965
DOI 10.1095/biolreprod.103.020552
Distinction of Sperm-Binding Site and Reactive Site for Trypsin Inhibition on P12
Secreted from the Accessory Sex Glands of Male Mice
1Ching-Wei Luo,
3,5Han-Jia Lin,
6S.C.B. Gopinath,
4,6and Yee-Hsiung Chen
2,5,6Institute of Biochemical Sciences,
5College of Science, National Taiwan University, Taipei 106, Taiwan
Taiwan Institute of Biological Chemistry,
6Academia Sinica, Taipei 106, Taiwan
ABSTRACT
Six variants of P12, a Kazal-type trypsin inhibitor in the se-cretion of male mouse accessory sexual glands, were made using single-site mutations including R19L, Y21V, D22G, R43G, K44S, and R45T, based on one-letter-code mutation of amino acids. The other two variants, Nd10 and Cd8, were made using the deletion of 10 and 8 residues from the N- and C-terminals, re-spectively. Their CD profiles revealed maintenance of the P12 conformation in the seven variants, excluding Cd8, which be-came unfolded. Only R19L entirely lost the ability while the other variants were as strong as P12 in inhibiting the trypsin digestion of N-benzoyl-Phe-Val-Arg 7-amido-4-methylcoumarin. The immunocytochemical results demonstrated that D22G and Cd8 failed to bind to sperm, Y21V very weakly did so, and the other variants retained their sperm-binding abilities. Concomi-tantly, the immunocytochemical stainability of each ligand was parallel to its inhibitory effect on125I-P12-sperm binding, and a
synthetic oligopeptide corresponding to residues 18–24 of P12 was able to inhibit P12-sperm binding. The data together con-cluded that R19 was essential for protease inhibition and D22
and/or Y21 mainly being responsible for the binding of P12 to
sperm. The steric arrangement of R19, Y21, and D22on the tertiary
structure of P12 is discussed.
gamete biology, male reproductive tract, male sexual function, seminal vesicles, sperm
INTRODUCTION
Protease inhibitors are ubiquitous in the genital tracts of
mammals [1–3]. It is believed that they are important for
the protection of genital tract epithelium against the damage
of proteolysis [4]. In addition, they have physiological
functions in regulating the fertilization processes [5, 6]. For
instance, the trypsin-like activity seems to involve the
bind-ing of mouse spermatozoa to zona pellucida [7]. Caltrin is
the rat seminal vesicle protein that gives an inhibitory effect
on acrosome protease and is able to suppress Ca
21uptake
by spermatozoa to prevent premature acrosome reactions
1Supported in part by grants (B001-076 and NSC91-2311-B002-049) from the National Science Council, Taiwan.
2Correspondence: Yee-Hsiung Chen, P.O. Box 23-106, Taipei, Taiwan. FAX: 886 2 23635038; e-mail: bc304@gate.sinica.edu.tw
3Current address: Stanford University School of Medicine, Department of Gynecology and Obstetrics, 300 Pasteur Dr., Room S385, Stanford, CA 94305-5317.
4Current address: Functional Nucleic Acids Group, Institute of Molecular and Cell Biology, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Ibaraki 305 8566, Japan.
Received: 23 June 2003. First decision: 23 July 2003. Accepted: 24 November 2003.
Q 2004 by the Society for the Study of Reproduction, Inc. ISSN: 0006-3363. http://www.biolreprod.org
far from the oviduct [8, 9]. Hence, the study of protease
inhibitors in the genital tract becomes an important subject
of molecular reproduction.
The cDNA of P12, a Kazal-type trypsin inhibitor, was
cloned from the mouse ventral prostate by Mills et al. [10].
Because P12 RNA message is detectable in the male
ac-cessory sexual glands of adult mice while its expression is
constitutive in the pancreas [10], substantial progress has
been made in establishing its genomic structure. As a result,
the DNA-binding sites for some transcription factors such
as GC2 and SP1 have been identified in this gene [11, 12].
