Polymorphisms of GA/GT Microsatellite Loci from
Anguilla japonica
Mei-Chen Tseng,1Chaolun Allen Chen,2Hsiao-Wei Kao,2 Wann-Nian Tzeng,1and Sin-Che Lee2,*
1Department of Zoology, National Taiwan University, Taipei 10764, Taiwan 2Institute of Zoology, Academia Sinica, Taipei 11529, Taiwan
Abstract: Six novel microsatellite loci, containing (GA)15–17or (GT)10–19perfect tandem repeats, were isolated
and characterized for the catadromous eel Anguilla japonica. The allelic size of the 6 loci ranged from 79 to 226 bp in length. All loci were polymorphic with a mean number of 14.7 alleles per locus and a mean heterozygosity of 0.67, suggesting higher polymorphism than that of freshwater and anadromous fishes, but lower than that of marine fishes. Genotype diversity of the 6 loci ranged from 0.22 to 0.61 with a mean value of approximately 0.5. Cross-species amplification showed that 5 of the 6 microsatellite primers proved to be useful in addressing questions of population genetics for all Anguilla species.
Key words: microsatellite, catadromous, Anguilla japonica, polymorphism, genotype diversity.
I
NTRODUCTIONAnguilla japonica Temminck and Schlegel, 1846 is a
tem-perate freshwater eel distributed in rivers of northeastern Asian countries including Taiwan, China, Japan, and Korea (Tesch, 1977). As a catadromous fish, it experiences an interesting life cycle of spawning in the sea and growing up in rivers. The spawning grounds were inferred to be to the west of the Mariana Islands, at 15°N, 140°E (Tsukamoto, 1992), evidently from the occurrence of freshly hatched leptocephali in the area. Taking 4 to 6 months to drift passively to the coasts of northeastern Asia by the Kuroshio Current, the leptocephali undergo a metamorphosis on the way to their destinations (Cheng and Tzeng, 1996). The metamorphosis into translucent elvers is completed before
the commencement of upstream migration. Eels live in riv-ers for 5 to 20 years until their gonads begin to mature in late autumn, when they are ready to migrate downstream to the ocean for spawning (Tesch, 1977).
Japanese eel is an important aquaculture species in Tai-wan, China, Korea and Japan. In order to meet the require-ments of eel farmers in these countries, large numbers of elvers are caught from estuaries during their upstream mi-gration from November to March (Tzeng, 1985). Because of the limited knowledge of their life history and genetic popu-lation structure, some researchers have attempted to estab-lish a complete cultivation system to ensure a continuous supply of elvers. However, as far as we know, this has not yet been successful. As to the population structure, the geo-graphical clines of glass eels based on allozymes (Chan et al., 1997) and the mitochondrial DNA sequences in the D-loop regions (Sang et al., 1994) do not show interpopulational genetic variation, suggesting that it is a panmictic popula-tion with low levels of genetic structuring due to high levels of gene flow. However, the above results remain somewhat
Received October 11, 2000; accepted December 29, 2000.
*Corresponding author: telephone +886-227899520; fax +886-227858059; e-mail sclee@gate.sinica.edu.tw
controversial. Other genetic markers with high polymor-phism are needed. Because microsatellite markers offer many advantages over allozyme electrophoresis and mtDNA sequencing, the present study may reveal finer de-tails of the population structure in these fishes.
Previous studies of life cycles and population structures of aquatic animals have been limited owing to the lack of proper markers. A possible solution to the questions about the Japanese eel is the use of repetitive DNA markers char-acterized by high variability. The consistencies of microsat-ellite sequences include unique DNA sequences and short tandem repeats of 1 to 5 bases in length (Beckmann and Weber, 1992). Because they tend to have higher variability, are inherited in a Mendelian fashion, and require the least amounts of tissue for assay, microsatellites have been re-garded as the most appropriate genetic markers for answer-ing many questions of biology (Wright and Bentzen, 1994; O’Reilly and Wright, 1995). For example, O’Connell et al. (1998) used 5 highly variable microsatellite loci to investi-gate the population structure among Alaskan Pacific her-ring populations. Nevertheless, the prospective value of the microsatellite is in the search for universal primers for use among closely related taxa. Recent developments in cloning and characterization of microsatellite loci in teleost fishes have enabled scientists to resolve many problems of popu-lation biology with these new markers (Crooijmans et al., 1997; Jones and Avise, 1997; Beacham and Dempson, 1998; Roques et al., 1999; Beheregaray and Sunnucks, 2000).
