Comparative Analysis of Plasmid-Encoded KPC-2 Isolated from Klebsiella pneumoniae and Escherichia coli in 2012: Genetic Basis Leading to the Confined Plasmid Spreading among K. pneumoniae Carrying KPC-2 in Taiwan Ying-Tsong Chen1,2,3, Jung-Chung Lin4, Chang-Phone Fung5, Po-Liang Lu6, 7
,Yin-Ching Chuang8,9 , Tsu-Lan Wu10* L. Kristopher Siu4, 11,12*
1Institute of Molecular and Genomic Medicine and 12National Institute of Infectious
Diseases and Vaccinology, National Health Research Institutes, Miaoli; 2Institute of
Genomics and Bioinformatics, National Chung Hsing University, 3Biotechnology
Center, National Chung Hsing University, Taichung;
4Division of Infectious Diseases and Tropical Medicine, Department of Internal
Medicine, Tri-Service General Hospital, National Defense Medical Center, Taipei;
5Section of Infectious Diseases, Department of Medicine, Taipei Veterans General
Hospital, Taipei;
6Department of Internal Medicine, Kaohsiung Medical University Hospital
and7College of Medicine, Kaohsiung Medical University, Kaohsiung:
8Department of Internal Medicine and Medical Research, Chi Mei Medical Center,
Tainan;
9Department of Internal Medicine, Chi Mei Medical Center, Liouying, Tainan; 10Department of Clinical Pathology, Linkou Chang Gung Memorial Hospital,
11Graduate Institute of Basic Medical Science, China Medical University, Taichung,
Taiwan
*Contributed equally
Corresponding: L. Kristopher Siu, National Institute of Infectious Diseases and Vaccinology, National Health Research Institutes, Miaoli; Taiwan. Email:
Background: Two KPC-2 plasmids isolated from carbapenem-resistant Klebsiella
pneumoniae (CR-KP) and Escherichia coli (CR-EC) were sequenced and analysed.
The CR-KP strain was selected from an outbreak in 2012, and the CR-EC strain was the first KPC-2-carrying EC identified in the same carbapenem resistance monitoring program in Taiwan.
Methods: Antimicrobial susceptibility tests, multilocus sequence typing (MLST), and the conjugal transfer of plasmids to recipient KP and EC were performed. Complete nucleotide sequencing of the KPC plasmids was performed using the 454
shotgun approach.
Results: The CR-KP and CR-EC strains in this study were determined to be ST-11 and ST-410, respectively, by MLST. The plasmid encoding KPC from CR-KP was unable to transfer conjugatively into recipient K. pneumonia and E. coli, while the KPC plasmid from CR-EC was able to transfer into recipient E. coli. CR-KP and CR-EC were resistant to almost all of the tested antibiotics except gentamicin, amikacin, tigecycline, and colistin. The pKPC-LK30 plasmid from CR-KP is 86.5 kb in length and carries the blaKPC-2 and blaSHV-11 genes on a backbone similar to the
IncFIIK plasmids but lacks one of the two replication origins. The pKPC-LKEc
plasmid from CR-EC is 145 kb in length and carries the blaKPC-2, blaCMY-2, blaCTX-M-3
chimera of three regions related to the replicon of the IncI, IncN and RepFIC groups. Conclusion: The genetic differences between the backbones of these two plasmids may reflect their differences in cross species transferability, which could have led to the confined KPC-2-carrying CR-KP outbreak in Taiwan. Although KPC-2-carrying CR-EC was the first isolate identified in 2012, the KPC-2 plasmid CR-EC, which has a relatively high transferability, should be closely monitored to avoid increasing
Introduction
The increasing incidence of carbapenem-resistant bacteria, which are resistant to almost all antibiotics, is of great concern. Mechanisms that cause carbapenem resistance, including over expression of the active efflux pump, concomitant AmpC production with outer membrane porin loss 5-7 and the production of carbapenemases, have been identified . Among these mechanisms, carbapenemases,
such as NDM or KPC, are frequently studied due to their rapid global spreading.