Meanwhile, we have purified the P12 cDNA-derived
pro-tein with an inhibitory constant (K
i) of 0.15 nM to trypsin
from mouse seminal vesicle secretions (SVS) [13] and
re-covered a recombinant P12 with full activity of the
natu-rally occurring P12 from a chimeric polypeptide of
gluta-thione-S-transferase and P12 (GST-P12) expressed in
Esch-erichia coli [14]. Moreover, we demonstrated a single-type
P12-binding site (1.49
3 10
6sites/cell) with a K
d
value of
70 nM on the anterior region of mouse sperm and showed
the inhibitory effect of P12 on the Ca
21uptake by mouse
sperm [15]. We conducted this study to have a better
un-derstanding of the structural feature for the multifunction
of P12.
MATERIALS AND METHODS
Purification of P12 and Preparation of Oligopeptides
Outbred CD-1 mice were purchased from the Charles River Labora-tories (Wilmington, MA) and were maintained and bred in the animal center at the College of Medicine, National Taiwan University. Animals were treated following the institutional guidelines for the care and use of experimental animals. They were housed under controlled lighting (14L: 10D) at 21–228C and were provided with water and NIH 31 laboratory mouse chow ad libitum. Adult mice (8–12 wk old) were humanely killed by cervical dislocation. Seminal vesicles were carefully dissected to free them from the adjacent coagulating glands, and the secretion collected from 50 mice was placed directly into 50 ml of ice-cold 5% acetic acid. P12 was purified from SVS according to a previously described procedure [13, 16].
The protected oligopeptides were synthesized using stepwise solid-phase methodology on an Applied Biosystems (Foster City, CA) 433A synthesizer with Fmoc chemistry and 4-hydroxymethyl phenoxyacetic acid preloaded resin. After each amino acid coupling, an acetylation step was introduced. After cleavage, the crude sample was purified using reverse phase-HPLC (RP-HPLC) on a C18 300A column (Waters prepacked cartridge 23 25 3 100 mm, 15mm; Waters Corporation, Bedford, MA) with a linear gradient of water and 20% aqueous acetonitrile, both containing 0.1% TFA (v/v), at a flow rate of 20 ml/h. The fractions corresponding to the main peaks were analyzed using RP-HPLC and electrospray ionization-mass spectrometry (ES-MS), pooled, and lyophilized.
Preparation of the Recombinant P12 Variants
For site-directed mutagenesis for P12, we generally followed the in-structions of the Promega Altered Sites II in vitro Mutagenesis Systems kit (Madison, WI). Based on P12 cDNA, the mutagenic oligonucleotides
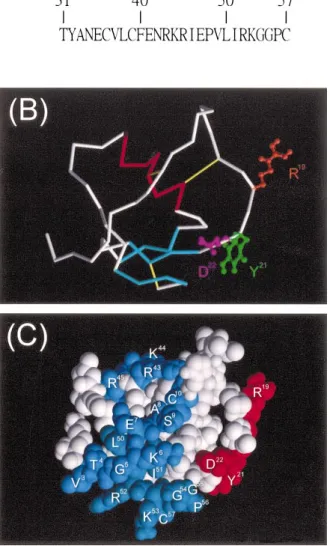
FIG. 1. Molecular structure of P12. The tertiary structure was generated by homology modeling, using the automated Swiss-Model service. A) The amino acid sequence of P12. B) The tubing diagram shows the polypep-tide backbone consisting of one helix (red), one antiparallel b sheet formed by three short strands (cyan), and other structures (white and gray). In addition, three disulfide bridges (yellow) and ball-and-stick structures for the side chains of R19(brown), Y21(green), and D22(purple) toward different directions are displayed. C) The space-filling structure depicts the distributions of R19, Y21, D22, R43, K44, R45, R52, and K53 on the protein surface. R19, Y21, and D22(red) are distant from the N-terminal 10 residues, 43RKR45, and the C-terminal 8 residues (blue).