The objective of this paper is to describe the character-ization of 6 polymorphic microsatellite loci in the Japanese eel genome. All primer sets of microsatellite loci may have a universal application to other freshwater eel species. These markers are expected to further unveil details of population structure, stock identification, and migratory behavior in life cycles of Anguilla species.
M
ATERIALS ANDM
ETHODSSampling
In total, 62 A. japonica elvers were collected alive in De-cember 1999 and February 2000 from two locations:
Sha-lung (121°25⬘E,25°10⬘N), in the estuary of the Tanshui
River, northwestern Taiwan, and Fangliao (120°38⬘E,
22°20⬘N), in southwestern Taiwan. Another 6 closely
re-lated congeneric species were collected: A. marmorata and
A. bicolor pacifica from Fangliao; and A. reinhardti, A. rostrata, A. nebulosa nebulosa, and A. australis australis
ob-tained abroad. All these freshwater eels were preserved in 95% ethanol.
DNA Extraction and Cloning of Microsatellites
Genomic DNA was isolated and purified from muscle tis-sues. Five hundred micrograms of tissues with 1 ml lysis buffer were digested with 55 µl proteinase K solution (10 mM Tris-HCl, pH 8.0, 2 mM ethylenediaminetetra-acetic acid [EDTA], 10 mM NaCl, 1% sodium dodecylsulfate [SDS], 10 mg/ml dithiothreitol [DTT], 0.5 mg/ml protein-ase K). DNA extraction was carried out twice with an equal volume of phenol, twice with phenol–chloroform–isoamyl alcohol (25:24:1), and once with chloroform-isoamyl (24:1) as described in Kocher et al. (1989). DNA was precipitated once with 95% ethanol, once with 70% ethanol, and then dissolved in TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) following a standard procedure (Sambrook et al., 1989).High molecular weight DNA from the genome of one adult individual was digested with AluI, HaeIII, and RsaI restriction enzymes (Stratagene, La Jolla, Calif.) according to the manufacturer’s instructions. Fragments of 200 to 800 nucleotides were gel-purified using the BIO101 Geneclean Kit (Perkin Elmer) and inserted into the SmaI blunt site of pUC 18 (Bayou Biolabs, Harahan, La.). Recombinants car-rying the fragments were transfected in the competent
Esch-erichia coli DH5␣ strain. The library was plated onto 2YT
medium plates, and colonies were lifted onto Whatman filters. The probes used for hybridization were oligo DNA
(GT)10 and (GA)10, which were labeled with 32P-␥-ATP
(Amersham). Hybridization was carried out overnight at 58°C with hybridization buffer containing 5× SSPE, 2× Denhardt’s, 0.5% SDS, and 100 µg/ml RNA (Sambrook et al., 1989). After hybridization, the filter was washed twice in primary washing buffer (1× SSC/0.2% SDS) at room tem-perature, washed once in secondary washing buffer (0.5× SSC/0.2% SDS) at room temperature, and washed twice in a third washing buffer (0.1× SSC/0.2% SDS) at the hybrid-ization temperature. The results were visualized by expo-sure of the filters to x-ray film for 3 hours at −70°C. Positive clones were sequenced on an Applied Biosystems (ABI, Fos-ter City, Calif.) automated DNA sequencer 377 (version 3.3) using a Bigdye sequencing kit (Perkin-Elmer).