One intriguing observation is that carbapenem resistance genes, such as KPC or the OXA type carbapenemase in Klebsiella 10 and Acinetobacter, respectively, are highly species specific 11-13. However, these species-specific carbapenemases are occasionally observed in other bacteria, such as E. coli, Enterobacter spp, etc.14 Previous reports indicated that the KPC-encoding plasmid could be conjugatively transferred within species of Klebsiella but not E. coli 9. The plasmid encoding KPC is lost unless co-cultured with K. pneumonia, suggesting that the plasmid is unstable outside of Klebsiella pneumoniae9. Although it is not as common as in K.
pneumoniae, KPC-producing E. coli can be isolated from patients15. However, little is known on the genetic backgrounds of the blaKPC-encoding plasmids in E. coli and K. pneumoniae. This information may lead to the understanding of how cross
species transmission is achieved. KPC-producing K. pneumoniae were first identified in 2011 in Taiwan16. In this report, we obtained the first bla
KPC-encoding E. coli isolated in 2012 and compared the plasmids in KPC-producing E. coli and K. pneumoniae isolated in 2012.
Methods and Materials
Isolates of KPC-encoding K. pneumoniae and E. coli
In 2012, a national surveillance of carbapenem resistance in K. pneumoniae (CR-KP) and E. coli (CR-EC) was supported by the Center of Disease Control, Taiwan. According to the collection, the plasmid encoding KPC-2 in the CR-KP outbreak was observed in the surveillance (data in submission). In KPC-2-positive strains, we observed only one CR-EC. To understand the mechanism underlying the low frequency of the plasmid encoding KPC-2 in CR-EC, we selected one outbreak strain of KPC-2 CR-KP for plasmid analysis and compared it to KPC-2 CR-EC. KPC-2 CR-KP obtained from the USA 9 was used as a control for the susceptibility tests.
Multilocussequencetyping(MLST)
MLST was performed according to Turton et al.17 and Keith Jolley18 for K.
pneumoniae and Wirth T et al.19 for E. coli. The sequences of seven housekeeping genes were obtained from liver abscess isolates from patients and carriers. The sequence information was compared with the sequences from the MLST website
(http://pubmlst.org/kpneumoniae/) and http://mlst.ucc.ie/mlst/dbs/Ecoli/ for K.
assigned accordingly. The sequences for the alleles that were not in the database were submitted to the curator, and a new allele number was obtained. A difference in two or more alleles indicated that the sequence types were unrelated.
Conjugation of the CR-KP and CR-EC plasmids into recipient E. coli or serotype K2 carbapenem susceptible K. pneumoniae
Two different conjugation experiments were performed according to previous reports . In the first set of experiments, the sodium azide-resistant strain E. coli J-53 or rifampin-resistant strain E. coli J-995 were used as the recipients 21. Neither the donor strains nor wild-type CR-KP or CR-EC with KPC grew on MacConkey agar with sodium azide or rifampin (100 g/ml), respectively. The recipient and donor strains were inoculated separately into brain heart infusion broth (Oxoid, Basingstoke, Hampshire, England) and incubated at 37°C for 4 hours. The strain broths were then mixed together at a ratio of 1:10 (by volume) and incubated overnight at 37°C. A 0.1-ml volume of the overnight broth mixture was then spread onto a MacConkey agar plate containing sodium azide or rifampin (100g/ml) and imipenem (2 g/ml). The transconjugants were then selected from the agar plate.
In the second part of the conjugation experiments between CR-KP and CR-EC with KPC to the recipient serotype K2 K. pneumoniae, CR-KP containing the KPC
plasmid, which was susceptible to streptomycin, was the donor. The serotype K2 K.
pneumoniae with intrinsic resistance only to ampicillin was used as the recipient.
The mating procedure was described as above using BIND (brilliant green agar containing inositol-nitrate-deoxycholate) for K. pneumoniae-specific selection. The BIND agar allows the growth of K. pneumoniae but not E. coli. Serotype K2 K.
pneumoniae transconjugants were selected using BIND agar supplemented with 2
g/ml of imipenem and 100 g/ml of streptomycin. For the conjugation of KPC from CR-EC to the recipient serotype K2 K. pneumoniae, serotype K2 K.
pneumoniae transconjugants were selected using BIND agar supplemented with 2
g/ml of imipenem. None of the donors, including CR-KP or CR-EC and serotype K2 K. pneumoniae, could grow on the specific selective medium, except the serotype K2 K. pneumoniae strain harbouring the imipenem-resistant KPC plasmid. The transconjugants from the experiments were sub-cultured onto MacConkey or Mueller Hinton agar (BBL, Sparks, MD) for 14 days to check for KPC plasmid stability in E. coli and K. pneumoniae after conjugation.
