Department of Mathematics, National Tsing-Hua University January 6, 2010y ,
Cancer as a mini-evolutionary process.
Yoh Iwasa
Evolution: slow change of organisms
mutation:
i
k
f
d
i
mistake of reproduction
natural selection:
mutant with a higher survival
and a faster reproductive rate
and a faster reproductive rate
will replace the old type.
Colon cancer arises in a crypt
Colon cancer arises in a crypt
Apoptosis
on top of crypt A crypt consists
of 1000-4000 cells.
36 hours
A small number of stem cells replenishes the whole crypt.
7
Tumorigenesis includes multiple steps of
t ti
f t
ll
l
f
mutations of stem cells:
z
loss of tumor suppressors
zoncogenes
g
z
angiogenesis (induction of blood vessels)
metastasis
z
metastasis
zetc.
Carcinogenesis is an Evolutionary Process
Carcinogenesis is an Evolutionary Process.
(1) Chromosomal Instability
(2) Tissue structure
(2) Tissue structure
Tumor Suppressors
p53
Tumor Suppressors
Rb
prevents cell division
d
i
APC
and causes apoptosis,
if
thi
i
Loss of Both Copies of a Tumor Suppressor Gene is
is
the First Step toward Cancer.
+ + / TSG TSG + /− TSG− /− u + p0 2u TSG TSG SG Cancer initiation E t i Escape apoptosis
Chromosomal Instability (CIN)
Chromosomal Instability (CIN)
Normal cells
Normal cells
C
Ch
l I t bilit
Can Chromosomal Instability
Effect of Chromosomal Instability (CIN)y ( )
+ + /
TSG TSG + /− TSG− /− Cancer initiationith t CIN
u + p0 2u TSG TSG TSG without CIN 2 2 Cancer initiation 2ncu 2ncu
fast
+/− TSG +/+TSG Cancer initiationwith CIN
p
fast
CIN 2u CIN − TSG CIN Loss Of Heterozygosity HeterozygosityFixation of mutant
1 0 1.0 0 1 fraction 0.0 0.0 cell generations 1 O(N) 1 Nuρ( )
r >>Moran Process
1 Select a cell for reproduction 1. Select a cell for reproduction 2. Cell division
4 Add the new cell 4. Add the new cell
Fixation probability
ρ
( )
r
Fixation probability
N l N llρ
( )
1.0 1.0 N: large N: small 1.0 1/N 0.0 0 1 r fitness 0.0 0 1 r fitness fitness fitnessDeleterious mutations can be fixed,,
if N is small.
Fixation of an
intermediate
mutant
1 0 1.0 0 1 2 0 0 0.0 cell generationsT
li
Tunneling:
The Second Mutation Spreads without the Fixation of p the First One
1.0
0 2
1
0.0
explicit formula for tunneling rate
⎡ ⎤
explicit formula for tunneling rate
Rtunnel ≈ Nu1 − 1− r( )+ 1− r( ) 2 + 2 1+ r( )ru2ρ( )a 1+ r − ρ( )r ⎡ ⎣ ⎢ ⎢ ⎤ ⎦ ⎥ ⎥ 1+ r ⎣ ⎢ ⎦ ⎥ +
deleterious mutation
Nu1 ru2 1− r ρ( )a − ρ( )r ⎡ ⎣ ⎢ ⎤ ⎦ ⎥ + if 1− r >> 2 u2ρ( )a ⎧ ⎪ ⎪deleterious mutation
≈ ⎣ ⎦+ N ⎡ ⎢ ( ) 1 ⎤ ⎥ if 1 2 ( ) ⎨ ⎪ ⎪ ⎪ Nu1 u2ρ( )a − N ⎣ ⎢ ⎦ ⎥ + if 1− r << 2 u2ρ( )a ⎩ ⎪neutral mutation
neutral mutation
Intermediate mutant is deleterious
Fixation probability of Type 2 mutant 0.008 0 006 X 0.004 2 0.006 tunneling 1 t 0.002 1-step process 0.0 0 20 40 60 N fixation of type 1 2-step process N 2-step processSmall compartment: 2-step evolution
Intermediate mutant is neutral
Fixation probability of Type 2 mutant 0.03 li 0.02 X2 tunneling 1-step process 0.01 fixation of type 1 0.0 0 20 40 60 80 N 2-step process NSmall compartment: 2-step evolution
) ( ) ( 2Nu u + p0 ρ a ) ( ) (u p0 a N + ρ 2u TSG + /+ TSG + /− TSG − /− 0 X X1 X2 ) (r Nucρ ) (r Nucρ ) ( 1 2 p ar r r u Nu c ρ − 1 r (ar) rp Nuc ρ − 2u TSG + /+ TSG + /− fast TSG − /− 0 Y Y1 Y2 TSG + /+ TSG + / TSG /
CIN CIN CIN
) ( 2Nu pρ a
r
<1 ) ( 1 2 p ar r r Nuuc ρ −C
Ch
l I t bilit
Can Chromosomal Instability
Chromosomal Instabilityy
/+ 2u / u + p0 / Cancer initiation
+ /
TSG TSG + /− TSG− /− Cancer initiationwithout CIN
p0
2ncu 2ncu
+/−
TSG
+/+
TSG Cancer initiationwith CIN
p
fast
CIN 2u CIN − TSG CIN Loss Of H i pCIN 2u CIN CIN
Heterozygosity
D
th Ti
St
t
Does the Tissue Structure
Change the
Cancer Risk?