Screening and Analysis of Microsatellite Loci
Pair primers of 18 to 25 nucleotides in length were designed using DNASTAR Primer Select software (version 4.0) for 12 clones containing perfect repeat sequences. Polymerasechain reaction (PCR) was performed in a volume of 5 µl including 0.1 µl genomic DNA (1–2 ng), 0.5 U Taq poly-merase (Takara), 0.125 pmol forward primer labeled with
0.3 µCi32P-␥-ATP, 1.125 pmol forward primer, 1.25 pmol
reverse primer, 5 mM dNTP, 0.05–0.1 mM MgCl2, and 0.5
µl 1% Tween 20. Amplification was performed in a PC-960G Microplate Thermal Cycler (Corbett Research) sched-uled under the following successive steps: 5 minutes at 94°C; 38 cycles of 30 seconds at 94°C, 30 seconds at T°C, and 30 seconds at 72°C, where T corresponds to the an-nealing temperatures in Table 1. In the cross-species am-plification experiment, the annealing temperature of PCR was decreased to 50°C. All species were double-checked with an additional 1 to 2 individuals.
After amplification, the PCR products were subjected to electrophoresis on an 8% denatured polyacrylamide gel and visualized by exposing the dried gel to x-ray film over-night at room temperature. The sizes of each allele were checked by comparison with a M13mp18 sequencing ladder (Amersham T7 sequenase sequencing kit). Observed values
of heterozygosity (Ho) were determined as the percentage of
observed hetgerozygous individuals divided by the number of all individuals. Expected heterozygosity was estimated as
(1 −∑Pi2), where Pi is the frequency of the ith allele
(De-Woody and Avise, 2000). The polymorphism of microsat-ellite loci was calculated as the frequency of all genotypes divided by the number of all individuals.
R
ESULTS ANDD
ISCUSSIONAmong 1362 clones screened, 52 had positive signals. After sequencing, only 38 different microsatellite sequences could be retained for use in this study. They include 4 false-positive clones, 7 GA, and 19 GT microsatellites. The re-mainder contained both GA and GT repeats. In fishes, GT repeats are more frequent than GA repeats (Estoup et al., 1993), as was found in this study. Primers were designed for only 12 of the 38 microsatellite sequences. The remaining microsatellites sequenced were not used owing to the oc-currence of a repeated site in one of the flanking regions, the short sequences (limited by the restriction site), and the imperfect and compound repeat sequences. Primer sets of 12 loci did not produce good amplifications which were probably affected by different molecular structures.
We tested different concentrations of MgCl2, annealing
temperatures, and other PCR conditions. After resolution on 4% agarose gels, the 5 loci gave PCR products of a
nonspecific amplification and an inaccurate size. However, Table