Antimicrobial susceptibility testing
The MICs of antimicrobial agents were determined using the broth microdilution test according to the recommendations from the Clinical and Laboratory Standards
Institute 22. The following antimicrobial agents were used: ampicillin, aztreonam, cefazolin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, ertapenem, imipenem, meropenem, doripenem, nalidixic acid, ciprofloxacin, levofloxacin, gentamicin, amikacin, colistin, tigecycline and trimethoprim-sulfamethoxazole
(SXT).
Sequencing and annotation of the plasmids
To determine the complete sequence of the KPC plasmid pKPC-LK30 from CR-KP
K. pneumoniae LK30, we performed shotgun sequencing using the 454 GS Junior
(Roche) on an 8-kb paired-end library constructed from the total genomic DNA prepared from LK30. The reads were assembled using Newber 2.7 (Roche). A scaffold carrying blaKPC-2 was identified, and gap filling between the contigs was
accomplished by adding Sanger reads with the aid of Consed.23 The sequence annotation of the contigs was performed using the RAST Server 24 followed by manual inspection and correction. The sequencing of another KPC plasmid, pKPC-LKEc, was performed using a 3-kb paired-end library prepared from the plasmid DNA extracted from CR-EC E. coli. The complete nucleotide sequences of pKPC-LK30 and pKPC-LKEc were submitted to GenBank and assigned the sequence accession numbers KC405622 and KC788405.
Results
Molecular typing and conjugation of the CR-KP and CR-EC plasmids into recipient E. coli or serotype K2 carbapenem-susceptible K. pneumoniae
MLST indicated that CR-KP and CR-EC were ST-11 and ST-410, respectively. The KPC resistance gene plasmid from CR-EC was able to transfer into recipient JP-995
E. coli but not J-53 E. coli. The plasmid encoding KPC from CR-KP was unable to
transfer into recipient serotype K2 K. pneumoniae, E. coli JP-995 or E. coli J-53.
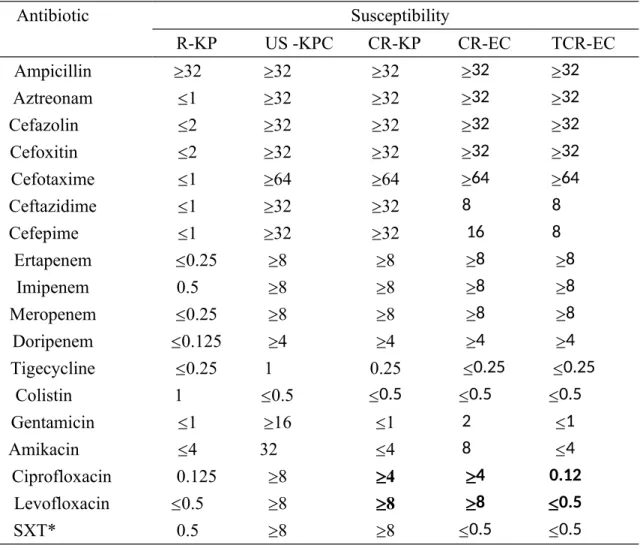
Antimicrobial susceptibility of the clinical KPC (CR-KP), recipient serotype K2 (R-KP), US-KPC control (US-KPC) and transconjugants (TCR)
Antimicrobial susceptibility tests indicated that the control US-KPC from the USA was resistant to almost all of the tested antibiotics except tigecycline and colistin. Although the resistance profile of KPC from Taiwan was similar, different susceptibilities were found in aminoglycoside resistance. The CR-KP and CR-EC isolates were susceptible to aminoglycosides, including gentamicin and amikacin
(Table 1).
