利用蛋白質聚合物達到訊號放大
全文
(2) Dedication. This thesis is dedicated to my family. I deeply appreciate my parents’ support to my pursue of master’s degree. When I was at the most depressing situation, they kept encouraging me to go ahead and they let me feel comfortable to focus on studying without worry. When I was confused, they gave me precious advice to strengthen my faith and led me to find my own direction. Without their spiritual support and love, I could have not finished the master program.. I.
(3) Acknowledgement. First of all, I would like to acknowledge the most special person, my advisor, Dr. James R. Carey. His enthusiasm for chemistry wins the respect of everyone in the academic society. He is not only a mentor but also a friend to me, he shares lots of chemistry knowledge as well as life experience to me. He makes my learning journey fulfilled with happiness. Sometimes he gives me the pressure to complete the study, sometimes he gives me a spacious room to try and to create experimental methods freely. He always teaches me to be not only a chemist but also a scientist. Examining each piece of evidence carefully and using different approaches to test the same question. Not satisfy with results easily, but pursuit the truth or perfection cautiously. Try to gather the information from various ways, and try to learn from different fields of people. He teaches me not only how to do the study but also how to treat people. He provides an excellent research environment that inspires me to self learning, problemsolving, and self development. I have grown up a lot. Thank you, Dr. Carey. I appreciate everything you have done for me. I owe you too much. Next, I would like to give my thanks to my friends. All the laboratory members give me lots of help, especially my undergraduate student, Chenhao Chen. He works extra hard from day to night voluntary and has never complained to me. He helps me to do experiments and to collect data that enables me to focus on my research. His contribution cannot be II.
(4) ignored. Thank you, Chenhao Chen. I also have to thank for all the PCBAMers, Hueishian Lin, Yuwei Chu and undergraduate student Boyao Wang. Yuwei Chu is good at thinking and creating an idea. The PCBAM proposal can develop further partially due to his contribution. His serious attitude toward research inspires me a lot. Additionally, his creative method stimulates me to develop the new method in our project. Hueishian Lin is an energetic girl who always brings happiness to us. She manages the lab well and gives useful advises about my study. We encourage and discuss the experiment design as well as the experimental results with each other. Boyao Wang is an easy-going man and has full of curiosity. He gives everyone in the laboratory a great help. A lot of work cannot be done without his contribution. Thank to lab manager, Binsyuan Huang. Although she is busy for doing her studies in organic fields, she still takes care of us. She does an excellent job on managing the labs that enables me to focus on my study. Finally, I would like to thank my committee members, Dr. Yaoyuan Chuang and Dr. Yeunghaw Ho. Their comments have been really useful in guiding me to understand the PCBAM project. Thanks to the National Science Council and National Kaohsiung University provides financial support on the research, so I can complete the study smoothly. I would like to acknowledge my parents and the younger sister who I sincerely thank and love.. III.
(5) Table of contents Dedication ................................................................................................... I Acknowledgement .....................................................................................II Table of contents ...................................................................................... IV List of Tables..........................................................................................VIII List of Figures .......................................................................................... IX 摘要...................................................................................................... XVII Abstract ...............................................................................................XVIII Chapter 1. Introduction ...............................................................................1 1.1 Research motivation .....................................................................1 1.2 Protein conglomeration based amplification method ...................3 1.3 SHRPSA method and biHRP method...........................................5 1.4 Properties of streptavidin and biotin.............................................7 1.5 References.....................................................................................9 Chapter 2. Literature Reviews ..................................................................12 2.1 Immunohistochemistry ...............................................................12 2.2 Enzyme-linked immunosorbent assay ........................................13 2.3 Peroxidase antiperoxidase complex technique ...........................15 2.4 Labeled avidin-biotin technique .................................................17 2.5 Bridged avidin-biotin technique .................................................19 2.6 Avidin biotin-peroxidase complex technique .............................20 2.7 Photopolymerization-based amplification..................................22 IV.
(6) 2.8 Dendritic amplification ...............................................................24 2.8.1 Layer by layer dendritic amplification ............................24 2.8.2 Laminar flow-assisted dendritic amplification................25 2.9 References...................................................................................27 Chapter 3. Materials and Methods............................................................28 3.1 Materials .....................................................................................28 3.1.1 ELISA kit .........................................................................28 3.1.2 Buffer ...............................................................................28 3.1.3 Reagents...........................................................................29 3.2 Methods for development of PCBAM........................................29 3.2.1 Synthesis of biotinylated BSA.........................................29 3.2.2 Quantify the amount of biotin on the bBSA and B-HRP 30 3.2.3 ELISA ..............................................................................31 3.2.4 ABC method ....................................................................31 3.2.5 BiHRP method .................................................................32 3.2.6 SHRPSA method .............................................................32 3.2.7 Poly-SHRPSA method.....................................................33 3.2.8 Spectrophometeric analysis .............................................34 3.3 References...................................................................................34 Chapter 4. Protein Conglomeration Based Amplification Method ..........35 4.1 Principles of PCBAM .................................................................35 4.2 Research of ELISA .....................................................................36 4.2.1 The calibration curves of IL-7 and IL-4 ..........................36 V.
(7) 4.2.2 The detection limit of IL-7 ..............................................40 4.2.3 The specific test of IL-4 and IL-7....................................42 4.3 Studies on PCBAM.....................................................................43 4.3.1 Prove the concept of PCBAM step by step .....................43 4.3.2 Detecting lower concentration of AG7 with PCBAM .....45 4.4 Summary .....................................................................................49 4.5 References...................................................................................50 Chapter 5. SDS-Polyacrylamide Gel Electrophoresis ..............................51 5.1 Research intention.......................................................................51 5.2 SDS-PAGE technique .................................................................51 5.3 Materials and methods................................................................52 5.3.1 Materials ..........................................................................52 5.3.2 Procedure of SDS-PAGE .................................................53 5.4 Results and discussion ................................................................55 5.4.1 The demonstration of sample marker ..............................55 5.4.2 Electrophoresis analysis of proteins ................................57 5.4.3 The effect of dithiothreitol...............................................58 5.4.4 Analysis of various antibodies and SA-DA complex ......59 5.4.5 Analysis of SA-DA7 complex in various ratios ..............61 5.5 Summary .....................................................................................62 5.6 References...................................................................................62 Chapter 6. Advance research in PCBAM .................................................63 6.1 Foreword .....................................................................................63 VI.
(8) 6.2 Strategy of development .............................................................65 6.3 Binding ability of reagents..........................................................68 6.3.1 Various reagents in various methods ...............................68 6.3.2 Control experiment of SHRPSA with or without bBSA .71 6.4 Interaction of reagents on the antibody ......................................73 6.4.1 Comparison with SHRPSA and biHRP ...........................73 6.4.2 Different cycles of operation ...........................................76 6.4.3 Incubation time ................................................................78 6.4.4 Various concentration of reagents....................................80 6.5 Apply in detecting antigen ..........................................................83 6.6 Poly-SHRPSA method................................................................85 6.7 References...................................................................................90 Chapter 7. Conclusion...............................................................................91 Chapter 8. Expansion of Currently Approach ..........................................93 8.1 Extension of SHRPSA method ...................................................93 8.2 References...................................................................................94 Biography..................................................................................................95 . VII.
(9) List of Tables. Table 1.. Absorbance in detecting IL-7 antigen with ELISA ................38. Table 2.. Absorbance in detecting IL-4 antigen with ELISA ................38. Table 3.. The detection limit test in low concentration of IL-7 ............40. Table 4.. The specific test of IL-4 and IL-7 ..........................................42. Table 5.. The absorbance of PCBAM in every step..............................44. Table 6.. The procedure of PCBAM .....................................................46. Table 7.. The coefficients of variation in various concentration...........47. Table 8.. Background of various reagents in control experiment .........48. Table 9.. Various conditions in various percentage of SDS-PAGE gel 55. Table 10. The binding ability of various reagents in various methods..69 Table 11. Control experiment of S-HRP with or without bBSA ...........73 Table 12. BiHRP method introduced to DA coated array......................74 Table 13. SHRPSA method introduced to DA coated array ..................74 Table 14. Different cycles of operation..................................................77 Table 15. The various incubation time in SHRPSA method .................79 Table 16. The various ratios of reagents in SHRPSA method...............80 Table 17. The various concentration of reagents in SHRPSA method ..81 Table 18. Apply SHRPSA method in detecting antigen ........................83 Table 19. Various incubation time conditions in poly-SHRPSA ..........88. VIII.
(10) List of Figures. Figure 1.. The presume scheme of forming SA-DA complex. By using. streptavidin-biotin system, Streptavidin and DA at selected ratio 1:3 to form a SA-DA complex. .............................................................................2 Figure 2. The scheme of protein conglomeration based amplification method (PCBAM). PCBAM is performing by using streptavidin bind with biotinylated antibodies to amplify chiefly. Step 1, the antigen is added (e.g. IL-7 or E. coli). Step 2, biotinylated antibody is added. Step 3, streptavidin is added. Step 4 and Step 5, more unbound biotin or secondary antigen is added. These steps will cause oligomerization and precipitation...............1 Figure 3.. The scheme of SHRPSA, biHRP and ABC method. ABC. method is using B-HRP and streptavidin to form the ABC mixture and then bind to the DA. While biHRP method is utilizing S-HRP bind to the B-HRP instead of streptavidin and were added these two reagents layer by layer. SHRPSA method is combining S-HRP and bBSA to form a polymer-like complex to enhance the signal. .............................................6 Figure 4. The structure of streptavidin binding with four biotins. Protein Data Bank (PDB) ID is 1STP......................................................................7 Figure 5. The structure of biotin. A two rings structure with a carboxyl acid side chain, and use this -COOH group reacts with -NH2 groups which on the protein cause biotinylation. Whereas the two rings structure. IX.
