reduced pressure using a vacuum pump. The Eppendorf tube with the dried probe was ready for the labeling reaction with protein samples.
Recombinant expression and purification of His-tagged PhaZ
Purification of wild-type or mutant PhaZ proteins in Escher-ichia coli JM109 was performed as previously reported [28]. The final eluted fraction from Ni-NTA beads was dialyzed twice against a buffer containing 50 mM Tris (pH 8) and 150 mM NaCl to remove excess imidazole. The resulting protein can be stored at 4 °C for up to 6 months with minimal loss of enzyme activity.
Labeling reaction
The purified wild-type or S102A mutant PhaZ was adjusted to 1μM (0.034 mg/mL) using a buffer containing 50 mM Tris– Cl (pH 8) and 150 mM NaCl. Proteome samples from mouse tissues were adjusted to 1 mg/mL using 50 mM Tris–Cl (pH 8) buffer. Protein solution was added into the probe-containing Eppendorf tube and allowed to react at 25 °C for a selected period for the concentration- or time-dependent experiments. SDS-PAGE sample buffer was then added to stop the reaction. For pH profiling experiment, each pH buffer (50 mM Tris, 50 mM CAPS, and 50 mM citrate) was adjusted by HCl or NaOH to a different pH value and then mixed with the protein stock solution to a dilution of at least ten fold.
Colloidal Coomassie brilliant blue staining
First, the gel was fixed in buffer containing 50 % methanol and 10 % acetic acid for at least 30 min. Next, the gel was stained with colloidal Coomassie solution (3 % acid
phosphoric, 17 % (NH4)2SO4, 30 % methanol, and 0.1 %
Coomassie brilliant blue G250) for 12–16 h. The resulting gel was immersed in distilled water until the background was clear.
Western blot analysis
Protein samples were separated by SDS-PAGE, transferred to a polyvinylidene fluoride membrane, and the membrane blocked in 3 % bovine serum albumin (BSA) in a solution of Tris-buffered saline/Tween 20 (TBST). After incubation with streptavidin–horseradish peroxidase (HRP; BD Sci-ence) in 3 % BSA/TBST, the membrane was washed three times with TBST and exposed to an X-ray film. Proteins were visualized by chemiluminescence. For the His-tagged rabbit antibody, 5 % non-fat dry milk is used as the blocking solution instead of BSA in the protocol.
In-gel fluorescence scanning
SDS-PAGE gel was soaked in distilled water and then scanned using a Typhoon Trio imager (GE Healthcare). Exci-tation was provided by a 30-mW Argon ion laser (488 nm), and a 520DF30 filter for fluorescein was used to detect the fluorescence signal. Voltage was set at 400–500 V.
In-gel digestion
Protein bands of interest on the gel were excised, destained by 50 % acetonitrile/ammonium bicarbonate (25 mM), and dehydrated with 100 % acetonitrile to give a gel pellet. After incubation with ammonium bicarbonate solution containing sequencing grade modified Trypsin (Promega), the gel piece was digested at 37 °C for 12 h. The resulting peptides were extracted by sonication in 50:50 acetonitrile/water (1 % trifluoroacetic acid).
Preparation of mouse tissue proteome
After washing in PBS buffer, mouse tissue samples were subjected to cell disruption using a motor-driven Potter– Elvehjem tissue homogenizer (Wheaton) in buffer contain-ing 50 mM Tris–Cl (pH 8) and sucrose (0.3 M). Next, the homogenate was centrifuged sequentially at 1,100×g (10 min), 13,000×g (20 min), and 13,000×g (30 min). The final supernatant was collected and quantified using a pro-tein assay rapid kit (Wako Chemicals) as the cytosolic fraction to be treated with FP probes.
Neutravidin pull-down for biotinylated proteins
After FP-edo-biotin reacted with the proteome (1 mg/mL) at 25 °C for 1 h, excess free probes were removed using a
YM-10 Microcon centrifugal filter device (Millipore). In addi-tion, proteins were denatured by the addition of SDS to the solution (final concentration, 0.2 %, w/v) with 10 min heat-ing at 95 °C and cooled to room temperature. Then, 100μL (50 % slurry) of Neutravidin agarose beads (Thermo Scien-tific) was added to 1 mg of the labeled proteome and incubated at 4 °C for 12–16 h. The beads were washed sequentially twice with PBS/SDS (0.2 %) and twice with PBS and eluted by heating in 2X SDS-PAGE sample buffers.