1. PCR Conditions, Sample Size, Number and Size of Alleles per Locus, Observed Heterozygosity (H o ), Expected Heterozygosity (H E ), and Polymorphism of A. japonica Microsatellite Loci Detected by PCR Amplification Locus Repeat motifs Primer sequence (5 ⬘-3 ⬘) Annealing temp. (°C) MgCl 2 (mM) No. o f samples No. o f alleles Range of allele sizes (bp) Heterozygosity Polymorphism EMBL accession no. (H o )( HE ) AJMS-1 (GT) 16 F: TCGAGACACCAGATAGTCAC 58 0.5 35 13 194–226 0.63 0.89 0.611 AJ297599 R: ACATCCTAGGCTCACACC AJMS-2 (GA) 15 F: ATTTCACGTCATCGGACCTGC 56 0.5 41 18 103–137 0.67 0.92 0.609 AJ297600 R: GCTGGGAGCGACGCTTTATC AJMS-3 (GT) 10 F: GGTATGAATGCAGGCGTTTATG 56 0.5 59 7 79–91 0.58 0.72 0.220 AJ297601 R: GCAACCGATTTGATCTCCAG AJMS-5 (GT) 15 F: CCTTCAGATTGCTAGCAC 58 0.5 28 17 117–153 0.89 0.96 0.525 AJ297602 R: CGGAGTCTAATTGTCTCCTC AJMS-6 (GT) 19 F: ACAGAGCCAGACAAACAGAC 56 1.0 52 13 85–111 0.54 0.84 0.442 AJ297603 R: GGTCAGCAAGCAAAACGAAC AJMS-10 (GA) 17 F: TGTCTAACACTAAGAAAAGGAGAGG 58 1.0 40 20 137–175 0.70 0.97 0.549 AJ297605 R: GGCTGCCAGTATCTTCTCAAAG Mean 14.7 0.67 0.88 0.50
we succeeded in testing for amplification on 7 loci that yielded a consistent PCR product corresponding to a single locus of the expected size. One of the 7 loci (AJMS-8) was monomorphic in the individuals screened (n = 20); the
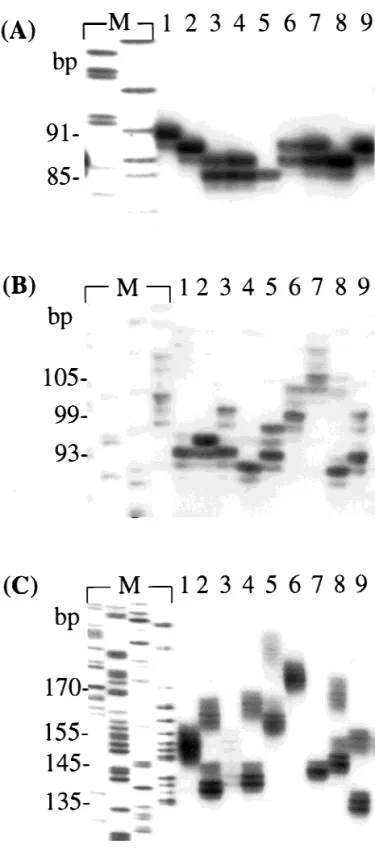
remaining 6 loci were highly variable. Primer sequences and PCR conditions of the above 6 polymorphic loci that were correctly amplified are given in Table 1. All 6 loci are vari-able, with the number of alleles ranging from 7 in AJMS-3 to 20 in AJMS-10. The sizes of the alleles in the 6 loci ranged from 79 bp in AJMS-3 to 226 bp in AJMS-1; AJMS-3 was the only locus to have an allele size shorter than 100 bp (Figure 1).
The sequence of identified microsatellites included three categories: perfect repeats, imperfect repeats, and compound repeats (Weber, 1990). These are represented in
(GT)nand (GA)nmicrosatellites cloned from the genome of
the Japanese eel. The microsatellites consist mainly of im-perfect repeat sequences (20 of 34 microsatellite sequences, or approximately 59%). The total number of repeated
mo-tifs of the core sequence ranges from 12 to 83 for (GT)n
microsatellites and from 11 to 70 for (GA)nmicrosatellites.
In total, 6 amplified loci are (GA)nor (GT)nperfect repeats.
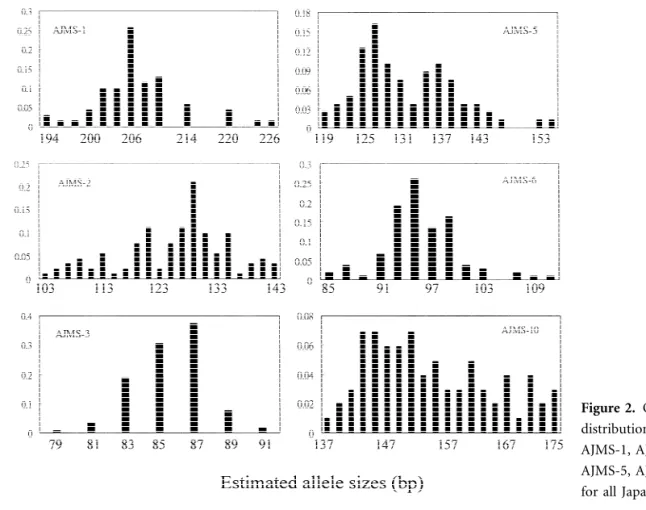
The mutations at these 6 microsatellite loci are due to the change of copy number in the 2-bp repeat unit (Figure 2). A high rate of mutation in the microsatellite loci is probably affected by the slip-strand mispairing (SSM) molecular mechanism. This model infers that the instability of the microsatellite is caused by errors derived from the elevated rate of SSM during DNA replication (Levinson and Gut-man, 1987; Goldstein and Schlotterer, 1999).
