Biochimica et Biophysica Acta. 1088 (1991) 3t5-318 ~ 1991 Elsevier Science Publishers B.V. 0167-4781/91/$03.50 A D O N I S 0167478191000933
BBAEXP 90213 B B A R e p o r t - S h o r t S e q u e n c e - P a p e r
315
Cloning and characterization of the carp prolactin gene
H u a n g - T s u C h e n k C h i e n - S h u n C h i o u ~ a n d W e n - C h a n g C h a n g ~': I Instttute of Btoh)gtcal Chemtstrt'. Academta Szni~ct and " Institute o[ B,)chemical S(wn(~;. Natt,mal Tamar I_/n:ver.~l(v.Tuipei (Tam'an. China)
(Received 21 November 1990)
Key words: Prolactin: Genomic ~quence: Palindrome: (Carp)
A carp genomic DNA clone containing the carp prolactin (Prl) gene was isolated with carp Prl cDNA as a probe. The organization of the carp Pd gene was determined by restriction nuelease mapping and nueleotide .sequencing. The Pd gene comprises approx. 2.8 kilobasepairs (kb) of DNA including the 5'-flanking region, five exons, four introns and the 3'-flanking region. Analysis of the 5'-flanking region reveals (I) the sequence TATATAAT at positions - 38 to - 3 1 upstream from the cap site which was found to he a guanine residue, and (2) the palindrome, CTCATrGCATA- TACAAATGAG zt positions - 7 9 to - 5 9 . The carp Pd gene matches with the reported cDNA except for one difference in coding region and five in the 3'-flanking region, while the encoded amino acid sequences are identical. The arrangemeat of exons and introns is very. similar to that .seen in carp GH as well as mammalian Pd, which, however, have much longer inm,,;z.
( A ) EMBL 3 B a m H I N h e l B a m H I Nhel Left arm
-
I
i/. ht a'm
( B ) pGEM4 BamH I Sac I a t - -Cla I Hind III Sac I P s i I H~nd III Nhe OXba I
I
I
I
I
.
-4--
w ~ v
v ~ v
Fig. 1. The restnction map of carp Prl gene in EMBL 3 (A) and the sequencing strateg~ for the subeloned tragment in pGEM 4 (B) The arrows show the direction of sequencing by the chain-terrmnation method. P. TA'IA box: MCS. multiple cloning site; solid box. coding region; open box.
untranslated reg:on; lines betv.een the boxes, introns.
The sequence data in this paper have been submitted to the EMBL/Genbank Data Lib .nes under the accession number X52881. Abbreviations: Prl. prolactin: GH. growth hormone.
Correspondence: W.C. Chang. Institute of Biological Chemist~, Academia Sinica, P.O. Box 23-106, Taipei, TaJ,,~ran.
Prolactin (Prt} is a polypeptide hormone produced and secreted by the anterior pituitary gland [1]. It is a member of a family of hormones that includes Pri, growth hormone (GH) and placental lactogen. This family is believed to have evolved from a single ancestral gene by duplication and sequence divergence [2].
3 1 6 a a t a g c t c a t a a a c a t t g a t c a g c c a a a c c c t c a a a t c a a t g t t t t t c a t t a g t t t c a a g g t t t t a c t a a t a c a c c t g g a g t g c a a g a c t - 7 8 +i Gattacatatacaaatgagaggaccaagaagtgcaggcctatataatgggaagaaaatgacagagagctcaagagag G A A A T C A G C T G Ii C A A A G A T T C A A G C G A G A G A A T A A A G A A A A C A G T T A A A A T G A C T C A A G G A T C T A G A C T A T A C T T T G C A G g t a a g c a c t c 89 M T Q G S R L Y F A ctccattcctcataacatccccttttttattttctgttcatacttattagacttcagatgactatactgttattttttgaacagtttatt 1 7 9 t t t c t t g t t g c t t t t c a g T G A C T G T T C T G A T G T G T G C G T T T G T C T C C A T C A A T G G T G T C G G T C T G A A T G A T 2 5 0 V T V L M C A F V S I N G V G L N D T T A C T G G A G C G A G C C T C T C A A C T T T C A G A C A A A C T C C A C T C T C T C A G C A C T T C T C T C A C C A A T G A C 3 1 6 L L E R A S Q L S D K L H S L S T S L T N D C T G gtcagtacccttgtaaacgtgtttggtgttatatgcacaatcacttatttgcagtatgtgacttgagttttatccaaacagatatt 4 0 5 L t a t t t c a t t g a t t t g a t t g