FIG. 2. Circular dichroism of P12 and its variants. Each protein (0.5 mg/ ml) was in PBS at pH 7.4 at room temperature. The spectra of several proteins are selectively represented. Nd10, R19L, Y21V, D22G, R43G, K44S, and R45T share a very similar CD profile with that of P12.
used for site-directed mutagenesis included 5 9-GGATCATAAATTA-GGGGACATCCCGC-39 (R19L), 59-AGGATCAACAATTCTGGGACAT-39 (Y21V), 59-CACACAGGACCATAAATT CTGG-39 (D22G), 59-GC GTTTCCCGTTTTCAAAGC-39 (R43G), 59-CTCTATGCGACTCCTGTT TTC-39 (K44S), and 59-GGCTCTATGGTTTTCCTGTTTT-39 (R45T), where the mismatched bases are underlined and the mutated proteins are indicated by a one-letter-code mutation of the amino acids in parentheses. The EcoRI/BamHI fragment of P12 cDNA was purified from an expres-sion vector (Gfp), which was previously constructed by the insertion of P12 cDNA into pGEX-2T [14]. The DNA fragment was inserted into pALTER-1. Using single-stranded DNA of the recombinant phagemid DNA as a template, the mutant strand containing each mutagenic oligo-nucleotide was completed according to the manufacturer’s recommenda-tions. The phagemid from each selected transformed colony was se-quenced to confirm the mutation. The procedures for construction of two truncated variants were based on PCR amplification. The cDNA of Nd10 in which the N-terminal 10 residues of P12 were deleted or Cd8 in which
the C-terminal 8 residues of P12 were deleted was amplified using Gfp as a template and the primer pairs of 5 9-CTCGGATCCCATGA-TGCAGTGGCGG-39 and 59-CTCGAATTCTCAGCAAGG CCCAC-39 or
59-CTCGGATCCGCTA AGGT GACTG-39 and 59-CTCGAATTCTC
AGACAGGCTCTATG-39, where the EcoRI or BamHI site is underlined. The EcoRI/BamHI fragment of each P12 variant cDNA was purified and ligated into pGEX-2T, and each constructed vector was transformed into the Escherichia coli strain JM109. Expression of the recombinant protein followed a previously described technique [14]. The transformed cells were harvested and resuspended in 10 ml thrombin reaction buffer (50 mM Tris-HCl, 150 mM NaCl, and 2 mM CaCl2at pH 8.0). Cells were then lysed by refreezing them three times in liquid nitrogen, followed by the addition of DNase I. After centrifugation, the supernatant was passed through an affinity column of glutathione agarose beads (Sigma, St. Louis, MO). According to a method modified from the manufacturer’s instruc-tions, the GST fusion protein in the column was mixed with an equal volume of thrombin reaction buffer containing thrombin (20 U/ml) at room temperature for 4 h. The nonbound fractions were eluted from the column, dialyzed against water, lyophilized to dryness, and redissolved in water for the RP-HPLC on a C4300A column (Waters). The major sample peak in each chromatogram was identified using SDS/PAGE to be a single 6.0-kDa band that was immunoreactive to rabbit antiserum against P12 using the Western blot procedure (data not shown), which indicated that each P12 variant was purified to homogeneity.