Colon cancer arises in a crypt
Colon cancer arises in a crypt
Apoptosis
on top of crypt A crypt consists
of 1000-4000 cells.
36 hours
A small number of stem cells replenishes the whole crypt.
7
Small Compartments Large Compartments Mutants with
Small Compartments Large Compartments
(gate-keeper) Mutants with higher fitness (gate keeper) Mutants with lower fitness (care-taker) S ll Small compartments
reduce the risk of mutants with higher fitness, but enhance the risk of CIN.
Moran Process
1 Select a cell for reproduction 1. Select a cell for reproduction 2. Cell division
4 Add the new cell 4. Add the new cell
1. Select a cell for reproduction
2. Cell division
Linear Process
3. Shift the others division
No Somatic Selection
CIN is important
(1) Compartmentalization
(1) Compartmentalization
(2) Stem cells/non-stem cells
Somatic Selection is Suppressed
Somatic Selection is Suppressed
Risk via High Fitness Mutants is Reduced Risk via High Fitness Mutants is Reduced Risk via Low Fitness Mutants is Enhanced
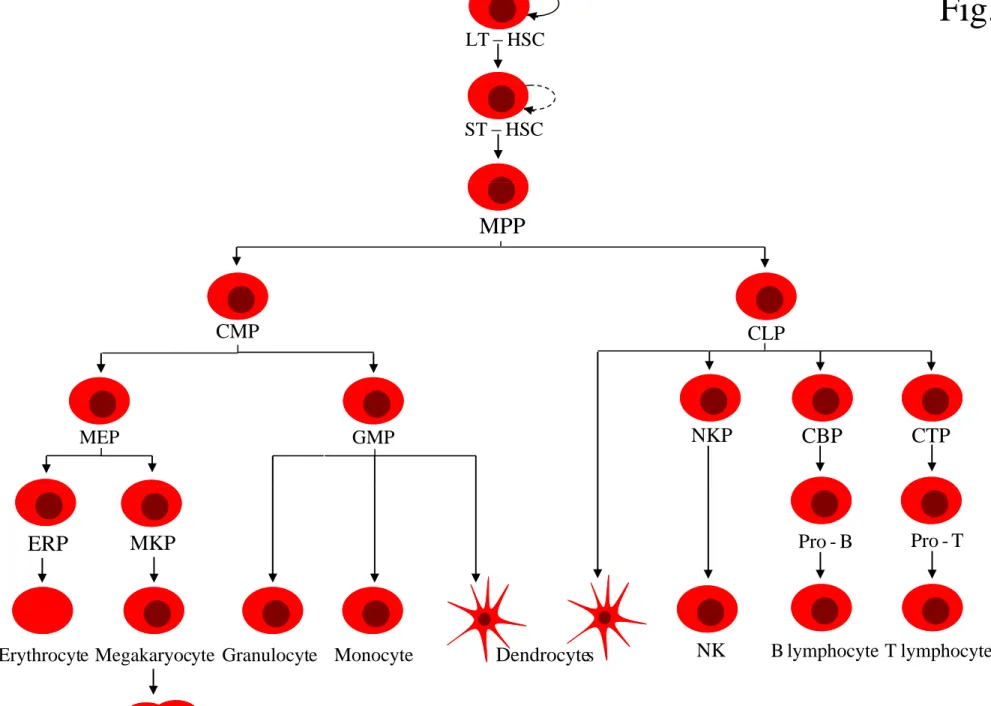
Dynamics of Chronic Myeloid
y
y
Leukemia (CML)
Fig. 1 HSC LT− HSC ST− MPP CMP CLP MEP GMP NKP CBP CTP
ERP MKP Pro-B Pro-T
s Dendrocyte
e
Erythrocyt Megakaryocyte Granulocyte Monocyte NK Blymphocyte Tlymphocyte
I
i ib
i
Imanitib
is a very
effective drug.
g
Under treatment
cancer cells
cancer cells
decrease
in two phases
After the stop of treatment, the number of cancer cells
quickly increases and exceeds the original level.g
Why?