g t t g t t a g a t a t t a a c c t t g c t t t c a t c c a c a g G A T T C T C A C T T T C C T C C T G T T G G T A G G 484 D S H F P P V G R G T A A T G A T G C C T C G T C C A T C G A T G T G C C A C A C T T C C T C T C T T C A G A T T C C C A A T G A C A A A G A C C A A 5 5 0 V M M P R P S M C H T S S L Q I P N D K D Q G C C T T G A A A A T A C C G g t a a g t a a t t t t a a c t c a t c c a t c t c t a t c a a a t g c a a a a a t t a t t t c t c c t a c a c t c t c a g a a a a t a a a 6 3 5 A L Q I P atg•atgaatgctgtcactggggcagtttattttcaaaaggtacacttttgtatctttcaggcgctaatgtcgacactttaggtactaat 7 2 5 atgtaggctacctttaaggtaccaatatgaaaagagaccgcccccgcgacagctttcgtacctttttttctgagagtgttttccagcttt 8 1 5 tttattattctattatttgatttgatttttataattaaaaatattaaatattatacagcatatttaaaaaaaaatagaaatagaaaaaat 905 acattacttaattgttgttataactattatattttatatacaacaaaataaacaaaaatacaaaaacgtgatactatgatgcaagctt~c 9 9 5 aaacatgtgaagttctcataggttttttttttcattcagtctcaatccaacctattttcagcatatttcacacaaaaatgaaaattctgt 1 0 8 5 catttactcaccattgatcaaacccattagactttagttgatcctcaaaacaaaaaagaagatatttttaatgaaccacaagggatttct 1 1 7 5 gtccctccattgaaagttcattccaccaaaactttgatccatataaatcgagctgtttaatccaagtcctctaaagaaacgtcatcactt 1 2 6 5 cacatgatgaacagatttaatttaggttttattcacatatgaacaatactcaaaacac•tatagagcacattaaacaataatatatatga 1 3 5 5 gctcagctgtacttcttgcagaatcacaaatgtgtgtgacctcattggttcaaagtcttatggatttggaacaacatgaatttacatatt 1 4 4 5 tatggggtgaactttcaatgtgaagtgacaaacatctcccacttgtgtcacatttgtagttgcagaagaaacagaccagtagtgagtgaa 1 5 3 5 g t t t t a t c t g a t t g t t t g t g c a g G A G G A T G A G C T G C T G T C T T T G G C T C G G T C T C T G C T G C T G G C G T G G T C C 1 6 0 5 E D E L L S ! A R S L L L A W S G A T C C C C T C G C C C T T C T C T C C T C T G A G G C A T C C A G C C T G G C A C A T C C A G A A C G C A A C A C C A T T G A C 1 6 7 2 G D P L A L L S S E A S S L A H P E R N T I D A G C A A G A C C A A A C A G C T G C A G G A C A A C A T C A A C A G C C T G G G A G C A G G T C T G G A G C A C G T C T T T C A A S K T K E L Q D N I N S L G A G L E H V F Q 1 7 3 8 A A G g t g a t t g t a c t c a c a g t g c a t a t a t t g t g a a t g t t g a t t g g a a g a g g g c a a t a t g a a g t t a t t c a a a a t a t t g a a a g a a a t c t t t t t 1 8 2 8 K aagcaaggttggtttatacatcgtccctgtccgtgttttcttctcLacag A T G G G C T C A T C T T C A C A C A A C C T G T C C T C T 1 9 0 8 M G S S S D N L S S C T C C C T T T T T A C A C C A G C A G C C T T G G C C A G G A Y A A A A C T T C T C G A C T T G T C A A T T T C C A T T T C C T G 1 9 7 4 L P F Y T S S L G Q D K T S R L V N F H F L C T G T C C T G T T T C C G C A G G G A C T C C C A C A A A A T T G A C A G T T T C C T C A A A G T T C T C C G C T G T C G G G C A 2 0 4 0 L S C F R R D 5 H K I D S F L K V L R C R A G C C A A G A A G A G A C C T G A G A T G T G T T A G A G T G G A A A T G C T G T G C T C T G C T T C T C T C A G T G T G G A T G T T A A G T T A A A A T G G C 2 1 2 0 A K K R P E M C * * A G A G • T G T G A G T G A T T T G A A A A T C A A A T G G T A A A A T A T G G T • T T T A T T G G T T G T T T C A A A G A T A T A G A T A T T T G A T T T A C T T G A A T A A A T 2 2 1 0 • C * * G A C C T G G A C C A A C A T C A T A G A T T A • T G T A A T C T C A A A G A T A A A A G A C A • • A A A A T A A A T A C A T T T T A A A A A t t a g a a a a t t g a a a c a c a g c 2 3 0 0 C T A caggttatgcaaaatgtctgtatgcagtgatattgtatgatgaataaatgcatcgtatattgaaatgtttgactagagtaactaaattgt 2 3 9 0 atttattaaggcagttatgttggatggttaacagttattttaacattatcttctgtaatgtgatatttaaagctttgcaccaatgtcaat 2 4 8 0 aaatttctataatgcagatttatattgactcatggggaaaaaaatgtatgcatgaaaaaggactgtgaaaaaaaaattctctaccacttc 2 5 7 0 c c a c a c a a a t g t g a a c a t a c a c a g a a t a a g t g t g t a g g c t a c a c a c a c a c a c a c a c a c a c a c a c a c a c a a a c a g t t 2 6 4 6