Mass spectrometry analysis (MALDI-TOF/MS)
For peptide mass fingerprinting of PhaZ, tryptic digests were spotted on Anchorchip with 2 mg/mL α-cyano-4-hydroxycinnamic acid as the matrix. Ultraflex II matrix-assisted laser desorption/ionization–tandem time-of-flight (MALDI-TOF/TOF) mass spectrometer (Bruker Daltonics) was used to analyze the peptide profile. The m/z peak list from each spectrum was generated using FlexAnalysis 2.0. Online MASCOT software (Matrixscience) with the NCBInr database was used for protein identification. Data were searched with a peptide mass tolerance of 200 ppm and trypsin with two maximum missed cleavage sites.
Mass spectrometry analysis (LC-MS/MS)
For labeling site verification and pull-down identifica-tion, tryptic digest was dried and dissolved in H2O
(0.1 % formic acid) for LTQ-Orbitrap hybrid tandem mass spectrometry (Thermo Fisher). The mass spectrom-eter was inline-coupled with an Agilent 1200 nanoflow HPLC system equipped with LC Packing C18 PepMap 100 (5-mm length, 300-μm internal diameter, and 5-μm beads) as the trap column and an Agilent ZORBAX XDB-C18 (5-cm length, 75-μm internal diameter, and 3.5-μm beads) as the separation column. An LC gradi-ent elution of peptide separation was set from 5 to 70 % acetonitrile in H2O (0.1 % formic acid) at a flow
rate of 300 nL/min for 30 min. The MS/MS spectra ranging from 200 to 2,000 m/z were acquired from the five most abundant ions in every MS scan. The mass spectra were processed by the Distiller software. Online MASCOT software (Matrixscience) with mouse-specific NCBInr database was used for protein identification and annotation. Data were searched with a precursor mass tolerance of 20 ppm and a fragment ion mass tolerance of 0.8 Da. The maximum of trypsin missed cleavage sites was set at 1. Variable modification was set to the oxidation of methionine and FP-edo-biotinylation of ser-ine. In addition, the fixed modification was set to the carbamidomethylation of cysteine.
Results and discussion
Characterization of FP probe labeling ability
A previous study showed that FP-peg-rhodamine exhibited better activity than FP-rhodamine in profiling the serine hydrolases of Arabidopsis leaf extracts [24], suggesting that the introduction of polyethylene glycol (peg) into the probe structure might increase its solubility and, thus, the contact frequency to targets. The application of FP-peg-rhodamine in profiling the serine hydrolase of proteome has been frequently reported [21, 23,25]. However, a hydrophobic pocket is usually preferred for the catalytic mechanism of serine hydrolases [30], and the hydrophilicity of polyethyl-ene glycol in the active region of the probe structure may cause high entropy during enzyme/probe interaction. We thus designed and prepared the biotin and FP-edo-fluorescein, which both contain an edo structure in the linker region, to maintain hydrophilicity without affecting the part that interacts with the hydrophobic pocket of the enzyme (Fig. 1 and ESM Fig. S1). The in vitro reaction of these probes with trypsin and other enzymes demonstrated their adequate ability toward serine hydrolases (data not shown). Among the diverse enzymes capable of reacting with FP-edo-probes, the PhaZ of B. thuringiensis [28] showed com-parable reactivity with trypsin. Notably, trypsin has been known to cause autocleavage of itself during probe labeling reaction and is not suitable to serve as a standard protein to monitor probe activity. Conversely, PhaZ showed great sta-bility during the probe labeling reaction and is further ex-amined for its reaction details with edo-biotin and FP-edo-fluorescein.