Why?
Resurgence
Resistant Resistant
normal cells
regulated at a constant level stem cells
transition is accompanied
b i i b
progenitor cells
by expansion in number (finite time cell divisions)
differentiated cells differentiated cells
observed cell number
terminally differentiated cells
leukemic cells normal cells stem cells stem cells progenitor cells progenitor cells differentiated cells differentiated cells differentiated cells differentiated cells terminally differentiated cells terminally differentiated cells without treatment
leukemic cells normal cells stem cells stem cells progenitor cells progenitor cells differentiated cells differentiated cells differentiated cells differentiated cells terminally differentiated cells terminally differentiated cells with treatment
P l i D i M d l Population Dynamic Model
? x 0 =
[
λ( )x0 − d0]
x0 y ?0 = r[
y(1− u)− d0]
y0 ? x 1 = axx0 − d1x1 y ?1 = ayy0 − d1y1 ? x 22 = bxxx11 − d22x22 y ?2 = byy1 − d2y2 ? x 3 = cxx2 − d3x3 y2 yy1 2y2 ? y 3 = cyy2 − d3y3 3 x 2 3 3 y3 yy2 3y3Two rates of Two rates of cancer cells decrease decrease correspond to turnover rates of PC and DC d2 = 0.05 d1 = 0.008 1 d2 = 20 days 1 =125 days d1 =125 days
After the stop of treatment, the number of cancer cells
quickly increases and exceeds the original level.g
S ll
Stem cells are
leukemic cells resistant tumor normal cells SC SC stem cells PC PC progenitor cells DC DC differentiated cells DC DC differentiated cells TC TC terminally differentiated cells with treatment
Population Dynamic Model ? x 00 =
[
[
λ( )( )x00 − d00]
]
x00 y ?0 = r[
y(1− u)− d0]
y0 z ?0 = r( z − d0)z0 + ryy0u ? d y0[
y( ) 0]
y0 0 ( z 0) 0 yy0 ? y a y d y ? d ? x 1 = axx0 − d1x1 y ?1 = ayy0 − d1y1 ? b d ? z 1 = azz0 − d1z1 ? x 2 = bxx1 − d2x2 y ?2 = byy1 − d2y2 z ?2 = bzz1 − d2z2 ? x 3 = cxx2 − d3x3 y ?3 = cyy2 − d3y3 z ?3 = czz2 − d3z3 i ll resistant cellsModel for the number of resistant cells
h iti ll b h M
when sensitive cell number reaches M
M
lls
the detectable number
M
em ce sensitive cells m ic st e u mutation rate euke m u mutation rate r of l e resistant cells u mbe r time n u
= 1 − exp[ −
R
• p
M −1∑
]
probability of
= 1 exp[
R
x• p
x x=1∑
]
resistance
expected number of new mutationssurvivorship of one lineage until the detection time
is the expected number of new mutations
R
is the expected number of new mutationswhen the number of sensitive cells is .