-lactam resistance in TCR-EC was found to be identical in CR-EC, TCR-EC and CR-EC, including all four carbapenems. The resistance to quinolones and trimethoprim-sulfamethoxazole was not transferred, making TCR-EC susceptible,
indicating that chromosomal resistance was likely the cause of quinolone and trimethoprim-sulfamethoxazole resistance in donor CR-KP (Table 1).
Sequencing and annotation of the 86-kb pKPC-LK30 plasmid from CR-KP We performed complete sequencing of the KPC plasmid pKPC-LK30 from CR-KP. The carbapenem-resistance plasmid is 86,518-bp in length and carries the blaKPC-2
and blaSHV-11 genes (Figure 1a). A repB gene was found at the putative replication
origin flanked by genes responsible for plasmid partitioning and stability, including
parA/parB, vagC/vagD, and stbA/stbB. An iteron sequence composed of 17.5x
imperfect repeats of a 36-bp repeat unit was identified immediately downstream of the repB gene. The 9.7-kb region flanking repB and the nearby genes, including the iteron, is similar to one of the putative replication origins reported in the antimicrobial resistance plasmid pK245 from K. pneumoniae (Figure 1a), which is a
chimera with multiple replicon types 25.
Two KPC-2 plasmids reported recently from K. pneumoniae also contain replication initiation regions similar to pKPC-LK30. These plasmids include the IncFIIK5
plasmid pKP048 and IncFIIK1 plasmid pKPN101-IT. Both pKP048 and pKPN101-IT
were classified as IncFIIK-type, which belongs to the prevalent group of IncFII
Enterobacteriaceae . pKP048 is a blaKPC-2-, blaDHA-1- and qnrB4-positive multidrug
resistance plasmid of 151,188-bp isolated from China in 2006. Two replication initiation regions, including a FIIK1 region and a repB region, were identified in this
plasmid 27. The plasmid pKPN101-IT carries bla
KPC-2 and blaTEM-1 as well as other
resistance genes. The 107,748-bp plasmid isolated from Italy also possesses two replication regions similar to pKP048. Surprisingly, the FIIK1 region in pKP048 and pKPN101-IT was missing in pKPC-LK30. Comparative analyses revealed several conserved regions among the three plasmids (Figure 2). These regions include the spanning region of the repB replication origin, the associated iteron sequences, several plasmid partitioning/stability genes, the blaKPC-2 region, and a
mercury-resistance gene cluster region (Figure 2).
Sequencing and annotation of the 145-kb pKPC-LKEc plasmid from CR-EC The KPC plasmid pKPC-LKEc from CR-EC was completely sequenced. Annotation
and comparative analyses revealed that the majority of the 145-kb plasmid was composed of the backbones from three replicons, which include an 82-kb region similar to the IncI plasmid R64, a 20-kb region similar to the IncN plasmid R46, and an 18.6-kb region similar to the RepFIC replicon (Figure 1b). The 82-kb R64-like region contained the repZ gene for autosomal replication, a shufflon for the adhesion
of R64 pilus, and the tra genes for conjugal transfer. Comparative analysis revealed that the 82-kb region, albeit inserted by several short non-homologous regions including an IS1294, is very similar to the backbone of the plasmid R64, a conjugative R plasmid originally isolated from Salmonella enterica 30. Another 20-kb region was found to contain the repA gene and the stbA stbB plasmid stability genes. The nucleotide sequence of this region is similar to the backbone of the IncN plasmid R46, which was also identified from S. enterica serovar Typhimurium 31. The 18-kb region that is similar to the F plasmid from E. coli contained the region from traF, trbA, and artA to finO together with the repA1 gene and other replication regulation genes that represent the RepFIC replicon of F32. The antimicrobial resistance genes blaKPC-2, blaCMY-2, blaCTX-M-3 and blaTEM-1 were found to be associated
with IS/transposable elements and located between the three conserved replicon backbones (Figure 1b).