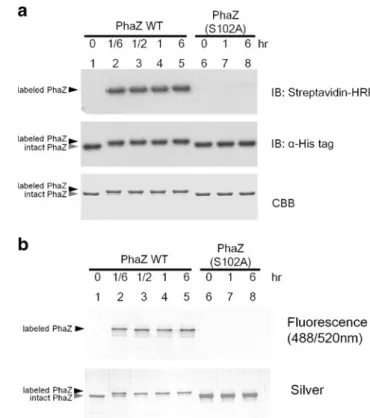
Labeling of PhaZ by FP-edo-biotin or FP-edo-fluorescein is dose-dependent
PhaZ was labeled with FP-edo-biotin at diverse ratios, in-cluding 1:1, 1:2, 1:4, and 1:8, for 30 min at 25 °C and analyzed in SDS-PAGE, as shown in Fig. 2a. Notably, a higher band shift of PhaZ on PAGE was observed upon its labeling with FP-edo-biotin, which could be used to monitor reaction progress. In addition, probe-induced biotinylation of PhaZ could be detected using streptavidin–HRP blotting. The results showed that the ratio of 1:8 PhaZ/probe reaction gave the strongest signal, indicating that probe labeling is dose-dependent. On the other hand, either using an activity-dead mutant PhaZ (S102A) or a preheating protocol prior to labeling both abolished the band shift and biotin intensity completely, indicating that such probe labeling of PhaZ only occurred on the catalytic serine residue and is activity-dependent. Collectively, we have demonstrated that FP-edo-biotin is able to label PhaZ and induces a band shift of the probe-labeled PhaZ. To our knowledge, this special
phenomenon has never been reported in serine hydrolase labeling with any FP-based probes. Similarly, FP-edo-fluorescein could also label PhaZ in the same way, as shown in Fig.2b.
Labeling of PhaZ by FP-edo-biotin or FP-edo-fluorescein is time-dependent
PhaZ was further labeled with FP-edo-biotin at a 1:4 ratio of protein/probe for 10, 30, 60, and 360 min at 25 °C and analyzed in SDS-PAGE, as shown in Fig.3a. The labeling started efficiently and gave a product (the shifted band) within 10 min. Conversely, the PhaZ (S102A) mutant exhibited a very weak signal after a 6-h incubation. Time-dependent experiments of FP-edo-fluorescein also showed similar results (Fig. 3b). In addition, the esterase activ-ity of PhaZ upon probe labeling was further determined using p-nitrophenylacetate (pNPA), a pan substrate of esterase. As shown in ESM Fig. S2, the esterase activ-ity of PhaZ was indeed blocked by FP-edo-biotin, whereas PhaZ without treatment showed normal ester-ase activity. In addition, S102A PhaZ, a previously known activity-null mutant, was used as a negative
control to represent the auto-hydrolysis of pNPA in the experiment. Since the preparation of PhaZ is rela-tively simple and easy, its efficient labeling by
FP-Fig. 3 Time course analysis of PhaZ labeling by FP-edo-biotin and
FP-edo-fluorescein. a Wild-type PhaZ or the S102A mutant (6μM)
was incubated with FP-edo-biotin (24μM) for various times, followed
by immunoblotting with anti-His antibody and streptavidin–HRP.
Coo-massie blue staining was performed in another gel with the same
loading. b Wild-type or S102A PhaZ (6μM) was incubated with
FP-edo-fluorescein (24μM) for various times, followed by fluorescence
scanning. The gel was further stained by silver staining Fig. 2 Concentration-dependent labeling of PhaZ by FP-edo-biotin
and FP-edo-fluorescein. a Wild-type PhaZ or the S102A mutant
(6 μM) was incubated with various amounts of FP-edo-biotin,
fol-lowed by immunoblotting with anti-His antibody and streptavidin–
HRP. Coomassie blue staining was performed in another gel with the
same loading. b Wild-type or S102A PhaZ (6μM) was incubated with
various amounts of FP-edo-fluorescein, followed by fluorescence scan-ning. The gel was further stained by silver staining
Fig. 4 Pull-down of probe-labeled PhaZ by Neutravidin agarose
beads. Wild-type PhaZ (6 μM) was incubated with FP-edo-biotin
(24μM) for 30 min, followed by affinity purification using
Neutravi-din agarose beads. SDS-PAGE analysis followed by Coomassie blue staining and Western blot showed that probe-labeled PhaZ was present in the eluted fraction, but not in flow-through
based probes makes it a suitable standard serine hydro-lase for measuring the kinetics of probe activity.