x
R
x f1( )0 =1, fx( )0 =0 for x =2, 3,...,M−1 Rx = rux 1 d / fx(t)dt ∞∫
df1 dt = 2df2 − (r + d) f1 1− d /r∫
0 dfx dt = r(x −1) fx−1 + d(x +1) fx+1 − (r + d)xfx df r: division rate df M−1 dt = r(M − 2) fM−2 − (r + d)(M −1) fM−1 d: death rate u: mutation rate the probability th t th iti ll t tit
generating function: g g g (ξ, t) = E[ξZ ( t ) | Z (0) = 1] ⇒ g (ξ,0) =ξ g (ξ, t + Δt) = aΔtg (ξ, t)2 + bΔt •1+ (1− (a + b)Δt)g (ξ, t) g(ξ ) g(ξ ) ( ( ) )g(ξ ) ⇔ ∂g ∂t = (a − bg )(1− g ) (ξ 1)(b a)e( a−b )t (ξ b a) ⇔ g (ξ, t)= (ξ −1)(b a)e ( ) − (ξ − b a) (ξ −1)e( a−b )t − (ξ − b a) 1 M M = x exp[(r − d)t] t = 1 r − d log( M x ) 1 b ⎛ ⎜ ⎞ ⎟ 1 b ⎛ ⎜ x ⎞ ⎟ a−b ( ) (r−d) ⎡ ⎢ ⎤ ⎥ px = 1− a ⎝ ⎜ ⎠ ⎟ 1− a M⎝ ⎜ ⎠ ⎟ ⎣ ⎢ ⎢ ⎦ ⎥ ⎥
Probability of Resistance
Probability of Resistance
⎡
⎤
P
=1− exp −
MuF
1 d
⎡
⎣
⎢
⎤
⎦
⎥
11
− d r
⎣
⎢
⎦
⎥
F = 1− b a 1− (b a)y(a−b) (r−d) dy 0 1∫
where ( )y 0 d i i iM : detection size u : mutation rate r, d : division/death rate (sensitive cells) a, b : division/death rate (resistant cells)
Simulation results fit the formula
Simulation results fit the formula.
0.5 0.4 M=104 a− b r− d = 2 P 0.3 0.4 0.3 P d=b=1 u=10−5 r d 0 1 0.2 ra= 2= 3 d = b = 1 u= 10−5 0.1 0.2 0.1 104 2×104 3×104 4×104 5×104 u= 10 1.2 1.4 1.6 1.8 2.0 M (detection size)
r
(division rate)Slow growth implies higher risk of resistant
Slow growth implies higher risk of resistant
cells
small r higher risk of
small r higher risk of
resistance
longer time until diagnosis longer time until diagnosis
more mutations more cell division
Mean number of Mean number of resistant cells
(conditional to one or (conditional to one or more resist. cells)
Distribution of
i t
t
ll
resistant cell
number
z: large Pr y[ 1 < Y ≤ y2]= 1 − 1 z: large Pr y[ 1 < Y ≤ y2] Fy1 ( )1α Fy2 ( )1α ( )2 1 ( )y 1 1 z: small Pr Y[ = y]= (1− b a) 2 αF z1α(1− z)y−1 1− b a( )z ( )y+1 dz 0 1 ∫Cancer Progression
Cancer Progression
is
is
Somatic Evolution
Somatic Evolution.
Collaborators
zMartin A. Nowak (PED, Harvard Univ)
zFranziska Michor (Sloan-Kettering Cancer Research Inst.)
zFranziska Michor (Sloan Kettering Cancer Research Inst.)
zSteven A Frank (UC Irvine)
zRobert May (University of Oxford)y ( y )
zDominik Wodarz (UC Irvine)
zNatalia L Komarova (UC Irvine)
zBert Vogelstein (Johns Hopkins Univ.)
zChristoph Lengauer (Johns Hopkins Univ.)
zTim. P. Hughes (Inst.Med.Vet.Sci. Adelaide)
zSusan Branford (Inst.Med.Vet.Sci. Adelaide)
zN il P Sh h (UCLA)
zNeil P. Shah (UCLA)
zCharles L. Sawyers (UCLA)
zHi hi H (K h U i i )
Probability of drug resistance
d
b
f
i
i
and number of resistance in CML
Poisson distribution of infected
ll
b
( i l i f
i
)
cell number (viral infection)
Drug resistance requires two mutations
Age distribution for CML incidence
g
Drug resistant virus:
Drug resistant virus:
• HIVHIV
• hepatitis B virus • influenza virus
• simple herpes virussimple herpes virus Pathogenic virus
Pathogenic virus ..
(1) produces enzymes decomposing the drug (2) changes the structure of target molecules (3) interrupt the drug delivery
http://web.uct.ac.za/depts/mmi/stannard/emimages.html http://www.howstuffworks.com/aids2.htm
When virus infection is diagnosed,
what is the probability for resistant
strain(s) already to exist in the host.