(11) can fit in streptavidin pocket through interact with tryptophan residues and other amino groups...............................................................................1 Figure 6.. The scheme of enzyme-linked immunosorbent assay (ELISA).. (A) Primary capture antibody was coated on the micro-immunoassay plate. (B) Capture antibody bound with specific antigen. (C) Secondary biotinylated antibody was added to bind to antigen. (D) Placed in enzyme-conjugated streptavidin (S-HRP) to seize biotin which is on the secondary antibody. (E) In the end, supply TMB substrate to react with HRP enzyme and produce blue solution...................................................14 Figure 7.. Illustration of peroxidase antiperoxidase complex (PAP). method. Primary antibody binds to antigen first and secondary antibody plays a role as a bridge to connect this antigen-antibody complexes and peroxidase antiperoxidase (PAP) complex. PAP complex can react with hydrogen peroxide and DAB•4HCl to be visualized................................16 Figure 8.. Illustration of PAP complex in pentagonal structure. PAP. complex was composed of 3 peroxidase molecules (PO) and 2 anti-peroxidase molecules and stables in pentagonal structure................17 Figure 9.. Illustration of labeled avidin-biotin (LAB) method.. Biotinylated antibody binds to antigen and provides several biotin to react with alkaline phosphatase conjugated avidin (AAP). The whole complex can be visualized by AAP react with staining chemical substrate............18 Figure 10.. Illustration of bridged avidin-biotin (BAB) method.. Employing biotin-labeled antibodies react with antigens first, then treat X.
(12) with avidin as cross-linker to link biotinylated antibodies and biotinylated enzymes. Finally, stain with chemical substrates to react with enzymes to be visualized..............................................................................................19 Figure 11.. Illustration of avidin biotin-peroxidase complex (ABC). method. Primary antibody is added to bind the antigen while biotinylated secondary antibody can detect and bind to the primary antibody. The antibody-antigen complex then combine with the ABC complex at last step. Avidin and B-HRP need to mix with selected ratio to form ABC complex before use. By using TMB, enzyme can be visualized..............21 Figure 12. Synthesis of dual-function molecule. Initiators were bound to -COOH groups through ester linkages of the copolymer. Avidin was bound to the acrylamide groups through amide linkages of the copolymer........22 Figure 13. Illustration of photopolymerization-based amplification. (A) Biotin labeled oligonucleotides was covalently bound to the surface. (B) Treat with dual-functional macromolecules into the surface. (C) The polymer products were formed by photopolymerization………………..23 Figure 14. The scheme of two modes of dendritic amplification. (A) Layer by layer dendritic amplification mode. (B) Laminar flow -assisted dendritic amplification mode. First, anti-CRP was physically adsorbed on the surface. CRP and B-anti-CRP were sequentially bound to anti-CRP to make the sandwich immunocomplex. Next, F-SA and B-anti-SA were alternately introduced to the immunocomplex formed a dendritic structure with n layers (n generations)…………………...……25 XI.
(13) Figure 15. The procedure of ELISA. Primary capture antibody was coated on the microarray plate. Specific antigen and secondary biotinylated antibody was added to form a sandwich complex. Supplied the S-HRP to seize the biotin which on the biotinylated antibody via streptavidin-biotin system. TMB substrate was added to react with HRP enzyme and produce blue solution at the final step..................................37 Figure 16. The calibration curves of (A) IL-7 and (B) IL-4. IL-7 antigen has regular calibration curve from 50.0 pg/mL to 3.1 pg/mL. The r square value is 0.9992. IL-4 antigen shows a linear calibration curve from 35.0 pg/mL to 0.1 pg/mL in ELISA method and r squared value is 0.9976.......1 Figure 17.. The detection limit test in low concentration of IL-7.. Normalize the curve on log-concentration and versus the absorbance. The range of IL-7 antigen concentration is from 1.0×102 pg/mL to 6.2×10-10 pg/mL. A flat line indicated the ELISA has reached the detection limit ..41 Figure 18. Absorbance of IL-7 analyte for each step with PCBAM. Step 1 treats with IL-7 antigen and DA7 whereas step 2 supplies SA-DA4. Step 3 is adding different types of antigen and antibody, AG4 and DA4. While step 4 and step 5 is repeating step 2 and step 3. Step 5 is off scale and not shown in the figure. According to table 5 directly operate the S-HRP and TMB steps after finish each step. ..........................................45 Figure 19. PCBAM linear dilution profiles. Normalize the curve on log-concentration and versus the absorbance. The range of IL-7 antigen concentration is from 6.2×10-3 pg/mL to 6.2×10-8 pg/mL. Absorbance is XII.
(14) increased with the concentration. .............................................................49 Figure 20.. The demonstration of HiMarkTM Pre-stained marker..........56. Figure 21.. Electrophoresis analysis of antibodies, SA and SA-DA4. The. sample at lane 1 and lane 2 are SA, lane 3 and 4 are DA4, lane 5 and 6 are DA7, lane 7 is CA7, and lane 8 and lane 9 are the SA-DA4......................57 Figure 22.. Various samples with or without dithiothreitol (DTT). Lane. 1 to lane 4 is adding DTT in the sample, and respectively adding SA, DA4, DA7, SA-DA7. Whereas lane 5 to 8 is without treating DTT, only adds SA, DA4, DA7, SA-DA7...................................................................................58 Figure 23. Analysis of various antibodies and SA-DA complex. From left to right (1 to 9) is loading DA4, SA-DA4, DA7, SA-DA7, PC7, SA-PC7, PD7, SA-PD7 and SA, independently. Stain with different dyes would obtain (A) coomassie blue staining gel and (B) silver stain gel. ................1 Figure 24.. Analysis of SA-DA7 complex in various ratios. From left to. right (1 to 9) is loading various SA:DA7 ratios. Loading DA7 at first and then 1:4, 1:3, 1:2, 1:1, 1:0.5, 1:0.25, separately. SA is in lane 8 and BSA is in lane 9. The gel was stain with coomassie blue at last.......................62 Figure 25. The definition of cycle operation. Just adding S-HRP which presents in light blue called 0 cycle. Placed the bBSA (light orange) and S-HRP (blue) step by step on the 0 cycle is called 1 cycle. Keep adding bBSA (orange) and S-HRP (purple) would form 2 cycles complex.........64 Figure 26.. The scheme of binding ability test in various methods.. Various methods from left to right are ELISA, ABC, SHRPSA and biHRP XIII.
(15) method. For detecting the bBSA, ELISA is just only adding the S-HRP one time. Whereas ABC method is treated with streptavidin first and then added B-HRP layer by layer. SHRPSA method is using the S-HRP binding to bBSA step by step. While the biHRP method is contained two different HRP reagents, S-HRP and B-HRP. ...........................................................65 Figure 27. The scheme of interaction of reagents on the antibody. Take SHRPSA method for an example, the DA can be detected by S-HRP. Next, the bBSA was added to connect the S-HRP. With layer by layer adding, the signal can be enhance and verify the binding capacity among DA, S-HRP and bBSA......................................................................................67 Figure 28. The scheme of apply method in detecting antigen. Apply the developing method in detect the antigen is the final stage of development. Take SHRPSA method for an example, the signal should be increased from the left to the right. After a series of steps, the result would become polymer-like complex and produced signal amplification. ......................68 Figure 29. Running cycles of various methods. The binding ability test in various reagents with various methods, test B-HRP and S-HRP in biHRP (), test bBSA and S-HRP in SHRPSA(), and test B-HRP and streptavidin in ABC(c) ............................................................................70 Figure 30. The SHRPSA method with or without bBSA. 5% BSA was blocked on the well to avoid other reagents to affect the test. Treating with bBSA is called experimental group; whereas without adding bBSA and use HRP buffer instead is called control group. Comparing the difference after XIV.
(16) running 5 cycles operation........................................................................72 Figure 31.. Comparison with SHRPSA method and biHRP method.. SHRPSA method has an increasing tendency along with operation cycles in detecting 100 pg/mL of DA (). While use biHRP method for detecting 100 pg/mL of DA (c) looks like a horizon line after subtract the background. Same thing happened to SHRPSA at detecting 10 pg/mL of DA (). The absorbance is closed to background and become minus after deduct the background. .............................................................................75 Figure 32. Different cycles of operation in detecting DA. 0 cycle (c) can regard as ELISA and gets a relative flat curve. Nonetheless, after running 3 cycles () or 4 cycles () of operation with SHRPSA method would generate the amplified signal and let the slope become sharper. Besides, the absorbance became distinguishable in various concentration and obtained good r square value at 0.9991 after 4 cycles of operation. ........78 Figure 33.. The various concentration of reagents in SHRPSA. The. absorbance was increased along with concentration of reagents and cycles of operation after remove the background. 5X (c) has higher absorbance than 2X () and original one () though the error bar and background is bigger than others......................................................................................82 Figure 34.. Apply SHRPSA method in detecting antigen. Various. operation cycles in SHRPSA method to detect antigen. 0 cycle, or can consider as ELISA (), 2 cycles (), and 4 cycles (c). The amplified result depends on operation cycles. ..........................................................84 XV.