Comparative analysis of the blaKPC-2 region
The blaKPC-2 gene in the plasmid pKPC-LK30 is located between an interrupted Tn3
and Tn1721, which is similar to pK048 (Figure 3). The immediate genetic environment of blaKPC-2 in pKPC-LK30 and pKPC-LKEc is the same: an interrupted
containing blaKPC-2 and part of the downstream ISKpn6-like gene is identical to
Tn4401, which is presumably the origin of the blaKPC-2 mobilisation to the
plasmids33. In these plasmids, the Tn3 is interrupted by the insertion of an ISKpn8 at an identical location, leaving a short segment of Tn3 IR upstream of the blaKPC-2. The
Tn3 in pKPC-LK30 and pKPC-LKEc is interrupted further by the insertion of an IS26 element at the Tn3 tnpR. A Tn1721 region is located at the other end of the conserved region between pKPC-LK30 and pKP048. In both of the plasmids, the Tn1721 was positioned next to an IS26 tnpA pseudogene. However, IS26 was truncated by the IR of Tn1721 at a different location. This finding suggests that the acquisition of the blaKPC-2 region to pKPC-LK30 and pKP048, presumably aided by
the conserved Tn1721, was mediated by similar but independent events. In pKPC-LKEc, the ISKpn6-like element downstream of the blaKPC-2 is truncated by an IS26.
The 4.8-kb region spanning the blaKPC-2 gene was thus flanked by two IS26 elements
and resembled a composite transposon. The sequences of the direct repeats flanking the IS26 elements are, however, different and suggest the entire blaKPC-2 region was
Discussion
In this report, we selected two different isolates carrying KPC-2. These isolates have been identified from a national carbapenem resistance monitoring program (manuscript submitted). KPC-producing bacteria in Taiwan were first described in 201116 and became an outbreak through the clonal spread of K. pneumoniae 34.
Non-K. pneumoniae carrying KPC-2 was not identified until 2012. The present KPC-2 E. coli was first identified in 2012 and was the first strain observed in E. coli in our
locality. According to the data from the monitoring program in 2012, the clonal spreading of K. pneumoniae carrying KPC-2 is continually isolated from different
hospitals throughout Taiwan and is becoming an epidemic strain.
The incidence of non-K. pneumoniae carrying KPC type -lactamase is far less than the incidence of K. pneumoniae carrying KPC type -lactamase according to Medline and reports from the US 35. Although reports from the US have studied KPC E. coli 35, few reports have described in detail the transferability of the plasmid. Reports analysing the complete plasmid sequence that leads to transferability is important to understand the epidemiology of KPC plasmid36. It is interesting to compare the KPC-encoding plasmids from K. pneumoniae and non-K. pneumoniae. The emergence of KPC-producing E. coli is of concern because the KPC-encoding
plasmid may carry resistance across species barriers. In this report, we compared the whole plasmid carrying KPC from E. coli and K. pneumoniae. The KPC-positive transconjugant showed resistance to all -lactams but not other antibiotics (Table 1), indicating that only the -lactamase-producing genes were carried on the plasmid (Figure 1a & 1b). On the other hand, resistance to quinolones and trimethoprim-sulfamethoxazole were not transferrable by conjugation.
Complete sequencing of the plasmids pKPC-LK30 and pKPCLKEc from CR-KP and CR-EC, respectively, revealed distinct plasmid replicon structures. The blaKPC-2
gene in these plasmids is not associated with an entire Tn4401. Only the regions immediately adjacent to the KPC-2 determinant on these plasmids, including the partial Tn3 region, blaKPC-2 gene, and ISKpn6-like element, were identical to Tn4401.
Similar structures associated with blaKPC-2 in pKPC-LK30 were reported in pKP048,
suggesting that it is very likely that the antimicrobial resistance gene was acquired in these plasmids from a common molecular ancestor through recombination events near the flanking mobile elements. Comparative genomics analyses revealed that the backbone of the pKPC-LK30 plasmid from CR-KP is very similar to two incFIIK1 KPC-2-encoding plasmids, pKP048 and pKPN101-IT26, which were also identified from K. pneumoniae (Figure 2). The plasmids pKP048 and pKPN101-IT contained
two replication initiation regions, one of the incFIIK1 type and one containing repB and iteron sequences very similar to the plasmid pK245 25. However, the FIIK1 replication origin that contained the repBA genes was missing in pKPC-LK30 (Figure 2). It seemed that the pK245-like repB replication origin alone on pKPC-LK30 could still promote plasmid replication in K. pneumoniae. It is also worth noting that a conserved 36-kb region containing the complete transfer operon (locus
tra-trb) embedded in pKP048 and pKPN101-IT is completely missing in
pKPC-LK30, and this might be the reason why pKPC-LK30 lost its ability to conjugate. It is also likely that the loss of the FIIK1 region may hinder the replication and stability of pKPC-LK30 in some hosts and is thus responsible for the confinement of
plasmid spreading.