Proliferation of Virus
target cell
viral particles from a single infected cell Virus
lti l i f t d
p g
can infect multiple cells.
an infected cell Viral particles
multiple infected cells
: mean number of mutants when the b f i f t d ll i
R
xnumber of infected cells is x
f f f x fx−1 fx fx+1 r r r r R = rux f (t)dt ∞
∫
d d d d Rx = 1− ?g∫
0 fx(t)dt f 0( )= 1 f ( )0 = 0 for x = 2 3 M 1 x cells x+1 cells x-1 cells probability of x infected cells at t f1( )0 = 1, fx( )0 = 0 for x = 2, 3, ..., M − 1 df1 dt = (d + r ⋅ e −λ )⋅ 2 ⋅ f2−{d + r(1−λe−λ)}⋅1⋅ f1 df x−1 λ(x−k +1) probability of extinction of infected cells dfx dt = r λ( ) (x− k +1)!e −λ⋅ k ⋅ f k + k=1 ∑ (d+ r ⋅ e−λ)⋅ (x +1) ⋅ fx+1 −{d + r(1−λe−λ)}⋅ x ⋅ fx df M∑−2 λ(M−k ) re−λ(1− ?g ) + d = (r + d)?g dfM−1 dt = r λ( ) (M− k)!e −λ⋅ k ⋅ f k k=1 ∑ −{d + r(1−λe−λ)}⋅ (M −1) ⋅ fM−1: probability for the lineage of resistant cell
q
p y gline until the onset of drug use.
q
x M q = ν i e−ν(
1− gi)
∞ ∑ = 1 − e−ν(1− gx ) qx i! e(
1 gx)
i= 0 ∑ 1 e 1 ⎛ ⎞ M ⎛ ⎞ x g b bilit f ti ti gx = g 1 r(λ − 1) − d log M x ⎛ ⎝ ⎜ ⎞ ⎠ ⎟ ⎛ ⎝ ⎜ ⎞ ⎠ ⎟ Rx q 1 i 0 gx : probability of extinction of resistant virus time qx t i 0 M = x exp[{r(λ −1) − d}t] t = 1 (λ 1) d log M ⎛ ⎝ ⎜ ⎞ ⎠ ⎟ dg dt = ae −ν(1− g ) + b − (a + b)g , g (0) = 0 r(λ−1) − d ⎝ ⎠ xProbability that one or more cells are
infected by drug resistant virus y g
at diagnosis
(1 point mutation is enough for resistance)
M 1
⎡
⎤
(1 point mutation is enough for resistance)
R
q
xP
=1−exp−
rux
?
f
x(t)dt
∞∫
M−1∑
⋅(1−exp−
(
ν
(1
−g
x)
)
⎡
⎣
⎢
⎤
⎦
⎥
R
xq
xp
1
−?g
∫
0f
x( )
x=1∑
(
p ( g
(
x)
)
⎣
⎢
⎦
⎥
M : population size at diagnosis u : mutation ratep p g
r, d : cell division/mortality (wild type) a, b (resistant strain)
Probability of drug resistance is high
when....,
i P r=1.5 • mutation rate u is high r=5• infected cell number
r : reproductive
rate of sensitve type
ec ed ce u be at diagnosis M is large P u(mutation t ) 1 5 is large d ti t i rate) P r=1.5 r=5 • reproductive rate r is small M (cell number)
Risk of drug resistance is high, when.. mutation rate is high
mutation rate is high
population size at diagnosis is large
p p g g
population growth rate of virus is slow
Risk of drug resistance is reduced
b id tif i ti t d t ti d
by identifying patients and starting drug treatment as early as possible.
When leukemic stem cell number reaches
M th ti t i di d CML
Current d l
Symptoms of CML
M, the patient is diagnosed as CML.
M
ls model: Symptoms of CML appearM
m cel l drug sensitive stem cells ic ste m stem cells u mutation rate u kem i u mutation rate of le u drug resistant stem cells m ber time Nu mA tissue grows exponentially in development. Mutant cells can be produced in the process
Alternative
Mutant cells can be produced in the process, and some descendent may exist at diagnosis.
interpretation: End of development
M
normal cells cells er of c u mutation rate N umb e mutant cells (later causing N (later causing disease) timeProbability of drug resistance
d
b
f
i
i
and number of resistance in CML
Poisson distribution of infected
ll
b
( i l i f
i
)
cell number (viral infection)
Drug resistance requires two mutations
Age distribution for CML incidence
g
Clinical problems caused by cells
p
y
with two specific mutations.
- Drug resistance
CML cells acquires resistance when it becomes
resistant to two different drugs (Imatinib and Dasatinib)
(L387M/F317I etc.)