Enrichment of FP-edo-biotinylated proteins using Neutravidin agarose beads
The FP-edo-biotinylated PhaZ was further examined to de-termine whether it can be pulled down by Neutravidin agarose beads. The results showed that the labeled PhaZ was mostly present in the elution buffer, whereas it was hardly detectable in the flow-through fraction, indicating that the biotin-mediated enrichment of PhaZ was effective (Fig.4). In addition, FP-edo-biotin was also investigated to determine whether it can enrich PhaZ from a complex protein mixture. Our results showed that probe-labeled PhaZ could be successfully pulled down from the PhaZ-overexpressing E. coli, while other E. coli serine hydrolases were not significantly enriched (ESM Fig. S3). Although at least 40 serine proteases are encoded in E. coli [31], the absence of these enzymes in the elution fraction is probably due to their relatively low abundance compared to the large-ly induced expression of PhaZ in E. coli. Collectivelarge-ly, our
data suggested that activity-based biotinylation by FP-edo-biotin provides a basis to further enrich its targeted serine hydrolases.
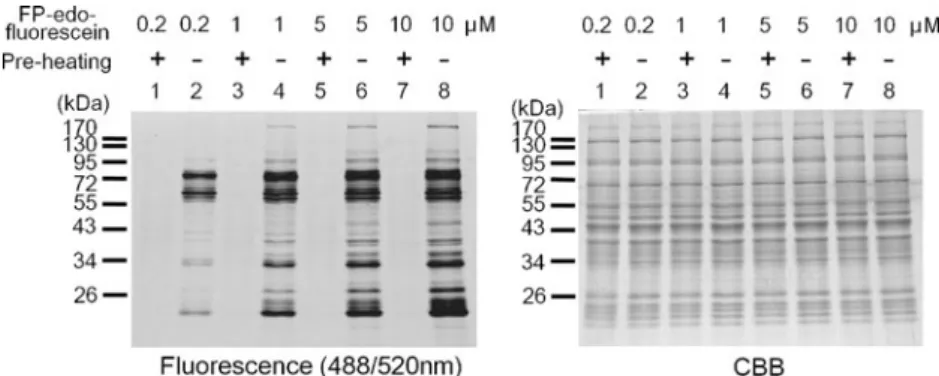
Establishment and optimization of serine hydrolase profiling by FP-edo-fluorescein
Activity measurement of diverse serine hydrolases in bio-logical samples provides a profile to reflect the enzymatic status. Accordingly, the highly sensitive FP-edo-fluorescein was used to establish the fluorescence-based profiles on various tissue samples. As shown in Fig. 5, the activities of serine hydrolases in mouse liver tissue were indicated by fluorescent signals from the labeling of FP-edo-fluorescein on protein targets within an hour, whereas preheating abol-ished these signals. In addition, increasing the dose of FP-edo-fluorescein resulted in more and stronger bands on SDS-PAGE, as anticipated. On the other hand, phenylme-thanesulfonyl fluoride (PMSF) is a broad serine protease inhibitor. We further determined whether PMSF can deacti-vate the FP probe labeling of serine hydrolase in liver proteome. Our results showed that PMSF indeed blocked
Fig. 5 Concentration-dependent labeling of mouse liver proteome by FP-edo-fluorescein. Cytosolic fraction of the liver tissue lysate was
respectively incubated with 0.2, 1, 5, or 10μM of FP-edo-fluorescein
for 60 min and resolved by SDS-PAGE. Fluorescent proteins were
detected by laser excitation at 488 nm and emission at 520 nm. The gel was further stained with Coomassie blue to indicate the equal quantity of protein loading in each lane
Fig. 6 pH-dependent labeling of mouse liver proteome by FP-edo-fluorescein. Cytosolic frac-tion of the liver tissue lysate was
incubated with 10μM
FP-edo-fluorescein at pH 6, 7, 8, 9, or 10 for 60 min and resolved by SDS-PAGE. Fluorescence signals were detected as previously de-scribed. Black arrowheads indi-cate the potential serine hydrolases with optimal activity at high pH, whereas gray arrowheads indicate proteins with preference at a low pH
the probe labeling of several serine hydrolases in liver proteome, as shown in ESM Fig. S4. The PMSF-resistant activity for the remaining serine hydrolase may be due to the presence of lipases and esterases rather than proteases.