One intriguing observation is that the KPC plasmid isolated from Taiwan was more susceptible to aminoglycoside compared to the US control KPC isolate, indicating the difference between the US and Taiwan KPC isolates. According to the sequence analysis, a conserved region containing the gene encoding the 16S rRNA methylase ArmA, which confers high levels of resistance to aminoglycosides, in pKP048 and pKPN101-IT is missing in pKPC-LK30. This may explain the differences in
Sequencing of the pLKEc plasmid from a recently identified E. coli KPC-producer revealed a chimera of three distinct replication regions, including IncI, IncN, and IncF. It is plausible that the incorporation of the different replication machineries may add to the versatility of the plasmid. The emergence of multiple origins of replication in this plasmid is of great concern, as it indicates not only that KPC finally breached the barrier but also that KPC, together with the other β-lactamase genes carried on pKPC-LKEc, may quickly spread to many non-K.
pneumoniae species. The major functional gene clusters responsible for the transfer
of the IncI plasmid pR64 include the traABCD regulatory gene cluster, pil genes in the type IV pilus gene cluster, tra/trb gene cluster, and oriT and nikAB gene cluster for DNA processing 37. A unique shufflon multiple inversion system may alter the C-terminal amino acid sequence of the PilV adhesion protein of the type IV pilus encoded by these plasmids, thus determining the specificity of the recipient cells during conjugation. 37 These findings suggest that the pKPC-LKEc replicon may be capable of conjugative transfer to specific hosts.
Although the KPC-2 plasmid from E. coli was isolated in the same carbapenem resistance monitoring program, and KPC-2-carrying K. pneumoniae clonal
spreading was observed, the plasmid from E. coli was very different from the KPC-2 plasmid from K. pneumoniae. Although the genes annotated on the plasmid suggested that the pKPC-LKEc plasmid was capable of conjugal transfer into different bacteria, and the transferability was confirmed in the laboratory using E.
coli JP-995, conjugal transfer was not successful in all occasions. This is most likely
because PilV adhesin, which determines the specificity of the type IV pilus, is encoded by a shufflon on the plasmid. By altering the C-terminus of the PilV adhesion domain that targets lipopolysaccharide on recipient cells, the specificity of the plasmid for the recipient can be modulated. However, the pKPC-LK30 plasmid from K. pneumoniae does not carry genes known for conjugal transfer and did not show a detectable conjugation ability even in recipients of the same species. The genetic differences between these two plasmids may reflect their differences in cross species transferability, which could have lead to the confined outbreak in Taiwan. The pKPC-LKEc plasmid isolated from E. coli has a relatively high potential of transferability and should be closely monitored to avoid increasing the incidence of plasmid spreading.
References
1. Kumarasamy KK, Toleman MA, Walsh TR et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular,
biological, and epidemiological study. Lancet Infect Dis 2010; 10: 597-602.
2. Walsh TR, Weeks J, Livermore DM et al. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 2011; 11: 355-62.
3. Davies TA, Marie Queenan A, Morrow BJ et al. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007-09. J Antimicrob Chemother 2011; 66: 2298-307. 4. Woodford N, Dallow JW, Hill RL et al. Ertapenem resistance among Klebsiella
and Enterobacter submitted in the UK to a reference laboratory. Int J Antimicrob
Agents 2007; 29: 456-9.
5. Ambretti S, Gaibani P, Berlingeri A et al. Evaluation of Phenotypic and Genotypic Approaches for the Detection of Class A and Class B Carbapenemases in
Enterobacteriaceae. Microb Drug Resist 2013 (online in press).