- Cancer
Retinobrastoma is caused when both alleles of Retinobrastoma is caused when both alleles of
tumor suppressor gene RB1 in a cell are inactivated during development
Model
r
1
a a2
Type-0 cell Type-1 cell
1 a Type-2 cell 2 a 1 u u2
Type 0 cell Type 1 cell
d
Type 2 cell
b1 b2
Type-0: Wild type cells Type 0: Wild type cells
Type-1: Cells with one mutation (intermediate state) (intermediate state)
Type-2: Cells with two mutations
( ll d i ll )
Parameters
number of mutants cell division rate cell death ratemutants rate rate
T 0 0
r
d
Type-0 1 u 0r
d
Type-1 1a
1b
1 u2 Type-2 2a
2b
2Model
cell number Type-0 mutation mutation Type-1 Type-2 time ypProbability that fhe first Type-1 cell that survives is producedwhen there are x Type 0 cells
P
x:
is producedwhen there are x Type-0 cells.
P −σR ′
(
1 −σR)
x−1∏
− x−1( )σR 1 −σR(
)
Px = e σRx ′(
1− e σRx)
′ x =1∏
= e ( )x 1σRx 1− e σRx(
)
rWhen the total cell number reaches M
Expected number of mutants
produced while there are x Type-0 cells:
ell numbe
Rx ≈ u1
1− d r
ce
Probability the linearge from a newly
x
σ ≈1− b1
formed Type-1can survive:
1
time t
Probability that type 1 cells produce type 2 cell(s)
b f th t t l ll b h th fi l l
before the total cell number reaches the final value
Q
=1−e
−u2y 1( −b1 a1)Q
x=1−e
The number of type 1 cells
m
ber
y
= e
(a1−b1)τxwhen total cell number reaches the final value
when cell number reaches the final number
cell nu
m
y
y
= e
:time from the production of a Type 1 cell to the final time
τ
xx xe(r−d)τx + e(a1−b1)τx = M
Type-1 cell to the final time
x
1
τ
x Expected number of mutationsuntil Type-1 cell number reaches y:
time t
u2y
Probability that one or more Type-2 cell exist
y
yp
at the final time
P
e
−β( )x−1(
1 e
−β)
(
1 e
−u2y 1( −b1 a1))
M −1∑
P
=
e
β( )(
1
−e
β)
(
1
−e
2y ( 1 1))
x=1∑
β =σRx = 1− b( 1 a1)(u1 (1− d r))where
y = e(a1−b1)τx xe(r−d)τx + e(a1−b1)τx = M τxis a solution of
and
y = e( )r, a1, a2: cell division rate
d b b li
x
d, b1, b2: mortality
Parameter dependence
Cells with two or more Cells with two or more
mutations exist more likely
– High mutation rates u1 and u2
– Fast cell division rate a1 of intermediate mutants
– Large final population size M – Large mortality d
Probability distribution of
y
Type-2 cell number
red line: prediction by
red line: prediction by the formula Pr z[ 1 < Z < z2]= Pr κz 1 <κZ <κz2
[
]
= L L = Lz 2 − Lz1Probability of drug resistance
d
b
f
i
i
and number of resistance in CML
Poisson distribution of infected
ll
b
( i l i f
i
)
cell number (viral infection)
Drug resistance requires two mutations
Age distribution for CML incidence
g
Step number
0
1
2
1
p
0
1
2
...
n-1
n
Cancer incidence occurs when n stochastic
events occurs to a patient
Probability of incidence on or before age t )
( )
Step number of CML incidence date is
close to three
If a single mutation is sufficient for CML, can we have step number three?
Moran process
1 choose a single cell randomly 1. choose a single cell randomly.
2. cell division 4 add one cell 4. add one cell
Diagnosis as CML (or detection) occurs
t t ti l t
at a rate proportional to
the number of CML stem cells...
ells em c e m ic st e euke m u mutation rate r of l e u mbe r
i
i
n uDiagnosis as CML
Probability for a patient to be judged as
leukemia on or before t
⎛ ⎞ ⎡ ⎤leukemia on or before t
P t( )
= 1− 1+ e cx −1 N 1 1 r(
)
⎛ ⎝ ⎜ ⎞ ⎠ ⎟ −qN c ⎡ ⎣ ⎢ ⎢ ⎤ ⎦ ⎥ ⎥ e −b t−x( ) bdx t∫
N 1(
−1 r)
⎝ ⎠ ⎣ ⎢ ⎦ ⎥ 0 b = Nu 1−1 r(
)
τ c = r −1(
)
τS b f
Step number from
incidence age distribution can be quite large.