(17) Figure 35. Illustration of using poly-HRP. Poly-HRP is a reagent that has lots of S-HRP conjugated on the polymer, allows a large number of biotinylated analytes attach to it at once...................................................86 Figure 36. Procedure of poly-SHRPSA method. Poly-HRP was treated into well for 20 min and put the plate on the orbital shaker at 160 rpm in every step. Adds bBSA directly into the well for reacting another 20 min without wash. The total volume in the well would become 200 µL. Wash 3 times and repeat adding poly-HRP and bBSA for 3 cycles. Supply the TMB at last step. .......................................................................................87 Figure 37.. Various incubation time conditions in poly-SHRPSA.. Normally, the more incubation time has more absorbance. While employing ELISA and prolong the incubation time to 40 min, the signal still cannot perform as good as poly-SHRPSA method............................89 Figure 38. The scheme of tyramide signal amplification. TSA method is utilizing tyramide activated by the HRP to covalently bind to proteins. Through the catalytic activity of HRP, tyramide can high-density attach to the electron rich moieties such as tyrosine residues in proteins resulting in signal amplification...................................................................................94. XVI.
(18) 利用蛋白質聚合物達到訊號放大. 指導教授:蘇楷立博士 國立高雄大學應用化學系. 學生:林伯烜 國立高雄大學應用化學系. 摘要 現今越來越多新的疾病被發現在世界上,且大多發現在落後的國家。目前雖 有適當的技術來檢測疾病,但對資源匱乏的環境而言,檢測仍然是一個挑戰。一 個理想的診斷測試其特點應包含廉價、靈敏、特異、操作簡單、快速且穩定(無 需冷藏),最重要的是,不需使用昂貴設備。在傳統的細菌檢測技術上,它是耗 時且勞動力密集的。例如平板計數法(plate count techniques),酶聯免疫吸附試 驗(ELISA) ,生化試驗和聚合酶鏈反應(PCR)等。這些技術依賴於精密儀器, 昂貴試劑和大量的技術工人。對落後的國家和資源貧乏的地區而言,滿足這些需 求仍然是一大問題。 蛋白質聚集放大法(PCBAM)計畫是一種以視覺檢測和鑑定人類樣本中病 原微生物的技術。此新方法的核心在於使用生物素標記牛血清蛋白(bBSA)與 鏈黴親和素連結辣根過氧化物酶(S-HRP)形成類聚合物。該假設是:此類聚合 物可藉化學,生化或物理方法來達到連續訊號放大進而可以肉眼觀測。其長期研 究目標是,檢測血液中極低濃度的細菌樣品,並能藉由簡單步驟診斷微生物感染。 本研究嘗試證明 SHRPSA 法的可行性。如同其名,此法利用 S-HRP 與 bBSA 結合使訊號放大。此外,研究包括探討 SHRPSA 法應用於 ELISA 的偵測極限, 培養時間與試劑濃度比例的優化等。目前我們能證實使用 SHRPSA 法能夠使 IL-7 抗原在低濃度 6.2 pg/mL 下的吸收度放大,且有規律的隨著操作圈數而放大。雖 然其背景值稍高,但與整體的放大結果比較後可忽略。現階段我們仍然需要酵素 免疫分析儀來檢測吸收度,但在往後的研究我們將嘗試使用 poly-HRP 解決背景 值的問題,降低操作時間,並建立檢測器系統以達到不需儀器即可檢測的目的。. 關鍵字:酶聯免疫吸附試驗、生物素標記、鏈黴親和素、生物檢測、訊號放大 XVII.
(19) Signal Amplification by the Generation of Protein Polymer Networks. Advisor: Dr. James R. Carey Department of Applied Chemistry National University of Kaohsiung. Student: Boshiuan Lin Department of Applied Chemistry National University of Kaohsiung. Abstract There are many new diseases that are found in the world today, especially in developing countries. An appropriate technology to detect disease for using in resource poor settings remains a challenge. An ideal diagnostic test is affordable, sensitive, specific, simple to perform with minimal training, rapid, robust (stable without refrigeration), and equipment-free. Traditional bacterial detecting technologies, such as plate count technique, Enzyme-linked immunosorbent assay (ELISA), biochemical tests, and the polymerase chain reaction (PCR) are time-consuming and labor-intensive. The main problem with these techniques being performed in developing countries and resource-poor settings is that they rely on instrumentation, expensive reagents, and are labor intensive. The protein conglomeration based amplification method (PCBAM) proposal is a visual detection technique and is apply to identification of samples that is pathogenic microorganisms from human. The core of this developing method is to utilize biotinyled bovine serum albumin (bBSA), and streptavidin-horseradish peroxidase conjugate (S-HRP) to forming polymer-like complexes. The hypothesis is that the complexes would through chemical, biological or physical reaction to achieve continuous signal amplification and can be visually detected in the end. The ultimate goal of PCBAM is to detect low concentration of bacteria in human blood and develop an easy-to-use pathway to determine the microbial infection. XVIII.
(20) In this study, we attempt to demonstrate the feasibility of a SHRPSA method. As its name, this method uses the combination of S-HRP and bBSA to amplify the signal. In addition, we explored the detection limit of ELISA with SHRPSA method, optimization of the incubation time of the conglomeration components. Now we can confirm that the SHRPSA method can enhance the ELISA signal at 6.2 pg/mL of human IL-7 antigen, and have a regular increase in absorbance with the increase of operation cycles. Though the background is an issue that it can be omitted by comparison of overall the amplification results with increasing absorbance. In current stage, we still need to use the enzyme immunoassay analyzer to detect the absorbance. In future studies, we will try to solve the background issue and reduce the total operation time by using poly-HRP and build up a sensor system to achieve the purpose that the analyte can be detected without instruments.. Keywords: ELISA, biotinylated, streptavidin, biological detection, signal amplification. XIX.
(21) Chapter 1 Introduction N. 1.1 Research motivation Numerous scientists are developed innovative techniques to detecting specific interactions between biological molecules have been demonstrated.1-13 Currently, as few as tens to hundreds of analyte molecules are detected as a result of signal amplification provided by microbial. culture,2. electrocatalysis,9,10. fluorescent. molecular. polymers,3-5. diagnostic,11. and. nanoparticles,6-8 new. enzymatic. strategies.12 However, a long detection time, expensive reagents and luxurious equipment in diagnose disease still needs to overcome. Although these methods are good at detection of pathogens, it is hard to apply in the developing world because they cannot afford using luxurious technology device. Therefore, rapid, sensitive and low-cost development of biosensor to detect pathogenic is warranted. In order to overcome this problem, we focus on Enzyme-linked immunosorbent assay (ELISA) to investigate and develop a method which base on ELISA’s concept but work much better than ELISA. The reason why to choose ELISA is this method have been well-studied and has cheaper reagent than other methods. Also, it has advantage at simple operation and less operation time makes ELISA more attractive. However, the detection limit of ELISA is not low enough to detect as few as tens to 1.
(22) hundreds of analyte molecules in blood. Thus, to design a method which has lower detection limit is needed. The procedure of present sandwich ELISA is use a capture antibody to seize the antigen, and the antigen would bind to biotinylated detection antibodies (DA). Subsequent adding streptavidin labeled with horseradish peroxidase (S-HRP) which is the key reagents of the whole experiment. Finally, the antibody-antigen complex is stained with 3,3',5,5'-tetramethylbenzidine (TMB) and quenched with sulfuric acid.. Biotinylated antibody (DA) Figure 1.. Streptavidin conjugated biotinylated antibodies. Streptavidin. The presume scheme of forming SA-DA complex. By using. streptavidin-biotin system, Streptavidin and DA at selected ratio 1:3 to form a SA-DA complex.. In ELISA procedure, S-HRP is the key reagent since it using streptavidin-biotin. system. and. enzyme. to. detect. the. signal.. Streptavidin-biotin system has high affinity property and is one of the most popular tools in biochemical research.14-19 According to streptavidin is tetravalent protein,20 it can bind to four biotinylated protein. Hence, the hypothesis is if we can control the proportion of the reagents between DA 2.
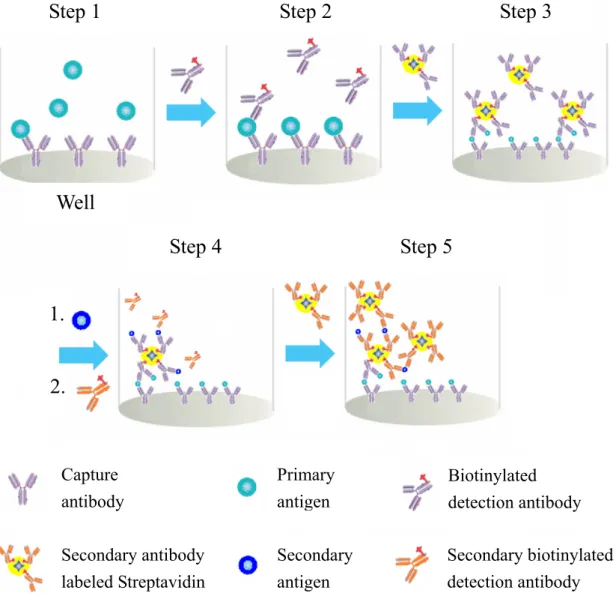
(23) and streptavidin to form a SA-DA complex which has molar ratio at 1:3, as shown in Figure 1. Then SA-DA complex would have 3 amplification sites while the remaining binding site in streptavidin can bind with another secondary DA. Subsequently, add a secondary antigen and secondary antibody, as shown in Figure 2. With layer by layer adding, it will form a gigantic biopolymer to achieve signal amplification. Hence, it has chance to use the naked eye to determine the presence of pathogenic.. 1.2 Protein conglomeration based amplification method Herein, protein conglomeration based amplification method (PCBAM) technique may change nowadays situation. The development of this new method builds up from the basic concept of ELISA procedures. As mentioned above, biotinylated antibody and streptavidin is the key point of new method. Our subject of study is to prove PCBAM is feasible and the results will be optimized. The process of presume scheme is show in Figure 2. Same as ELISA, captured antibodies were pre-coated on the microtiter plate. Antigen (such as IL-7 or E. coli) was added and it would bind to the capture antibodies in step 1. The detection antibodies were following added in step 2. In step 3, the biotin-streptavidin complex with the best ration was added into the well. The step 3 is the key points of PCBAM. In step 4, the secondary antigens and antibodies were added. Finally, step 3 3.