The frequency percentages of the major genotype across the above 6 loci screened ranged from 34% (AJMS-1 and AJMS-2) to 64% (AJMS-3). The mean number of al-leles per locus in the catadromous Japanese eel was esti-mated to be 14.7, which is higher than that of other fresh-water fishes (9.1) and other anadromous fishes (10.8), but lower than that of marine fishes (19.9) on average (De-Woody and Avise, 2000).
Observed heterozygosity (Ho) ranged from 0.54
(AJMS-6) to 0.89 (AJMS-5), with a mean value of 0.67,
which is smaller than the expected heterozygosities (HE) of
0.72 (AJMS-3) to 0.97 (AJMS-10), with an average of 0.88. The result shows strong heterozygote deficiencies when tested with Hardy-Weinberg equilibrium. By contrast, the significantly small mean heterozygosity (0.54) assayed in freshwater fishes and a high value of 0.77 in marine fishes (DeWoody and Avise, 2000) reveals that congeneric
An-guilla species are intermediate with respect to mean levels of
microsatellite variation. Polymorphism ranged from 0.22 (AJMS-3) to 0.61 (AJMS-1) (mean, 0.50). Six loci display high polymorphism despite only 7 alleles being found at the microsatellite AJMS-3 locus alone.
Figure 1. Genetic variability for microsatellite loci AJMS-3 (A),
AJMS-6 (B), and AJMS-10 (C). The right lanes numbered 1–9 indicate each Japanese eel sample; the left lanes marked with M represent the M13 marker.
Previous studies of microsatellite primers proved that they have a wide taxonomic range; for instance, those iso-lated from Carassius auratus may well work across a wide range of a portion of the genera in the Cyprinidae (Zheng et al., 1995). With the exception of AJMS-1, the other 5 microsatellite primers identified in A. japonica have been amplified for broad use in other congeneric species such as
A. marmorata, A. reinhardti, A. rostrata, A. nebulosa nebu-losa, A. bicolor pacifica, and A. australis australis (Table 2).
The above-mentioned 5 microsatellite loci are universally conserved in all Anguilla species. The amplification of the AJMS-1 primers did not fit in well with A. rostrata and A.
bicolor pacifica since the unique DNAs of the microsatellite
loci were deleted or degenerated by point mutations. In the long-term pursuit of excellence in microsatellite study of Anguilla eels, we provide a preliminary description of the characteristics of 6 novel microsatellite loci in the Japanese eel genome and their advantage as markers to
Table 2. Cross-Species Amplification with six Pairs of Japanese Eel (A. japonica) Microsatellite Primers*
Species
Locus
AJMS-1 AJMS-2 AJMS-3 AJMS-5 AJMS-6 AJMS-10
Anguilla japonica + + + + + + A. marmorata + + + + + + A. reinhardti + + + + + + A. rostrata − + + + + + A. nebulosa nebulosa + + + + + + A. bicolor pacifica† − + + + + + A. australis australis† + + + + + +
*Plus (+) indicates the amplification was successful; minus (−) indicates the failure of amplification (determined by agarose gel electrophoresis using ethidium bromide staining).
†Short fin eels; others are long fin eels.
Figure 2. Observed frequency distribution of alleles at loci AJMS-1, AJMS-2, AJMS-3, AJMS-5, AJMS-6, and AJMS-10 for all Japanese eel samples.
study genetic variation. The subsequent approach for us is to compare the polymorphism of microsatellite variations in several fish species with respect to a tendency of changing genetic variability in the progression among freshwater, anadromous, catadromous, and marine fishes. We have also investigated the potential value of microsatellite primers for extensive use in related taxa. The results confirm that these primers are proper genetic markers for further addressing questions of population genetics and life cycles of Anguilla species.
A
CKNOWLEDGMENTSThe authors thank Mr. C.W. Chang and Mr. Y.T. Wang for collecting specimens and other persons who provided over-seas eel samples. We are indebted to the technicians in the central laboratory for handling sequencing procedures. This study was financially supported by the National Science Council of Taiwan for a grant to S.C.L. (NSC89-2311-B-001-221).