Fig. 2. The complete nucleotide sequence of carp Prl gene. Exons are shown by capitals and introns by lower case letters. The numbering is given in the right margin and begins at the cap site ( + 1). Encodc~ amino acid residues are represented by one-letter codes below the codons. The bases in mRNA [17} are shown below the genomic sequence at positions of discrepancy indicated by asterisks. The upstream palindrome, TATA box and
317
Prl gene expression is under complex hormonal con-
trol. Many studies had shown that thyroid hormone, glucocorticoids, dopamine, etc. [3-7] directly regulate the levels of Prl a n d / o r G H mRNA. Prl and G H transcription is generally regulated in opposite direction by most of the above-mentioned hormones, providing a very interesting system to study the mechanisms of hormonal control of gene expression.
In vertebrates, Prl is involved in many physiological functions, the best known being lactogenesis [8]. In teleosts, the hormone is implicated in osmoregulation, electrolyte balance and many other functions [9,10]. Prunet et al. [11] found that Prl expression in rainbow trout might be effected by changes in water salinity. However, there is still little information concerning the role of Prl in fish adaptation to new osmotic environ- ments [12,13]. These features make Prl an interesting gene to study, especially with respect to the potential control sequence that may be located in the 5'-flanking region of this gene [14,16]. Analysis of the Prl gene could provide some insight into the structures which are involved in the gene expression. Furthermore, lifts may
o
"%j.
Fig. 3. Deternunation of the transcription start site. Lanes G. A. T and C indicate the size markers from dideoxy sequencing reaction products of vector mpl8 as template with a suitable printer. Lane P is the primer-extension products generated using synthetic oligonucleo- tide primer and mRNA. The asterisk indicates the guanine residue at
transcription start site.
contribute to an understanding of mechanisms which facilitate the differential expression and regulation of
Prl gene.
As an integral part of our studies on the structure, function and molecular biology of fish pituitary hormones [17-19], the carp Prl gene was cloned, se- quenced and characterized. These data will be essential for further studies on the regulation and functions of
Prl gene in fish.
Eggs collected from common carp (Cyprinus carpio) were immediately frozen in liquid nitrogen and stored at - 7 0 o C. Genomic DNA was prepared from the carp eggs and partially digested with BamHl. Fragments of 10-20 kb were recovered from 0.5% agarose gel and ligated with h phage EMBL3 arms. In vitro package and transfection were carried out according to the pro- toco!s of the commercial supplier (Promega).
The carp genomic library was screened with 32p. labeled carp Frl e D N A [17] as probe. The positive clones were digested with suitable restriction enzymes and subcloned in plasmid pGEM 4. Recombinant phage and plasmid were purified by the standard procedures [20]. D N A sequence was determined by chain-termina- tion method using denatured ptvsmid templates [21].
The transcription start site was determined by primer extension using carp pituitary mRNA ~s template. The m R N A was prepared as previously described [19] and annealed to a synthetic primer 5'-GTCTAGATCCT- T G A G T C A T - 3 ' which is complementary to the first six codons of exon 1 "ha the genomic sequence (Fig. 2). The primar extension was carried out w~th the mouse Molo- ney leukemia virus reverse transcriptase. The 32 P-labeled reaction products were ana137.ed by electrophoresis in 10% polyacrylamide sequencing gel.