(24) and step 4 were repeated sequentially. The amplification will be achieved by using this method that can produce precipitation or color change so that we can visually detect the pathogen.21 Step 1. Step 2. Step 3. Well Step 4. Step 5. 1. 2.. Capture antibody. Primary antigen. Biotinylated detection antibody. Secondary antibody labeled Streptavidin. Secondary antigen. Secondary biotinylated detection antibody. Figure 2. The scheme of protein conglomeration based amplification method (PCBAM). PCBAM is performing by using streptavidin bind with biotinylated antibodies to amplify chiefly. Step 1, the antigen is added (e.g., IL-7 or E. coli). Step 2, biotinylated antibody is added. Step 3, streptavidin is added. Step 4 and Step 5, more unbound biotin or secondary antigen is added. These steps will cause oligomerization and precipitation. 4.
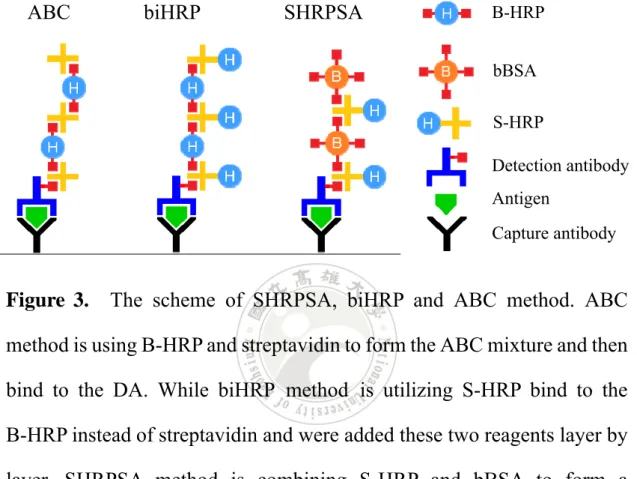
(25) The development is considered possible, because the ideas and principles are close to ELISA. Since PCBAM proposal just adding additional biotinylated antibody and the utilization of streptavidin and detection antibodies is same as ELISA, therefore, the amplification on the surface is predictive and practical.. 1.3 SHRPSA method and biHRP method The degree of popularities in streptavidin-biotin system can be observed in the methods of invention at biochemical field. The original thought of PCBAM proposal is also employ this system which describe in detail above. According to this powerful streptavidin-biotin system, PCBAM can derivative to various detection methods. Here we would like to introduce other methods which derivative from PCBAM, SHRPSA method and biHRP method. SHRPSA is a complex name that is combined with S-HRP and bBSA (biotinylated bovine serum albumin). Another meaning of SHRPSA is streptavidin-horseradish peroxidase polymer-like signal amplification. As its name, this method is combining S-HRP and bBSA to form a polymer-like complex to enhance the signal, as shown in Figure 3. While bi-horseradish peroxidase (biHRP) method is including two kinds of HRP, S-HRP and B-HRP (biotin-horseradish peroxidase). These two methods were inspired by avidin biotinylated peroxidase complex (ABC) method 5.
(26) and PCBAM.22 The principle of ABC method (Figure 3, left) is similar to the PCBAM, both of them are using the tetravalent protein to linkage biotinylated proteins. ABC. biHRP. SHRPSA. B-HRP bBSA S-HRP Detection antibody Antigen Capture antibody. Figure 3.. The scheme of SHRPSA, biHRP and ABC method. ABC. method is using B-HRP and streptavidin to form the ABC mixture and then bind to the DA. While biHRP method is utilizing S-HRP bind to the B-HRP instead of streptavidin and were added these two reagents layer by layer. SHRPSA method is combining S-HRP and bBSA to form a polymer-like complex to enhance the signal.. According to ABC method is use B-HRP and streptavidin to form the polymer-like complex and then bind to the DA, while ELISA uses S-HRP to bind the DA. Here comes out an idea of biHRP which is using S-HRP bind to the B-HRP instead of streptavidin. In this way, the amount of HRP is more than ABC method and the signal can be amplified more. Another idea for amplification of the signals is to introduce bBSA in the system. 6.
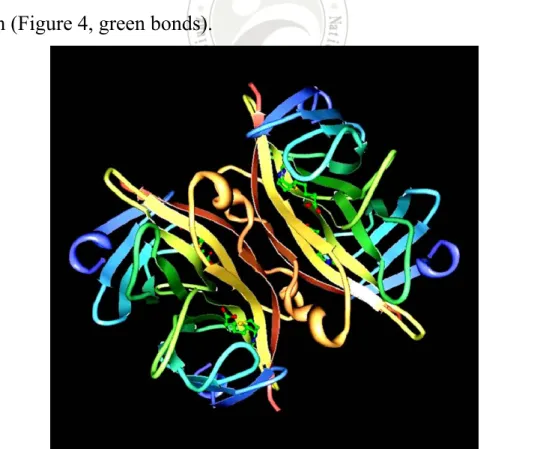
(27) The presumption is there are numerous of biotins on bBSA so that the binding sites of S-HRP are increasing. Hence, combining S-HRP and bBSA might perform a huge amplification on the initial signal.. 1.4 Properties of streptavidin and biotin Streptavidin is a type of protein that is separated from bacteria Streptomyces avidinii; its molecular weight is about 60~65 kDa. It has two binding sites on each of two opposing faces of the structure. As shown in Figure 4, streptavidin consist of four identical peptide chains,20 and each monomer can use tryptophan residues to bind to one molecule of biotin (Figure 4, green bonds).. Figure 4. The structure of streptavidin binding with four biotins. Protein Data Bank (PDB) ID is 1STP. 7.
(28) Isoelectric point of streptavidin is almost at pH 7, so there are few amount of strong charge group at about pH 7. In addition, streptavidin is a slightly acidic protein, and do not have any glycoside. So streptavidin is not glycoprotein and will not bind with lectins that would cause nonspecific binding. Therefore, the immunocytochemical detection system nowadays is using streptavidin more often than avidin. Biotin, which is also known as vitamin H or cofactor R (Coenzyme R), is a small molecule with 244 g/mol molecular weight.23 As shown in Figure 5, biotin is composed of an ureido (tetrahydroimidizalone) ring fused with a tetrahydrothiophene ring with a carboxyl acid side chain. In biochemistry studies, biotin usually reacts with protein as chemical reaction, and this process is called biotinylation.. Figure 5. The structure of biotin. A two rings structure with a carboxyl acid side chain, and use this -COOH group reacts with -NH2 groups which on the protein cause biotinylation. Whereas the two rings structure can fit in streptavidin pocket through interact with tryptophan residues and other amino groups.. 8.
(29) Streptavidin and biotin interaction has extraordinarily high binding ability with dissociate constant (Kd) at 10-15 M. The interaction between steptavidin and biotin is known to be one of the strongest non-covalent interactions. Thus, biotin has priority forming bond with streptavidin and can be detected by streptavidin from the biotinylated sample. In nowadays, streptavidin-biotin system is one of the popular tools used in immunocytochemical research.14-19. 1.5 References (1) Lee, H. J.; Li, Y.; Wark, A. W.; Corn, R. M. Anal. Chem. 2005, 77, 5096-5100. (2). Van der Valk, J.; Brunner, D.; De Smet, K.; Fex Svenningsen, Å.; Honegger, P.; Knudsen, L. E.; Lindl, T.; Noraberg, J.; Price, A.; Scarino, M. L.; Gstraunthaler, G. Toxicology in Vitro 2010, 24, 1053-1063.. (3) Dore, K.; Dubus, S.; Ho, H. A.; Levesque, I.; Brunette, M.; Corbeil, G.; Boissinot, M.; Boivin, G.; Bergeron, M. G.; Boudreau, D.; Leclerc, M. J. Am. Chem. Soc. 2004, 126, 4240-4244. (4) Miranda, O. R.; You, C. C.; Phillips, R.; Kim, I. K.; Ghosh, P. S.; Bunz, U. H. F.; Rotello, V. M. J. Am. Chem. Soc. 2007, 129, 9856-9857. (5) Zeytun, A.; Jeromin, A.; Scalettar, B. A.; Waldo, G. S.; Bradbury, A. R. M. Nat. Biotechnol. 2003, 21, 1473-1479. 9.