R
EFERENCESBeacham, T.D., and Dempson, J.B. (1998). Population structure of Atlantic salmon from the Conne River, Newfoundland as deter-mined from microsatellite DNA. J Fish Biol 52:665–676. Beckmann, J.S., and Weber, J.L. (1992). Survey of human and rat microsatellites. Genomics 12:627–631.
Beheregaray, L.B., and Sunnucks, P. (2000). Microsatellite loci isolated from Odontestheses argentinensis and the O. perugiae spe-cies group and their use in other South American silverside fish. Mol Ecol 9:629–631.
Chan, I.K.K., Chan, D.K.O., Lee, S.C., and Tsukamoto, K. (1997). Genetic variability of the Japanese eel Anguilla japonica (Tem-minck & Schlegel) related to latitude. Ecol Freshwat Fish 6:45–49. Cheng, P.W., and Tzeng, W.N. (1996). Timing of metamorphosis and estuarine arrival across th dispersal range of the Japanese eel Anguilla japonica. Mar Ecol Prog Ser 131:87–96.
Crooijmans, R.P.M.A., Bierbooms, V.A.F., Komen, J., Van der Poel, J.J., and Groenen, M.A.M. (1997). Microsatellite markers in common carp. Anim Genet 23:129–134.
DeWoody, J.A., and Avise, J.C. (2000). Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56:461–473.
Estoup, A., Presa, P., Krieg, F., Vaiman, D., and Guyomard, R. (1993). (CT)n and (GT)n microsatellites: a new class of genetic markers for Salmo trutta L. (brown trout). Heredity 71:488–496.
Goldstein, D.B., and Schlotterer, C. (eds.). (1999). Microsatellite: Evolution and Applications. Oxford, U.K.: Oxford University Press. Jones, A.G., and Avise, J.C. (1997). Microsatellite analysis of ma-ternity and the mating system in the Gulf pipefish Syngnathus scovelli, a species with male pregnancy and sex-role reversal. Mol Ecol 6:203–213.
Kocher, T.D., Thomas, W.K., Meyer, A., Edwards, S.V., Paabo, S., Villablabca, F.X., and Wilson, A.C. (1989). Dynamics of mito-chondrial DNA evolution in animals: amplification and sequenc-ing with conserved primers. Proc Natl Acad Sci USA 86:6196–6200. Levinson, G., and Gutman, G.A. (1987). Slipped-strand mispair-ing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221.
O’Connell, M., Dillon, M.C., Wright, J.M., Bentzen, P., Merkouris, S., and Seeb, J. (1998). Genetic structuring among Alaskan Pacific herring populations identified using microsatellite variation. J Fish Biol 53:150–163.
O’Reilly, P., and Wright, J.M. (1995). The evolving technology of DNA fingerprinting and its application to fisheries and aquacul-ture. J Fish Biol 47(suppl A):29–55.
Roques, S., Duchesne, P., and Bernatchez, L. (1999). Potential of microsatellite for individual assignment: the North Atlantic redfish (genus Sebastes) species complex as a case study. Mol Ecol 8:1703– 1717.
Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press.
Sang, T.K., Chang, H.Y., Chen, C.T., and Hui, C.F. (1994). Popu-lation structure of the Japanese eel, Anguilla japonica. Mol Biol Evol 11:250–260.
Tesch, F.W. (1977). The Eel: Biology and Management of Anguillid Eels. London: Chapman and Hall.
Tsukamoto, K. (1992). Discovery of the spawning area for Japa-nese eel. Nature 356:789–791.
Tzeng, W.N. (1985). Immigration timing and activity rhythms of the eel, Anguilla japonica, elvers in the estuary of northern Taiwan with emphasis on environmental influences. Bull Jpn Soc Fish Ocean 47/48:11–28.
Weber, J.L. (1990). Informativeness of human (dC-dA)n䡠 (dG-dT)npolymorphisms. Genomics 7:524–530.
Wright, J.M., and Bentzen, P. (1994). Microsatellites: genetic markers for the future. Rev Fish Biol Fish 4:384–388.
Zheng, W., Stacey, N.E., Coffin, J., and Strobeck, C. (1995). Iso-lation and characterization of microsatellite loci in the goldfish Carassius auratus. Mol Ecol 4:791–792.