From a library of 2-105 plaques, one clone was found to hybridize strongly with the radiolabeled e D N A probe. Restriction enzyme mapping and hybrid~ation with the e D N A probe revealed that the positive clone contained a Prl gene in an insert of about 15 kb (Fig. 1). The exon-containing fragments were identified by further digestion and hybridization and were subeloned in p G E M 4 vector and sequenced (Fig. 1). The complete nucleotide sequence was shown in Fig. 2 together with the encoded amino acid sequence of carp Prl. Compari- son of the genomic sequence with that of the eDNA reveals identical amino acid sequence of the gene prod- uct and only one base-change in the coding region and 5 more in the 3'-untranslated region of the last exon (Fig. 2). Certainly this 2.7 kb DNA fragment contains the full-length Prl gene which would give rise to the e D N A previously cloned [17]. A TATA box was found in the 5'-flanking region from - 3 8 to - 3 1 (Fig. 2). Further upstream was a 21 bp palindrome CTCAT- TCrCATATACAAATGAG from - 5 9 to - 7 9 . The
C A A T box was not found in this region. All the intron- exon boundaries followed the G T / A G rule [221.
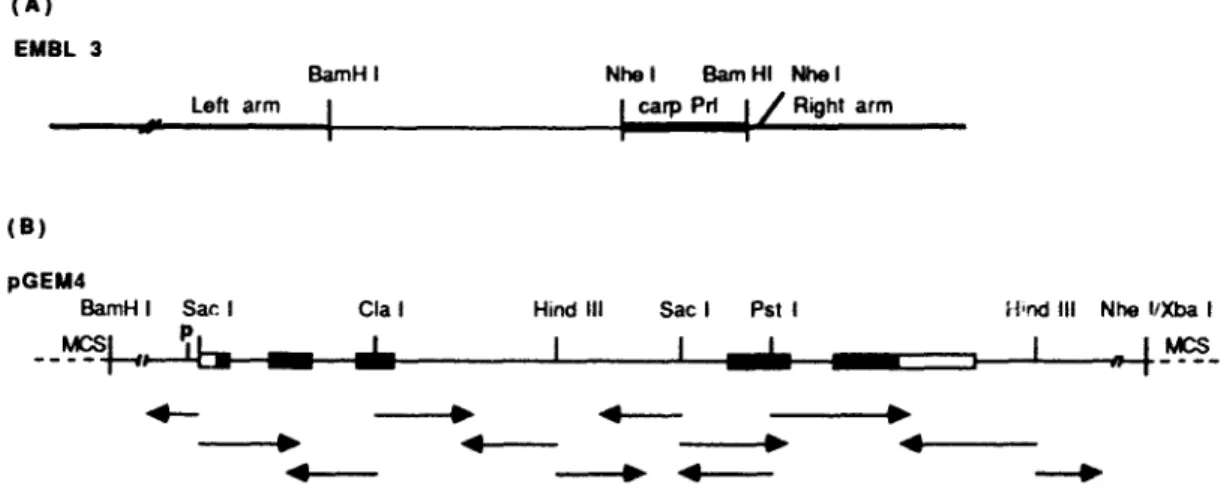
318 Carp GH 500 bp
I I
Carp Prl m H u m a n P r l ~ i I i 1 i Rat Prl --¢: m : BEFig. 4. Comparison of structures of the Prl genes of carp (this study), human 116] and rat [6] and of the carp GH gene [lg I. Solid boxes, coding regions: open boxes, untranslated regions: lines between the boxes, introns.