6. Chia JH, Su LH, Lee MH et al. Development of high-level carbapenem resistance in Klebsiella pneumoniae among patients with prolonged hospitalization and carbapenem exposure. Microb Drug Resist 2010; 16: 317-25.
carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol 2012; 61: 1270-9.
8. Chen YT, Lin AC, Siu LK et al. Sequence of closely related plasmids encoding bla(NDM-1) in two unrelated Klebsiella pneumoniae isolates in Singapore. PLoS
One 2012; 7: e48737.
9. Siu LK, Lin JC, Gomez E et al. Virulence and plasmid transferability of KPC
Klebsiella pneumoniae at the Veterans Affairs Healthcare System of New Jersey. Microb Drug Resist 2012; 18: 380-4.
10. Chmelnitsky I, Shklyar M, Hermesh O et al. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J Antimicrob Chemother 2013; 68: 74-83.
11. Chen TL, Lee YT, Kuo SC et al. Emergence and Distribution of Plasmids Bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother 2010;
54: 4575-81.
12. Huang LY, Lu PL, Chen TL et al. Molecular characterization of beta-lactamase genes and their genetic structures in Acinetobacter genospecies 3 isolates in Taiwan.
Antimicrob Agents Chemother 2010; 54: 2699-703.
13. Zarrilli R, Pournaras S, Giannouli M et al. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 2013;
41: 11-9.
14. Chen LF, Anderson DJ, Paterson DL. Overview of the epidemiology and the threat of Klebsiella pneumoniae carbapenemases (KPC) resistance. Infect Drug
Resist 2012; 5: 133-41.
15. Bratu S, Brooks S, Burney S et al. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin
Infect Dis 2007; 44: 972-5.
16. Chung KP, Tseng SP, Huang YT et al. Arrival of Klebsiella pneumoniae carbapenemase (KPC)-2 in Taiwan. J Antimicrob Chemother 2011; 66: 1182-4. 17. Turton JF, Perry C, Elgohari S et al. PCR characterization and typing of
Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat
and virulence gene targets. J Med Microbiol 2010; 59: 541-7.
18. Jolley KA. Internet-based sequence-typing databases for bacterial molecular epidemiology. Methods Mol Biol 2009; 551: 305-12.
19. Wirth T, Falush D, Lan R et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006; 60: 1136-51.
20. Yeh KM, Lin JC, Yin FY et al. Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis
21. Ma L, Lin CJ, Chen JH et al. Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents
Chemother 2009; 53: 104-11.
22. CLSI. Clinical and Laboratory Standards Institute. Performance standards for
antimicrobial susceptibility testing; 20th informational supplement M100-S17.
Wayne, PA: Clinical and Laboratory Standards Institute, 2010.
23. Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing.
Genome Res 1998; 8: 195-202.
24. Aziz RK, Bartels D, Best AA et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9: 75.
25. Chen YT, Shu HY, Li LH et al. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents
Chemother 2006; 50: 3861-6.
26. Frasson I, Lavezzo E, Franchin E et al. Antimicrobial treatment and containment measures for an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced blaKPC-2 plasmid. J Clin Microbiol 2012; 50: 3768-72.
pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4,
and armA. Antimicrob Agents Chemother 2010; 54: 3967-9.
28. Chen L, Chavda KD, Melano RG et al. Complete sequence of a blaKPC-2
harboring IncFIIK1 plasmid from a Klebsiella pneumoniae sequence type 258 strain.
Antimicrob Agents Chemother 2013. (online in press)
29. Villa L, Garcia-Fernandez A, Fortini D et al. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother
2010; 65: 2518-29.
30. Hedges RW, Datta N. Plasmids determining I pili constitute a compatibility complex. J Gen Microbiol 1973; 77: 19-25.
31. Hall RM. pKM101 is an IS46-promoted deletion of R46. Nucleic Acids Res 1987; 15: 5479.
32. Saadi S, Maas WK, Hill DF et al. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol 1987; 169: 1836-46.
33. Naas T, Namdari F, Bogaerts P et al. Genetic structure associated with bla
OXA-18, encoding a clavulanic acid-inhibited extended-spectrum oxacillinase. Antimicrob Agents Chemother 2008; 52: 3898-904.