Fur-thermore, protein labeling of FP-edo-fluorescein in mouse liver tissue was performed at diverse pH conditions. As shown in Fig. 6, the activity profiles at diverse pH values were slightly different, given that a few enzymes favored
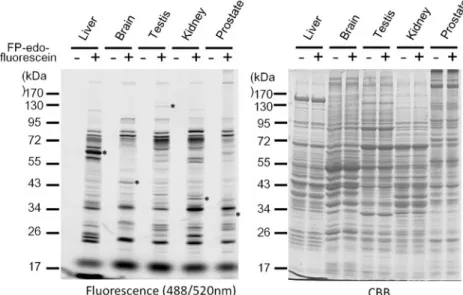
Fig. 7 Tissue profiling of serine hydrolases by FP-edo-fluorescein. Protein samples from diverse mouse tissues (1 mg/mL) were incubated with
10μM FP-edo-fluorescein for
60 min and resolved by SDS-PAGE. Fluorescent signals were detected as previously de-scribed. Asterisks indicate the tissue-specific fluorescent signals
Fig. 8 Identification of FP probe-labeled peptides in PhaZ. a Unlabeled PhaZ was digested and analyzed by MALDI-TOF/ MS. A total of 12 peptides in the spectrum were matched to the PhaZ of B. thuringiensis (58 % coverage). b The upper, middle, and lower panels re-spectively represent the spec-trum of PhaZ before and after treatments of FP-edo-biotin or FP-edo-fluorescein. The signal of the active site peptide
(resi-dues 96–121) was significantly
decreased after both FP probe labeling. c Peptide labeled by either biotin or FP-edo-fluorescein was identified as m/ z3,448.66 and 3,580.81, re-spectively. The intensity of these peptides was normalized based on another PhaZ peptide (residues 64–95)
acidic pH whereas several enzymes exhibited an increased activity upon the elevation of pH. However, the signals observed at a high pH may be due to the increase of nonspecific labeling caused by alkaline conditions as such nonspecific labeling persisted even after the preheating pro-cedure. In summary, our results demonstrated that the use of FP-edo-fluorescein could easily and rapidly establish a fluorescence-based profile of serine hydrolase activities. Moreover, it is anticipated that certain enzymes may not have optimal activity at physiological pH. Activity labeling of FP-edo-fluorescein at diverse conditions helps identify such serine hydrolases for their physiological preferences. Finally, FP-edo-fluorescein was applied to establish the individual serine hydrolase profile of diverse mouse tissues at pH 8. As shown in Fig.7, such FP probe not only could display cellular serine hydrolases in diverse tissues but also was able to recognize tissue-specific enzymes. Interestingly,
a comparison among these profiles indicated that several serine hydrolases are particularly active in certain tissues, which might not be revealed by a traditional expression-based study. Since the physiological functions of diverse tissues are different, the identification of tissue-specific ser-ine hydrolases provides a strategy to illustrate functional regulation in these tissues.
Verification of the probe labeling of protein
It has been reported that S102A mutation of PhaZ lost its enzymatic activity [28], and so was its labeling with FP probes in our experiments (Figs.2 and 3). However, point mutation of the conserved serine residue other than the catalytic serine can also lead to the abolishment of the enzyme activity as well as the ability to react with the probe [32]. To rule out this possibility and verify whether the FP
Table 1 List of the identified serine hydrolases from the mouse liver proteome by FP-edo-biotin
Description Accession no. Mass (Da) Gene name Mascot score Peptide no.