T h e t r a n s c r i p t i o n s t a r t site w a s d e t e r m i n e d b y t h e p r i m e r e x t e n s i o n m e t h o d (Fig. 3) a n d f o u n d t o be a g u a n i n e residue, l o c a t e d 38 b p d o w n s t r e a m f r o m t h e 1 A T A b o x (Fig. 2). A s s h o w n in Fig. 4, t h e Prl a n d G H [18] g e n e s in c a r p are v e r y s i m i l a r in t e r m s o f size, n u m b e r a n d l e n g t h o f e x o n s a n d i n t r o n s . H o w e v e r , t h e y a r e m u c h s h o r t e r as c o m p a r e d to m a m m a l i a n g e n e s , d u e to t h e s m a l l e r i n t r o n s in t h e c a r p g e n e s as is t h e c a s e for c a r p -f-crystallin g e n e s ( u n p u b l i s h e d d a t a ) . T h e c a r p Prl a n d G H also e x h i b i t 50,'~ h o m o l o g y in t h e i r c o d i n g se- q u e n c e , a p p r e c i a b l y h i g h e r t h a n rat a n d h u m a n , w h i c h h a v e a r o u n d 40% s e q u e n c e h o m o l o g y in t h e Prl a n d G H c o d i n g r e g i o n s ( d a t a n o t s h o w n ) . T h i s s t u d y w a s s u p p o r t e d b y a g r a n t f r o m t h e N a - tional Science C o u n c i l o f t h e R e p u b l i c o f C h i n a . References
1 Niall, H.D., Hogan, M.L., Sauer, R., Ro~enblum, I.Y. a , d Greenwood, F.C. (1971) Proc. Natl. Acad. Sci, USA 68, 866-869. 2 Li, CH. (1980) in Hormonal Proteins and Peptides (Li. CH.. ed.).
Vol. 8, pp. 1-36, Academic Press. New York.
3 Tashjian, A.H., Bancroft, F.C. and Levine. L. (1979) J. Cell Biol. 47, 61-70.
4 Gourdfi, D, Tougard, C. and Tixier-Vidal, A. (1982) in Frontiers in Neuroendocrinology (Ganong, W.F. and Martini, L, eds.), Vol. 7. pp. 317-357, Raven Press, New York.
5 Maurer, R.A, (1981) Nature 294. 94-97.
6 Maurer. R.A., Ermin, CR. and Donelson, J E (1981) 1. Biol. Chem. 256. 10524-10528.
"/ Murdoch, G.H., Potter, E., Nicolaiser' A.K., Evans, R.M. and Rosenfels, M.G. 0982) Nature 300, 192-194.
8 Emmerman, J.T., Enami. J., Pitelka. D.R. and Nandi, S (1977) Pro(:. Natl. Acad. Sci. USA 74, 4466-4470.
9 Clarke, W.C. and Bern, H.A. (19801 in Hormonal Proteins and Peptides, (Li. C.H., ed.L Vol. 8, pp. 105-197, Academic Press. New York.
10 Ensor, D.M. 0978) in Comparative Endocrinology of Prolactin, pp. 15-4.4, Chapman and Hall, London.
11 Prunet, P.. Boeuf, G, and Houdebine, LM. 0985) J. Exp. Zool. 235. 187-196.
12 Loretz. C.A. and Bern, H.A. (1982) Neurnendocrinology 35, 292- 304.
13 Hirano, T. (1986) in Comparative Endocrinology Developments and Directions (Ralph, C.L, ed.), Alan R. Liss, New York. 14 Gubbins, E.I., Maurer, R.A.. Lagrimini, M., Ermin, C.R. and
Donelson. J.E. (198G) J. Biol. Chem. 255, 8655-8662.
15 Camper, S.A., Luck, D.N, Yao. Y., Woychi, R.P., Goodwin. RG., Lyons, R.H. and Rottman, F.M. (1984) DNA 3, 237-249. 16 Truong, A.T.. Ducz. C.. Belayew, A., Renard, A., Pictet. R., Bell,
G. and Martial. J.A. (1984) EMBO J, 3, 429-437.
17 Chao, S.C., Pan, F.M. and Chang. W.C. (1988) Nucleic Acids Res, 16, 9350-9350.
18 Chlou, C.S., Chen, H.l. and Chang. W&'. 11990) Btochim. Bio- phys. Acta 1087. 91-94.
19 Chao, S.C.. Pan, F.M. and Chang, W.C. (1989) Biochim. Biophys. Acta 1007. 233-236.
20 Maniatis, T., Fritsch, E.F. and Sambrook, J, (1982) Molecular Cloning, A laboratory manual, Cold Spring Harbor Laboratory. Cold Spring Harbor. Nev, York.
21 Hattori, M. and Sakaki, Y. (1986) Anal. Biochem. 152. 232-238. 22 Breathnach, R a , d Chambon, P. (19811 Annu. Rev. Biochem 50,


![Fig. 4. Comparison of structures of the Prl genes of carp (this study), human 116] and rat [6] and of the carp GH gene [lg I](https://thumb-ap.123doks.com/thumbv2/9libinfo/8780570.215601/4.810.122.683.100.288/fig-comparison-structures-prl-genes-carp-study-human.webp)