(30) (6). Nam, J. M.; Thaxton, C. S.; Mirkin, C. A. Science 2003, 301, 1884-1886.. (7) Sia, S. K.; Linder, V.; Parviz, B. A.; Siegel, A.; Whitesides, G. M. Angew. Chem. Int. Ed. 2004, 43, 498-502. (8) Wu, S.; Zhong, Z.; Wang, D.; Li, M.; Qing, Y.; Dai N.; Li, Z. Microchim. Acta 2009, 166, 269-275. (9) Liu, G.; Wan, Y.; Gau, V.; Zhang, J.; Wang, L.; Song, S.; Fan, C. J. Am. Chem. Soc. 2008, 130, 6820-6825. (10) Das, J.; Aziz, M. A.; Yang, H. J. Am. Chem. Soc. 2006, 128, 1602216023. (11) Wang, Y.; Rollin, J. A.; Zhang, Y. H. P., Mol. Cell. Probe 2010, 24, 15-19. (12) Goodrich, T. T.; Lee, H. J.; Corn, R. M. J. Am. Chem. Soc. 2004, 126, 4086-4087. (13) Liu, H.; Gao, J.; Lynch, S. R.; Saito, Y. D.; Maynard, L.; Kool, E. T. Science 2003, 302, 868-871. (14) Gonzalez, M.; Bagatolli, L. A.; Echabe, I.; Arrondo, J. L. R.; Argarana, C. E.; Cantor, C. R.; Fidelio, G. D. J. Biol. Chem. 1997, 272, 11288-11294. (15) Numajiri, K.; Kuzuya, A.; Komiyama, M. Bioconjugate Chem. 2010, 21, 338-344. (16) Vermette, P.; Gengenbach, T.; Divisekera, U.; Kambouris, P. A.; Griesser, H. J.; Meagher, L. J. Colloid Interface Sci. 2003, 259, 13-26. 10.
(31) (17) Bontempo, D.; Maynard, H. D. J. Am. Chem. Soc. 2005, 127, 6508-6509. (18) Sano, T.; Cantor, C. R. Natl. Acad. Sci. U.S.A. 1995, 92, 3180-3184. (19) Schweitzer, B.; Wiltshire, S.; Lambert, J.; O’Malley, S.; Kukanskis, K.; Zhu, Z.; Kingsmore, S. F.; Lizard, P. M.; Ward, D. C. Natl. Acad. Sci. U.S.A. 2000, 97, 10113-10119. (20) Pei, R.; Cheng, Z.; Wang, E.; Yang, X. Biosens. Bioelectron. 2001, 16, 355-361. (21) Liang, R. Q.; Tan, C. Y.; Ruan, K. C. J. Immunol. Methods 2004, 285, 157-163. (22) Hsu, S. M.; Raine, L; Fanger, H. J. Histochem. Cytochem. 1981, 29, 577-580. (23) Valimaa, L.; Pettersson, K.; Vehniainen, M.; Karp, M.; Lovgren, T. Bioconjugate Chem. 2003, 14, 103-111.. 11.
(32) Chapter 2 Literature Reviews. 2.1 Immunohistochemistry Immunohistochemistry is a technology applies the feature of antibody which can specific bind to antigen. This technology uses a chemical reaction to let the reagents (fluorescent, enzyme, metal ions, isotopes, etc.) attach to the antibodies, and then staining with another chemicals to detect the tissue section or cells whether have target antigen (peptides, proteins, nucleic acids, polysaccharides, pathogens, etc.) exist or not. Therefore, through the antibody is specific to the target antigen, it can be qualitative or quantitative detecting the analyte in situ. In nowadays, there has many methods has been devise via the immunocytochemistry. Various marker substances have been used, such as fluorescent dyes, radioisotopes, ferritin, colloidal gold, enzymes (alkaline phosphatase or horseradish peroxidase), etc. Depending on the types of markers used in these methods, it can be categorized into immunofluorescence, radioimmunoassay, immunogold -silver technique, and enzyme linked immunosorbent assay. While in accordance with operation steps can be divided into the direct (also known as one step) method and indirect (two step, three step or multi step) method; classified according to the binding mode can be divided into antigen-antibody system and streptavidin-biotin system. In this study, we focus on the methods by using enzyme-labeled protein and 12.
(33) streptavidin-biotin system. The following describes the principles of operation in various methods which used in immunohistochemistry.1. 2.2 Enzyme-linked immunosorbent assay The most commonly used of technology in clinical diagnosis is still utilizing immunoassay method.1 Through the antibody has specificity and high affinity to the antigen, the specific antigen or antibody can be detect by quantitative or qualitative analysis. Immunoassay method has the characteristics of diversity and specificity makes it so powerful that stills a perfect detection technology which withstands the test in current biomedical research and analysis applications. Among in various immunoassay methods, the most widely used is enzyme-linked immunosorbent assay (ELISA). In 1971, Engvall’s group first applied in enzyme-linked immuno -sorbent assay for quantitative of Immunoglobulin G (IgG), and named the method as ELISA.2 The characteristics of ELISA is highly sensitive, solid phase analysis, and low cost. It is characterized by the use of enzymes as a marker. When the enzyme combined with antibody, it can react with colorless substrate which produces a colored product.3 The enzyme which frequently used in nowadays ELISA technique are alkaline phosphatase (AP), horseradish peroxidase (HRP) and β-galactosidase.. 13.
(34) (A). (B). (D). (C). (E). Figure 6. The scheme of enzyme-linked immunosorbent assay (ELISA). (A) Primary capture antibody was coated on the micro-immunoassay plate. (B) Capture antibody bound with specific antigen. (C) Secondary biotinylated antibody was added to bind to antigen. (D) Placed in enzyme-conjugated streptavidin (S-HRP) to seize biotin which is on the secondary antibody. (E) In the end, supply TMB substrate to react with HRP enzyme and produce blue solution.. Traditional ELISA is utilizing the antibody which made by affinity chromatography and using non-covalent adsorption to immobilize in stationary phase such as glass chips, plastic chips, microsphere or metal nanoparticles, etc. Take sandwich ELISA for an example, the procedure 14.
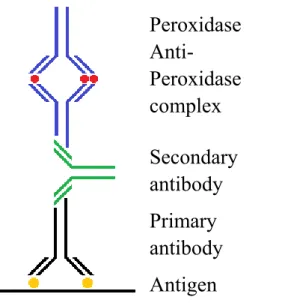
(35) can be brief described as following: Let the primary antibody coated on the micro-immunoassay plate first which has shown in Figure 6. The coated antibody will binding with specific antigen. After secondary antibody bind to antigen, labeled with enzymes or fluorescent chemical and finished in chemical staining.. 2.3 Peroxidase antiperoxidase complex technique In 1970, Sternberger's group established one of non-labeled antibody enzyme methods, peroxidase antiperoxidase complex (PAP) method.4 The principle of PAP staining technique is shown in Figure 7. It is by using secondary antibody as a bridge to connect antigen-antibody complexes and PAP complex. The PAP complex is visualized through utilize of hydrogen peroxide as an enzyme substrate, and an electron donor such as 3,3' -diaminobenzidine tetra-hydrochloride (DAB•4HCl), cause the deposition of colorific product. Studies have shown that the PAP method is 1000 times more sensitive than radioimmunoassay techniques. PAP method has been applied since the early 1990s and has high sensitivity at that time, but the problem is primary antibody and tertiary antibody must all belong to the same animal. Such as primary antibody is belonging to mice, then tertiary antibody has to belong to mice. Furthermore, PAP complex consists of three peroxidase molecules (PO) and two anti-peroxidase molecules and stables in pentagonal structure (see Figure 8). Therefore, PAP complex molecule is too huge to move freely. 15.
(36) Peroxidase AntiPeroxidase complex Secondary antibody Primary antibody Antigen Figure 7.. Illustration of peroxidase antiperoxidase complex (PAP). method. Primary antibody binds to antigen first and secondary antibody plays a role as a bridge to connect this antigen-antibody complexes and peroxidase antiperoxidase (PAP) complex. PAP complex can react with hydrogen peroxide and DAB•4HCl to be visualized.4. Moreover, the overlong incubation time of antibody can produce several adverse consequences such as prone to obtain high background. It would affect the target staining result is not obvious. Also, over incubation might lead to tissue slice slices depart from the slides and impact the success rate of dyeing. Therefore, lots of improvement methods come after PAP method such as ABC metheod.. 16.
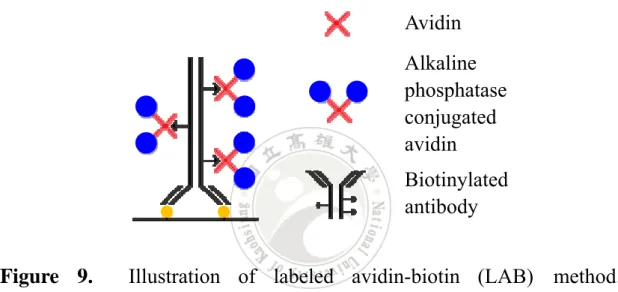
(37) Figure 8.. Illustration of PAP complex in pentagonal structure. PAP. complex was composed of 3 peroxidase molecules (PO) and 2 anti-peroxidase molecules and stables in pentagonal structure.4. 2.4 Labeled avidin-biotin technique Labeled avidin-biotin (LAB) method and the bridged avidin-biotin (BAB) method were established by Guesdon’s group in 1979.5 These two methods are major use biotinylated antibody as primary antibody to detect the antigen, as shown in Figure 9. Besides, LAB method utilize enzyme labeled avidin as secondary antibodies. This method is applied in detecting immune cells, demonstrates the specificity to the immunoglobulin. The operation of LAB method is very simple. As shown in Figure 9, just 17.
(38) involve two adding steps before stain, which are treating biotinylated antibody and alkaline phosphatase conjugated avidin (AAP), individually. In current days, the sensitivity of this technique is relative lower than other methods and has been improved by introducing streptavidin instead of avidin called labeled streptavidin biotin (LSAB) method. Using the characteristics of streptavidin can improve the sensitivity problems.. Avidin Alkaline phosphatase conjugated avidin Biotinylated antibody Figure 9.. Illustration of labeled avidin-biotin (LAB) method.. Biotinylated antibody binds to antigen first and provides several biotin to react with alkaline phosphatase conjugated avidin (AAP). Then the whole complex can be stained by AAP react with staining chemical substrate.5 To compare with ELISA, the principles of these two methods are very similar. The only difference is ELISA introducing another antibody to the detection system. Through secondary biotinylated antibody binds to primary antibody makes ELISA more specific and sensitive. Besides, replace AAP to S-HRP can reduce the background issue makes ELISA has more advantages than LAB method. 18.