Fatty acid synthase 148702861 276,695 FASN 141 2
Dipeptidyl peptidase 9 26347125 99,013 DPP9 30 1
Prolyl endopeptidase 6755152 81,669 PREP 307 7
Acylpeptide hydrolase 19343726 80,767 APEH 234 4
Butyrylcholineesterase 416798 68,990 BCHE 149 1
Fatty-acid amide hydrolase 13905142 64,206 FAAH 68 1
Liver carboxylesterase 31 29476863 63,790 ES31 1,251 18
Liver Carboxylesterase 1 148679152 63,170 CES1 386 10
Carboxylesterase 2 G 20072612 62,980 CES2G 231 7 Carboxylesterase 2 21704206 62,714 CES2 878 15 Carboxylesterase 5 27370126 62,676 CES5 824 13 Carboxylesterase 6 19527178 62,356 CES6 554 8 Carboxyesterase 2B 37718991 62,172 CES2B 434 9 Carboxylesterase 3 14269427 62,133 CES3 697 12 Carboxylesterase ML1 21450339 61,972 CES1F 1,132 23
Liver carboxylesterase 22 19526804 61,828 ES22 329 8
Sialate O-acetylesterase 1373055 61,522 SIAE 60 1
Liver carboxylesterase N 192854 61,387 ES1 449 9
Liver carboxylesterase 31-like 81915140 58,529 CES3B 656 12
Dipeptidyl-peptidase 2 13626390 56,804 DPP7 87 2
Cathepsin A 12860234 54,423 CTSA 62 1
Lysosomal Pro-X carboxypeptidase 20072291 51,454 PRCP 141 2
Carboxylesterase 2 F 109730703 49,194 CES2F 112 3
Arylacetamide deacetylase 13184050 45,392 AADAC 163 2
S-formylglutathione hydrolase 12846304 35,569 ESD 97 3
Kynurenine formamidase 21746157 34,322 AFMID 90 2
Monoglyceride lipase 6754690 33,708 MGLL 83 2
Isoamyl acetate-hydrolyzing esterase 1 27754071 28,412 IAH1 242 6
Platelet-activating factor acetylhydrolase 1Bβ subunit 1373363 25,647 PAFAH1B2 60 1
probe directly labeled to active site serine, the MALDI-TOF/MS analysis was used to identify and compare the peptide contents of proteins from the shifted and non-shifted PhaZ bands. As shown in Fig.8a, the tryptic pep-tides were identified with a 58 % sequence coverage of PhaZ. In Fig. 8b, c, the parental peptide 96–121 (m/z 2,830.39) decreased upon probe labeling, but new peptides whose mass matched the addition of probe to the parental peptide (m/z3,448.66 for FP-edo-biotin and m/z3,580.81 for FP-edo-fluorescein) were identified. Furthermore, labeling of FP-edo-biotin on the Ser102 residue of PhaZ was con-firmed from the peptide fragmentation pattern generated by LC-MS/MS analysis (ESM Fig. S5). To determine whether other serine residues in PhaZ were labeled by FP-edo-biotin, a systematic search of serine-containing peptides with a modification of FP-edo-biotin was conducted using LC-MS/MS. A total of 36 peptides covering all 17 serine resi-dues of His-tagged PhaZ upon probe labeling were identi-fied (p<0.05), covering 98 % sequence coverage. However, only serine 102-containing peptides were found with the labeling of FP-edo-biotin, as shown in ESM Fig. S6. By far, up to 50 % of the serine hydrolase family are unknown for their substrates and functions [11]. Mass spectrometric identification of the catalytic serine responsible for probe labeling of serine hydrolases in biological samples may increase the understanding of their enzyme chemistry and putative function.