Antimicrob Agents Chemother 2012; 56: 5016-22.
35. Kim YA, Qureshi ZA, Adams-Haduch JM et al. Features of infections due to
Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of
sequence type 131. Clin Infect Dis 2012; 55: 224-31.
36. Naas T, Bonnin RA, Cuzon G et al. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother 2013.
(online in press)
37. Sampei G, Furuya N, Tachibana K et al. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 2010; 64: 92-103.
38. Gyohda A, Zhu S, Furuya N et al. Asymmetry of shufflon-specific recombination sites in plasmid R64 inhibits recombination between direct sfx sequences. J Biol Chem 2006; 281: 20772-9.
39. Komano T, Kubo A, Nisioka T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic
Acids Res 1987; 15: 1165-72.
Funding
This work was funded by the project grants, DOH101-DC-1024 and DOH102-DC-1508, from Taiwan Centers for Diseases Control and National Health Research Institutes.
Transparency declarations None to declare
Table 1. Antimicrobial susceptibility tests in recipient serotype K2 K. pneumoniae (R-KP), control KPC-2 K. pneumoniae from the US (US-KPC), KPC-2 isolated from K. pneumoniae in Taiwan (CR-KP), KPC-2 isolated from E. coli in Taiwan (CR-EC) and the E. coli transconjugant (TCR-EC).
Antibiotic Susceptibility R-KP US -KPC CR-KP CR-EC TCR-EC Ampicillin 32 32 32 32 32 Aztreonam 1 32 32 32 32 Cefazolin 2 32 32 32 32 Cefoxitin 2 32 32 32 32 Cefotaxime 1 64 64 64 64 Ceftazidime 1 32 32 8 8 Cefepime 1 32 32 16 8 Ertapenem 0.25 8 8 8 8 Imipenem 0.5 8 8 8 8 Meropenem 0.25 8 8 8 8 Doripenem 0.125 4 4 4 4 Tigecycline 0.25 1 0.25 0.25 0.25 Colistin 1 0.5 0.5 0.5 0.5 Gentamicin 1 16 1 2 1 Amikacin 4 32 4 8 4 Ciprofloxacin 0.125 8 4 4 0.12 Levofloxacin 0.5 8 8 8 0.5 SXT* 0.5 8 8 0.5 0.5
*SXT, trimethoprim-sulfamethoxazole and MIC is indicated as trimethoprim. Bold, significant changes in susceptibility between the donor strain and plasmid recipient.
Figure Legends
Figure. 1(a) Schematic maps of the pKPC-LK30 and (b) pKPC-LKEc plasmids Black blocks on the outer circle are the CDS in the positive strand, and the black blocks on the inner circle are the CDS in the negative strand. The remarkable features are named. The regions similar to the other plasmids/replicons are marked with thick black arcs inside the circles. The non-consensus sections between the conserved regions are shown by thin lines.
Figure. 2 Comparison of the blaKPC-2 region between pKPC_LK30, pKPC_LKEc,
and pKP048
The regions spanning the blaKPC-2 gene on the three plasmids are shown, and the genes are depicted as arrows according to the direction of transcription. The blaKPC-2 genes are shown in black, the pseudogenes in grey, and the IRs are indicated by the vertical bars.
Figure. 3 Comparative views of the major features present in the pKPC_LK30 (GenBank accession number KC405622), pKP048 (FJ628167) and pKPN101-IT (JX283456) plasmids
In this linear diagram, the circular plasmids are depicted with their repB loci at the leftmost ends. The conserved regions of the three plasmids, with reference to
pKPC-LK30, are shown in black and linked by thin lines. These regions include the corresponding conserved regions containing the repB parAB genes, blaKPC-2 and
nearby IS genes, mercury resistance genes (mer), and vagC vagD genes (vag). Three conserved regions between pKP048 and pKPN101-IT but not pKPC_LK30, including a large segment of tra genes, the FIIK1 repB repA replication origin region, and a region carrying the armA resistance gene, are shown in grey. The corresponding regions on pKP048 and pKPN101-IT are inverted and indicated by crossed dotted lines. Mer2 is a second mercury resistance locus in pKPN101-IT.