(39) 2.5 Bridged avidin-biotin technique Bridged avidin-biotin technology (BAB), also known as BRAB method,5 is use avidin as a bridge to link biotinylate antibodies and biotinylate enzymes, as shown in Figure 10. With avidin as crosslinker makes the antigen which binds to biotinylated antibody can be identify by biotinylate enzymes. As shown in Figure 10, the procedures of detecting the antigen including four adding steps which are using biotin-labeled antibodies react with antigens (Figure 10, yellow spot on the bottom) of cells (or tissue) first, then wash away unbound antibody and treat with avidin, wash away the unbound avidin afterwards and adding biotinylated enzyme. Finally, stain with chemical reagents after wash. Avidin Biotin Biotinylated peroxidase Biotinylated antibody. Figure 10.. Illustration of bridged avidin-biotin (BAB) method.. Employing biotin-labeled antibodies react with antigens first, then treat with avidin as cross-linker to link biotinylated antibodies and biotinylated enzymes. Finally, stain with chemical substrates to react with enzymes to be visualized.5 19.
(40) The BAB method is more sensitive than the LAB method. In the BAB method, employing avidin provides plenty of biotin binding sites (one avidin molecule has four biotin binding sites). Hence the amount of enzyme is more than LAB method and performs better than LAB method. The scientists are always unsatisfied. Some of improvement method comes after BAB method such as ABC metheod (described in the following section).. 2.6 Avidin biotin-peroxidase complex technique ABC method was published by Hsu’s group in 1981.6-7 It is an improved method base on the BAB and LAB method. As like BAB method, the principle is use avidin, which is similar to streptavidin, connected to the biotinylated secondary antibody and biotinylated HRP (B-HRP). The feature is described in Figure 11, avidin and B-HRP need to mix with fixed ratio to forming ABC complex, and primary antibody is not labeled with biotin. Wait till the antigen and antibody are binding together, subsequent adding biotinylated secondary antibody and combine with the ABC complex at last step. The preparation of ABC complexes is to make biotin binding to the HRP, and react with excess of avidin afterwards. This polymer network structure with a large number of enzyme molecules, as long as the avidin has not been filled with biotin, the biotinylated antibodies can bind to the complex through avidin. ABC method is a simple method characterized by 20.
(41) sensitivity and specificity. This is because the biotin and avidin have strong binding force between each other and an avidin molecule has four biotin active sites, as shown in Figure 11. One biotinylated antibody can combined with multiple ABC complex networks, thus the sensitivity greatly enhanced. As the high sensitivity, the concentration of primary antibody and secondary antibody can be diluted to as much as possible, reducing non-specific staining and the total operation time. However, ABC method contains potential problems. The pre-formed avidin and biotin complex is a bulky structure limit the mobility. Besides, the usage of avidin may causes background due to the characteristic of avidin such as contain glycoside, and isoelectric point of about 10. Avidin Biotin conjugated peroxidase AvidinBiotin-peroxidase Complex Biotinylated secondary antibody Figure 11.. Illustration of avidin biotin-peroxidase complex (ABC). method. Primary antibody is first added to bind the antigen while biotinylated secondary antibody can detect and bind to the primary antibody. The antibody-antigen complex then combine with the ABC complex at last step. Avidin and B-HRP need to mix with selected ratio to form ABC complex before use. By using TMB substrates, enzyme can be visualized.6-7 21.
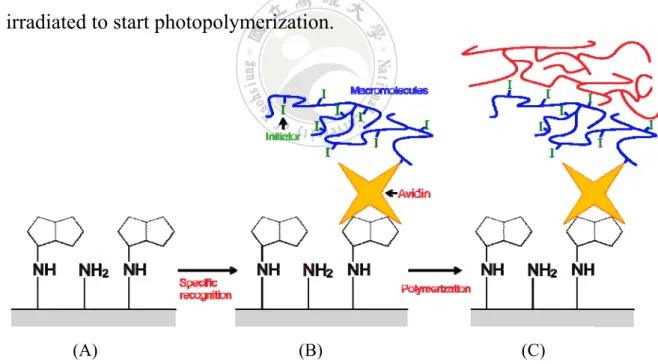
(42) 2.7 Photopolymerization-based amplification Photopolymerization-based amplification was present by Bowman’s group in 2008.8-9 It is via the light to produce a macroscopically visible polymer to amplify signals. The result can be easily observed by naked eye without any detection equipment. The core of the method is by means of using a dual-functional macromolecule that is able to initiating a polymerization reaction and selective recognition to achieve high sensitivity. The scheme of synthesis the dual-functional macromolecule is shown in Figure 12. The avidin-biotin interaction has been widely used in several biochemical fields, so as this technique. With using avidin that has high binding affinity to form the macromolecule, this method also applies in detect the biotinylated protein.. Figure 12. Synthesis of dual-function molecule. Initiators were bound to the -COOH groups through ester linkages of the copolymer. Avidin was bound to the acrylamide groups through amide linkages of the copolymer.8. As author mentioned, in the photoinitiated polymerization, free carbon-based radicals that are derived from organic initiator molecules react with the carbon-carbon double bonds of acrylate monomers, and 22.
(43) polymers are formed via a chain-growth mechanism. The concept of amplification is inherent in chain-growth polymerization reactions owing to the extremely large number of propagation steps that result from a single initiation event. Optical thin-film biosensor surfaces were prepared and biotinylated oligonucleotides were covalently coupled to the surfaces through hydrazone linkages. As shown in Figure 13, the surfaces were placed in contact with a dual-functional macrophotoinitiator which contains the avidin. The avidin binds only to the biotin which on the oligonucleotide and does not bind elsewhere. Then treated with a monomer solution and irradiated to start photopolymerization.. (A). (B). (C). Figure 13. Illustration of photopolymerization-based amplification. (A) Biotin labeled oligonucleotides was covalently bound to the surface. (B) Treat with dual-functional macromolecules into the surface. (C) The polymer products were formed by photopolymerization. 8 23.
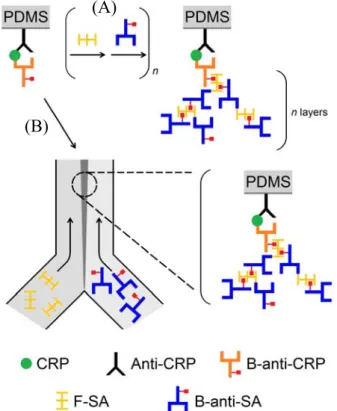
(44) 2.8 Dendritic amplification 2.8.1 Layer by layer dendritic amplification Dendritic amplification method was reported by Hosokawa’s group in 2007.10-11 It can divide into two methods depends on operation: Layer-by-layer dendritic amplification (LBLDA) and laminar flow -assisted dendritic amplification (LFDA). As shown in Figure 14, alternately supplied FITC-labeled streptavidin (F-SA) and biotinylated anti-streptavidin (B-anti-SA) for layer-by-layer growth up to n layers (Figure 14A). The reactions are occurring on the PDMS surface. First, anti-human CRP (anti-CRP) was physically adsorbed onto the surface. After blocking, human C-reactive protein (CRP) and biotinylated anti-human CRP (B-anti-CRP) were sequentially reacted with the surface to make the sandwich immunocomplex, (anti-CRP)-CRP-(B-anti-CRP). Next, F-SA and B-anti-SA were alternately supplied to the immunocomplex. Because both F-SA and B-anti-SA are multivalent, n times repetition of the reaction cycles should yield a dendritic structure with n layers (n generations). However, the author judged that three-layer is the practical upper limit. The LBL amplification has improved the limit of detection (LOD) of CRP by 3 orders of magnitude, from 0.11 nM (13 ng/mL) to 0.21 pM (24 pg/mL) with increasing number of layers (n = 1 to n = 3).. 24.
(45) (A). (B). Figure 14. The scheme of two modes of dendritic amplification. (A) Layer by layer dendritic amplification mode. (B) Laminar flow -assisted dendritic amplification mode. First, anti-CRP was physically adsorbed on the surface. CRP and B-anti-CRP were sequentially bound to anti-CRP to make the sandwich immunocomplex. Next, F-SA and B-anti-SA were alternately introduced to the immunocomplex formed a dendritic structure with n layers (n generations).10. 2.8.2 Laminar flow-assisted dendritic amplification In order to reduce the incubation steps, the Laminar flow-assisted dendritic amplification (LFDA) was devised. LFDA was design by treating two reagents, F-SA and B-anti-SA simultaneously and continuously and 25.
(46) supplied from two laminar streams formed by a Y-shaped microchannel (Figure 14B). As author describe, laminar flow means fluid flow without transverse velocity, when two different solutions (F-SA and B-anti-SA) are pumped into a microchannel, they are mixed only by diffusion through the interface between the two streams. At the contact line among the two streams and the channel floor, the components in the two streams are simultaneously and continuously supplied onto the solid surface. During the flow, the unbind molecule was taken away by the stream, hence needs no washing step and shorten the operation time. The assay can be performed through minimum incubation steps with the LFDA, which exploits the advantage inherent to the microchip format and provides a new opportunity for ultrasensitive point-of-care testing. From this reference, we find that LFDA is extremely similar to our idea, PCBAM. The PCBAM is using a streptavidin-biotinylate antibody complex and antigen to generate the signal, while LFDA using FITC-labeled streptavidin (F-SA) and biotinylated anti-streptavidin (B-anti-SA) to make amplification. The interaction between B-anti-SA and F-SA is not only use streptavidin-biotin system but also use antibody-antigen sytem (SA and anti-SA). LFDA encourages us that PCBAM is a promising proposal.. 26.