Identification of serine hydrolases in mouse liver
To evaluate the ability of FP-based probes in the iden-tification of endogenous serine hydrolases, mouse liver tissue was used as a model proteome. As shown in Table 1, a total of 30 cytosolic serine hydrolases were identified using LC-MS/MS, most of which were among the previously reported 47 enzymes [29]. Nota-bly, the identified dipeptidylpeptidase 7 (DPP7) in this study has not been reported in liver. However, it is anticipated that a slight change in the chemical struc-ture of FP-based probes might alter their targeting specificity. About half of 30 serine hydrolase targets are carboxylesterases (EC 3.1.1.1), which are involved in xenobiotic detoxification and prodrug activation. The broad reactivity of carboxylesterase has been stated and may account for the abundant presence of these enzymes in the targets of FP-based probes in this study and others [29].
Conclusions
In the present study, we have shown that PhaZ reacted efficiently and effectively with FP-based probes and
exhibited a gel shift on SDS-PAGE. These features make this E. coli-produced esterase suitable as a standard serine hydrolase for evaluating the activity and kinetics of FP-based chemical probes, which makes it possible to compare the activities among diverse probes for establishing the basis to develop better probes. In addition, two new hydrophilic FP-based probes, FP-edo-fluorescein and FP-edo-biotin, were prepared and demonstrated to be capable of labeling as well as the pull-down of their serine hydrolase targets. The better solubility of these probes is anticipated to provide more identification of serine hydrolase targets. Moreover, the activity profiles of serine hydrolases in diverse mouse tissues were established using FP-edo-fluorescein, which provides a comparative basis for the enzymatic activity among different samples. Finally, the pull-down of FP-edo-biotin-labeled proteins followed by mass spectrometric analysis were shown to be feasible for identifying active serine hydrolases in mouse liver. Since a large portion of serine hydrolases remained uncharacterized, the use of such an identification strategy between normal and diseased sam-ples is expected to discover abnormally regulated serine hydrolases which might be associated with diseases and should be further investigated.
Acknowledgments This work was supported by the National
Sci-ence Council of Taiwan, R.O.C. (NSC 97-2113-M-010-001 and NSC
98-2311-B-010-003-MY3) and the Ministry of Education for“Grant of
Aim for Top University Project.”
References
1. Cravatt BF, Wright AT, Kozarich JW (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu
Rev Biochem 77:383–414
2. Barglow KT, Cravatt BF (2007) Activity-based protein profiling
for the functional annotation of enzymes. Nat Methods 4(10):822–
827
3. Jessani N, Cravatt BF (2004) The development and application of methods for activity-based protein profiling. Curr Opin Chem Biol
8(1):54–59
4. Berger AB, Vitorino PM, Bogyo M (2004) Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am J Pharmacogenomics 4
(6):371–381
5. Evans MJ, Cravatt BF (2006) Mechanism-based profiling of en-zyme families. Chem Rev 106(8):3279–3301
6. Fonovic M, Bogyo M (2008) Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev Proteomics
5(5):721–730
7. Hagenstein MC, Sewald N (2006) Chemical tools for
activity-based proteomics. J Biotechnol 124(1):56–73
8. Jeffery DA, Bogyo M (2003) Chemical proteomics and its
appli-cation to drug discovery. Curr Opin Biotechnol 14(1):87–95
9. Kozarich JW (2003) Activity-based proteomics: enzyme chemistry
redux. Curr Opin Chem Biol 7(1):78–83
10. Uttamchandani M, Li J, Sun H, Yao SQ (2008) Activity-based protein profiling: new developments and directions in functional
11. Simon GM, Cravatt BF (2010) Activity-based proteomics of en-zyme superfamilies: serine hydrolases as a case study. J Biol Chem
285(15):11051–11055
12. Liu Y, Patricelli MP, Cravatt BF (1999) Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A 96 (26):14694–14699