(47) 2.9 References (1) Schmitt, O.; Preuße, S.; Haas, S. J. P. Brain. Res. Protoc. 2004, 12, 157-171. (2) Engvall, E.; Perlmann, P. Immunochem. 1971, 8, 871-874. (3) Gosling, J. P. Clin. Chem. 1990, 36, 1408-1427. (4) Sternberger L. A.; Hardy P. H.; Cuculis J. J.; Meyer H. G. J. Histochem. Cytochem. 1970, 18, 315-333. (5) Guesdon, J. L.; Ternynck, T.; Avrameas, S. J. Histochem. Cytochem. 1979, 27, 1131-1139. (6) Hsu, S. M.; Raine, L; Fanger, H. J. Histochem. Cytochem. 1981, 29, 577-580. (7) Avrameas, S.; Uriel, J. C R Acad. Sci. Hebd. Seances. Acad. Sci. D 1966, 262, 2543-2545. (8) Sikes, H. D.; Hansen, R. R.; Johnson, L. M.; Jenison, R.; Birks, J. W.; Rowlen, K. L.; Bowman, C. N. Nat. Mater. 2008, 7, 52-56. (9) Sikes, H. D.; Jenison, R.; Bowman C. N. Lab Chip 2009, 9, 653-656. (10) Hosokawa, K.; Omata, M.; Maeda, M. Anal. Chem. 2007, 79, 6000-6004. (11) Hosokawa, K.; Omata, M.; Sato, K.; Maeda, M. Lab Chip 2006, 6, 236-241.. 27.
(48) Chapter 3 Materials and Methods. 3.1 Materials 3.1.1 ELISA kit Human IL-7 and IL-4 kit which contain IL-7 capture antibody B-N18 (CA7) and detection biotinylated antibody B-S16 (DA7), IL-7 and IL-4 antigen standard, IL-4 capture antibody B-R14 (CA4) and IL-4 detection biotinylated antibody B-G28 (DA4), streptavidin-horseradish peroxidase conjugate (S-HRP), 3,3',5,5'-tetramethylbenzidine (TMB) were purchased from Abcam and Gene-probe. All ELISA reagents were used without further purification. The spectrophotometer measurements were acted by Thermo Scientific Multiskan EX microreader.. 3.1.2 Buffer 0.010 M phosphate-buffered saline (PBS) (Sigma-Aldrich) consisted of 0.137 M NaCl, and 0.270 mM KCl (pH = 7.4) which is known as coating buffer. Another recipe of PBS buffer is 0.100 M Na2HPO4 and 0.100 M NaH2PO4 mixed together to reach pH 7.4. Blocking buffer is consisted of a PBS with 5% (w/v) bovine serum albumin (BSA) (Sigma-Aldrich). Washing buffer is consisted of a PBS solution with 0.05% (v/v) Tween 20 (pH = 7.4). Standard diluent buffer is PBS 28.
(49) contained 1% BSA. PBS-BSA binding buffer is consisted of 1.00 g BSA and 0.88 g NaCl in 100 mL PBS buffer. HRP-streptavidin diluent buffer is contained PBS with 0.1% tween 20 and 1% BSA (w/v). Tris-buffered saline (TBS) is consisted of 0.025 M Tris (Bio-Rad) and 0.150 M NaCl. The pH of TBS is adjusted to 7.2 with 12 N HCl (Sigma-Aldrich).. 3.1.3 Reagents Streptavidin (SA) was purchased from Jackson Immuno Research. Biotin quantitation HABA kits, biotinylated horseradish peroxidase (B-HRP), Poly-HRP, NHS-PEG4-biotin, and NHS-PEG12-biotin were purchase from Thermo Scientific.. 3.2 Methods for development of PCBAM 3.2.1 Synthesis of biotinylated BSA Biotinylated BSA (bBSA) is an important reagent in our methods. It combines protein and biotin together and plays a role to bind to streptavidin or S-HRP. The procedure as following: Add 170 µL of DI water into NHS-PEG4-biotin or NHS-PEG12-biotin, the concentration would become 0.020 M. Took 100 µL of this reagent and mixed with 2.0 mg/mL of BSA. The more NHS-PEG-biotin reagent reacts with BSA would obtain more biotin on the BSA surface. Synthesis was at 4 °C for 29.
(50) overnight. One day later, filter the unreacted NHS-PEG4-biotin or NHS-PEG12-biotin with spin column (cutoff 10 kDa). Loading the bBSA in the spin column and centrifuge at 10000 gravity (g) for 20 min. Discard the filtrate and add PBS into spin column then centrifuge at 10000 g for 20 min again. Operate the filter step 3 times and then reverse the spin column to spin down the bBSA. Centrifuge at 1000 g for 30 min. Reconstitute the bBSA with PBS at last.. 3.2.2 Quantify the amount of biotin on the bBSA and B-HRP Equilibrate the HABA/Avidin Premix to room temperature first, and then add 100 μL of DI water to dissolve the powder and mix with pipette tip. Pipette 800 μL of PBS into a 1 mL cuvette. Use this cuvette with PBS to zero the spectrophotometer. Add the 100 μL of the HABA solution from to the cuvette. Mix by inversion, pipettes it up and down. Measure the absorbance of the solution in the cuvette at 500 nm. Add 100 μL of bBSA, to the cuvette and mix well. Use B-HRP as positive control. Measure the absorbance at 500 nm again. The result shows there has 6 or 7 biotins on each bBSA while B-HRP has 2 biotins.. 30.
(51) 3.2.3 ELISA Took 50 µL of IL-7 capture antibody (1 mg/mL) into 10 mL PBS and dispense 100 µL in each well and coating at 4 °C for overnight. Wash 3 times with wash buffer and block with 5% BSA for 2 h, each well needs 250 µL. Dump the solution out and tap the plate on paper towel after 2 h and put the plate dry on the bench at rt for 24 h. Then the plate is ready to use. Incubated with 100 µL of IL-7 antigen at various concentrations and 50 µL of IL-7 biotinylated detection antibody (DA, 1.2×10-3 µg/mL) incubated for 2 h to form the putative complexes which shown in Figure 6C. After 3 times wash, add 100 µL of S-HRP (1.5×10-4 mg/mL) into well incubated for 20 min. Treat with 100 µL TMB and put in the dark place for 15 min in final step.. 3.2.4 ABC method The protocol of ABC method in this study is a little different to original protocol which describe in the paper.1-2 The reagents are included streptavidin and B-HRP. Therefore, this method is applied in detecting the biotinylated protein such as bBSA or DA. For detecting the bBSA, it should coating the bBSA on the plate first, each well need 100 µL, and the concentration range is from 1.0×103 pg/mL to 3.0×10 pg/mL. Put the plate at 4 °C overnight. Wash 3 times and then add the 100 µL of streptavidin (1.5×10-4 µg/µL) incubate for 20 min. After 20 min, wash 3 times and then 31.
(52) add 100 µL of B-HRP (1.5×10-4 µg/µL) for 20 min. Repeat adding streptavidin and B-HRP until final step. The final step is wash 3 times and adds 100 µL of TMB in each well develops for 15 min in dark place.. 3.2.5 BiHRP method For detecting the bBSA, dispense 100 µL of bBSA (1.0×102 pg/mL) in each well surface first. Put this 96 wells plate at 4 °C for overnight. Dump out the solution and wash 3 times with wash buffer. Tap the plate on paper towel and then dispense 100 µL of S-HRP (1.5×10-4 mg/mL) in each well for 20 min. After incubated the S-HRP, wash 3 times and then add 100 µL of B-HRP (1.5×10-4 mg/mL) incubate for 20 min. Repeat adding S-HRP and B-HRP layer by layer until final step. Wash 3 times then adds 100 µL of TMB in each well and put in the dark place develops for 15 min.. 3.2.6 SHRPSA method SHRPSA method is using S-HRP and bBSA as reagents. In detecting bBSA experiment, dispense 100 µL of bBSA (1.0×102 pg/mL) in each well first; while in DA experiment, dispense various concentration of DA (From 2.0×102 pg/mL to 3.0×10 pg/mL) 100 µL in the well. Put the plate at 4 °C refrigerator for overnight coating. Dump out the bBSA and wash 3 times with wash buffer. Then dispense 100 µL of S-HRP (1.5×10-4 µg/µL) 32.
(53) incubate for 20 min. After 20 min, wash 3 times and then add 100 µL of bBSA (1.5×10-4 µg/µL) for 20 min. Repeat adding S-HRP and bBSA about 3 to 5 cycles. 1 cycle means the operation include adding S-HRP and bBSA. Wash 3 times, treat with 100 µL of TMB and put in the dark place for 15 min in final step.. 3.2.7 Poly-SHRPSA method The significant reagent in this method is use poly-HRP, a polymer conjugate with plentiful S-HRP. In detecting DA experiment, coating the plate as following: Dispense various concentration of DA (From 8.0×102 pg/mL to 1.2×10 pg/mL) 100 µL in the well and put in the 4 °C refrigerator for overnight. Dump out the DA and wash 3 times with wash buffer. Subsequent dispense 100 µL of poly-HRP (1.0×10-4 µg/µL) incubate for 20 min. Put the plate on the orbital shaker at 160 rpm in every step. After 20 min, add 100 µL of bBSA (1.5×10-4 µg/µL) directly into the well for reacting 20 min without wash, the total volume in the well would become 200 µL. After another 20 min, dump out the reagents and wash 3 times. Treat with 100 µL of poly-HRP again. Repeat adding poly-HRP and bBSA for 3 cycles. The cycle means the operation includes adding poly-HRP and bBSA. The final step is wash 3 times and adds 100 µL of TMB in each well develops for 15 min in dark place.. 33.
(54) 3.2.8 Spectrophometeric analysis After added colorless TMB substrate into the well and incubated for 15 min in dark place or wrap with aluminum foil, the color of TMB reagent would turn into blue. The degree of blue color depends on how many HRP on the well surface. Generally, the more concentrated analyte in the well, the color would get darker. Quench with 100 µL of sulfuric acid, the color would change again and become yellow. At meantime this yellow solution were be measured at 450 nm by using an ELISA reader called Thermo Scientific Multiskan EX microreader.. 3.3 References (1) Hsu, S. M.; Raine, L; Fanger, H. J. Histochem. Cytochem. 1981, 29, 577-580. (2) Avrameas, S.; Uriel, J. C R Acad. Sci. Hebd. Seances. Acad. Sci. D 1966, 262, 2543-2545.. 34.
(55) Chapter 4 Protein Conglomeration Based Amplification Method. 4.1 Principles of PCBAM The development of protein conglomeration based amplification method (PCBAM) may improve the current detection technology. The concept of this method is based on the procedure of ELISA. As noted above, biotinylated antibodies and streptavidin (SA) are the core of this method.1 According to the affinity constant (Ka) between streptavidin and biotin is about 1015 M-1, it is like covalently attach to each other in the solution. Once they bind together, the interaction would be an irreversible reaction essentially. Hence, this reaction regards as a high affinity and specific interaction. In addition, the streptavidin-biotin complex is very stable in a wide range of pH value and temperature. Therefore, the streptavidin-biotin system can be widely used in immunocytochemistry such as cell sorting or immunoprecipitation.2 We can take advantage of the feature of streptavidin-biotin system to synthesize the streptavidin-detection antibody complex (SA-DA). Based on SA has four biotin binding sites, utilize one binding site to attach the biotinylated antibody which was on the surface of wells. While other three sites are bind to another types of detection antibody (DA) (see Figure 2, step 3). Then the secondary antigen was added to attach the SA-DA and the secondary antibody was added again at the following step 35.
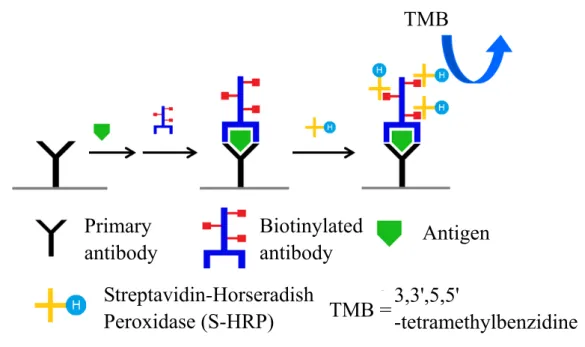
(56) to seize antigen. Subsequent go back to add the SA-DA again. These operation steps were repeated layer by layer, and the large protein polymers would form to achieve signal amplification.. 4.2 Research of ELISA 4.2.1 The calibration curves of IL-7 and IL-4 Since PCBAM is based on the procedure of the sandwich ELISA, realization of the principle of the ELISA is warranted. We exercised the human interleukin-7 (IL-7) ELISA by following to the procedure of the protocol. Steps are described in Figure 15. The first step was supplying 100 μL of various concentration of IL-7 antigen and 50 μL of IL-7 detection antibody (DA7) in the capture antibody pre-coated micro-well plate and incubated at room temperature (rt) for 120 min. Washed three times by washing buffer afterwards. The second step is adding 100 μL of S-HRP to incubate for 20 min. Then washed three times again and supply the 100 μL of TMB in the dark environment (e.g., warp with aluminum foil or put it in the drawer) to develop for 15 min at third step. Finally, 100 μL of 1 M sulfuric acid is use for quench the reaction. Measured the micro-plate with ELISA reader at wavelength of 450 nm. The data was shown in Table 1 and the calibration curve of IL-7 as shown in Figure 16A.. 36.
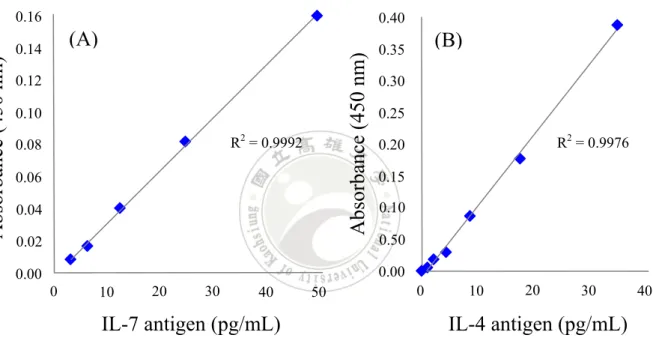
(57) TMB. Primary antibody. Biotinylated antibody. Antigen. Streptavidin-Horseradish 3,3',5,5' TMB = Peroxidase (S-HRP) -tetramethylbenzidine Figure 15. The procedure of ELISA. Primary capture antibody was coated on the microarray plate. Specific antigen and secondary biotinylated antibody was added to form a sandwich complex. Supplied the S-HRP to seize the biotin which on the biotinylated antibody via streptavidin-biotin system. TMB substrate was added to react with HRP enzyme and produce blue solution at the final step.. Table 1 is simply shown the absorbance of concentration of IL-7 antigens ranging from 50.0 pg/mL to 3.1 pg/mL after subtracting the background. The absorbance of IL-7 antigen decreased along with concentration. The differential value of absorbance was relative low at 6.2 pg/mL. It started to close to zero showing ELISA at this concentration is near to the detection limit.. 37.
(58) Table 1.. Absorbance in detecting IL-7 antigen with ELISA. IL-7 antigen (pg/mL). Absorbance. 50.0 25.0 12.5 06.2 03.1. 0.160 0.082 0.041 0.017 0.009. The procedure of human interleukin-4 (IL-4) ELISA is same as IL-7, whereas the antibodies and antigens are using IL-4 detection antibody (DA4) and IL-4 antigen instead. During operate the IL-4, it must be noted that the concentration of IL-4 antigen is different to IL-7. The recommended concentration in the IL-4 protocol is 35.0 pg/mL. As shown in Table 2, the data shows the absorbance which is deducted by background. The concentration ranged from 35.0 pg/mL to 0.1 pg/mL. Observe in Table 2, when the concentration at about 2.2 pg/mL, the absorbance is beginning not clearly to distinguish the difference. Especially between the 1.1 pg/mL and 0.1 pg/mL is extremely closed. Implies that the detection limit of IL-4 is 2.2 pg/mL, approximately.. Table 2.. Absorbance in detecting IL-4 antigen with ELISA. IL-4 antigen (pg/mL). Absorbance. 35.0 17.5 08.8 04.4 02.2 01.1 00.1. 0.387 0.177 0.087 0.031 0.019 0.007 0.000 38.
(59) The absorbance of IL-7 antigen decreases along with concentration and has regular calibration curve in Figure 16A. The r squared value is 0.9992, which is indicating that the test has a high accuracy. IL-4 also shows linear calibration curve in ELISA method which shows in Figure 16B. The calibration curve in IL-4 has r squared value at 0.9976.. 0.40. (A). 0.14. Absorbance (450 nm). Absorbance (450 nm). 0.16. 0.12 0.10 R2 = 0.9992. 0.08 0.06 0.04 0.02. (B). 0.35 0.30 0.25. R2 = 0.9976. 0.20 0.15 0.10 0.50 0.00. 0.00 0. 10. 20. 30. 40. 50. IL-7 antigen (pg/mL) Figure 16.. 0. 10. 20. 30. IL-4 antigen (pg/mL). The calibration curves of (A) IL-7 and (B) IL-4. IL-7. antigen has regular calibration curve from 50.0 pg/mL to 3.1 pg/mL. The r square value is 0.9992. IL-4 antigen also shows a linear calibration curve from 35.0 pg/mL to 0.1 pg/mL in ELISA method and r squared value is 0.9976.. 39. 40.
數據
相關文件
In the fourth quarter of 2003, 4 709 acts of deed were notarized on sales and purchases of real estate and mortgage credits, representing a variation of +19.6% in comparison with
6 《中論·觀因緣品》,《佛藏要籍選刊》第 9 冊,上海古籍出版社 1994 年版,第 1
develop a better understanding of the design and the features of the English Language curriculum with an emphasis on the senior secondary level;.. gain an insight into the
For the more able students, teachers might like to ask them to perform their play to an intended audience as an extended activity. The intended audience might be a primary
Consequently, Technology Education is characterized by learning activities which provide students with authentic experiences in various technological areas such as
The superlinear convergence of Broyden’s method for Example 1 is demonstrated in the following table, and the computed solutions are less accurate than those computed by
Falling prices in outbound package tours, hairdressing and air tickets after the Lunar New Year coupled with the continuous sale on clothing pushed down the indices of OTHER GOODS
On the other hand, rising prices in new arrivals of summer clothing, men’s and women’s footwear and the expiry of waiver of welfare housing rentals by the Housing Institute after