13. Everley PA, Gartner CA, Haas W, Saghatelian A, Elias JE, Cravatt BF, Zetter BR, Gygi SP (2007) Assessing enzyme activities using stable isotope labeling and mass spectrometry. Mol Cell
Proteo-mics 6(10):1771–1777
14. Aminoff D, Bochar DA, Fuller AA, Mapp AK, Showalter HD, Kirchhoff PD (2009) Research into selective biomarkers of eryth-rocyte exposure to organophosphorus compounds. Anal Biochem
392(2):155–161
15. Kidd D, Liu Y, Cravatt BF (2001) Profiling serine hydrolase
activities in complex proteomes. Biochemistry 40(13):4005–4015
16. Gillet LC, Namoto K, Ruchti A, Hoving S, Boesch D, Inverardi B, Mueller D, Coulot M, Schindler P, Schweigler P, Bernardi A, Gil-Parrado S (2008) In-cell selectivity profiling of serine protease inhibitors by activity-based proteomics. Mol Cell
Proteomics 7(7):1241–1253
17. Pan Z, Jeffery DA, Chehade K, Beltman J, Clark JM, Grothaus P, Bogyo M, Baruch A (2006) Development of activity-based probes for trypsin-family serine proteases. Bioorg Med Chem Lett 16 (11):2882–2885
18. Tuin AW, Mol MA, van den Berg RM, Fidder A, van der Marel GA, Overkleeft HS, Noort D (2009) Activity-based protein profil-ing reveals broad reactivity of the nerve agent sarin. Chem Res
Toxicol 22(4):683–689
19. Gershater MC, Cummins I, Edwards R (2007) Role of a carbox-ylesterase in herbicide bioactivation in Arabidopsis thaliana. J
Biol Chem 282(29):21460–21466
20. Tully SE, Cravatt BF (2010) Activity-based probes that target functional subclasses of phospholipases in proteomes. J Am Chem
Soc 132(10):3264–3265
21. Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR 3rd, Mueller BM, Cravatt BF (2004) Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A 101 (38):13756–13761
22. Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR 3rd, Jeffrey SS, Cravatt BF (2005) A
streamlined platform for high-content functional proteomics of
primary human specimens. Nat Methods 2(9):691–697
23. Jessani N, Liu Y, Humphrey M, Cravatt BF (2002) Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A 99(16):10335–10340 24. Kaschani F, Gu C, Niessen S, Hoover H, Cravatt BF, van der
Hoorn RA (2009) Diversity of serine hydrolase activities of un-challenged and botrytis-infected Arabidopsis thaliana. Mol Cell Proteomics 8(5):1082–1093
25. Baxter SM, Rosenblum JS, Knutson S, Nelson MR, Montimurro JS, Di Gennaro JA, Speir JA, Burbaum JJ, Fetrow JS (2004) Synergistic computational and experimental proteomics approaches for more accurate detection of active serine hydrolases in yeast. Mol Cell
Proteomics 3(3):209–225
26. Wang P, Shim E, Cravatt B, Jacobsen R, Schoeniger J, Kim AC, Paetzel M, Dalbey RE (2008) Escherichia coli signal peptide peptidase A is a serine-lysine protease with a lysine recruited to the nonconserved amino-terminal domain in the S49 protease
family. Biochemistry 47(24):6361–6369
27. Schopfer LM, Voelker T, Bartels CF, Thompson CM, Lockridge O (2005) Reaction kinetics of biotinylated organophosphorus toxi-cant, FP-biotin, with human acetylcholinesterase and human butyr-ylcholinesterase. Chem Res Toxicol 18(4):747–754
28. Tseng C-L, Chen H-J, Shaw G-C (2006) Identification and char-acterization of the Bacillus thuringiensis phaZ gene, encoding new intracellular poly-3-hydroxybutyrate depolymerase. J Bacteriol
188(21):7592–7599
29. Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF (2010) Superfamily-wide portrait of serine hydrolase inhibition achieved by
library-versus-library screening. Proc Natl Acad Sci U S A 107(49):20941–20946
30. Dugas H (1996) Bioorganic chemistry: a chemical approach to enzyme action, 2nd edn. Springer, New York
31. Rawlings ND, Barrett AJ, Bateman A (2012) MEROPS: the data-base of proteolytic enzymes, their substrates and inhibitors.
Nucleic acids research 40:D343–D350 (Database issue)
32. Patricelli MP, Cravatt BF (2000) Clarifying the catalytic roles of conserved residues in the amidase signature family. J Biol Chem 275(25):19177–19184
33. Patricelli MP, Giang DK, Stamp LM, Burbaum JJ (2001) Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1(9):1067–1071