Immunocytochemical Staining and Binding Assay
The rabbit antiserum against P12 was prepared [15]. Mouse sperma-tozoa were from the caudal epididymis and were prepared according to a method previously described [17, 18]. The spermatozoa were air dried on a glass slide and fixed with methanol. The slides were washed twice with PBS and preincubated in a blocking solution (3% nonfat skim milk in
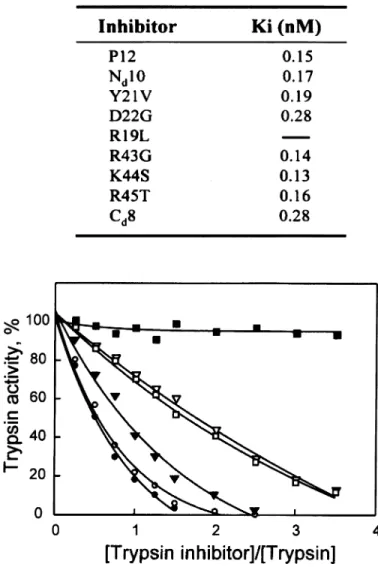
FIG. 3. The inhibitory effects of P12 variants on trypsin kinetics. Trypsin at 1.0 nM and each inhibitor at 0–4.0 nM were incubated at room tem-perature for 3 min before adding the substrate of N-benzoyl-Phe-Val-Arg 7-amino-4 methyl coumarin to a final concentration of 10.0mM. Hydro-lysis of the substrate was measured after 3 min of incubation. The trypsin activity was expressed using the activity measured in the absence of an inhibitor as 100%. Symbols:●, P12; C, Nd10; ., Y21V; □, D22G; ¹, Cd8;m, R19L. Except for R19L being unable to inhibit the enzyme, the kinetic data for trypsin activity in the presence of substrate at 5.0, 10.0, and 25.0mM were analyzed by Dixon plot to determine the inhibitory constant (Ki, at the top of the figure).
PBS) for 30 min at room temperature. Cells were further incubated with 10.0mM P12 or its variant for another 30 min. Alternatively, cells were incubated with each ligand in PBS for 30 min before their fixation on a glass slide for the immunocytochemical analysis. For oligopeptide com-petition, slides were preincubated with 2.0 mM oligopeptides for 30 min before further incubation in the presence of 10.0mM P12 for another 30 min. Slides were washed with PBS three times and then immunoreacted with the P12-induced rabbit antiserum diluted 1:200 in the blocking so-lution for 30 min. Slides were washed three times with PBS to remove excess antibodies before further reacting them with fluorescein-conjugated donkey anti-rabbit IgG (Pharmacia, Uppsala, Sweden) diluted 1:100 in blocking solution for 30 min. All slides were washed with PBS, covered with 50% (v/v) glycerol in PBS, and photographed under a microscope equipped with epifluorescence (AH3-RFCA; Olympus, Tokyo, Japan).
A method modified from that of Markwell [19] was followed to pre-pare125I-P12 according to our method previously used [15]. Spermatozoa (2.53 106cells/ml) and 100 nM125I-P12 in PBS at pH 7.4 were incubated under specified conditions. According to our previously described proce-dure [20], cells were collected on a Whatman GF/C glass microfiber filter (Whatman, Maidstone, Kent, UK), and the filter was counted with ag counter.
Analytical Methods and Spectral Measurement
The kinetic data from the digestion of N-benzoyl-Phe-Val-Arg 7-ami-do-4-methylcoumarin by bovine pancreatic type III trypsin (Sigma) in the presence of an enzyme inhibitor were analyzed using Dixon plots to de-termine the Kiof a tightly binding enzyme inhibitor according to a method previously described [13].
Protein concentration was determined using the BCA protein assay (Pierce, IL) according to the manufacturer’s instructions. The homogeneity of the recombinant polypeptide was determined using SDS/PAGE on a gel slab (10.03 8.0 3 0.075 cm) according to the method of Laemmli [21]. The proteins on the gel were stained with Coomassie brilliant blue or transferred to a nitrocellulose membrane. After the transfer, protein blots were immunodetected using Western procedures with P12-induced rabbit antiserum as the primary antibody and goat anti-rabbit IgG conjugated with horseradish peroxidase (Pharmacia) as the secondary antibody.
The circular dichroism (CD) spectra were measured with a Jasco J-700 spectropolarimeter under constant flushing with N2at room temperature. The mean residue ellipticity,u, was estimated from the mean residue weight, which was calculated from the primary structure.
The cDNA sequence of each variant was read using the dideoxynu-cleotide chain termination method with a primer designed for each DNA concerned. Each base was determined at least three times using an ABI PRISM 377-96 DNA sequencer with an ABI PRISM Big Dye Terminator cycle sequencing ready reaction kit (PE Applied Biosystem).
Molecular Modeling
Following the first approach model reported in the homology-modeling method by Schwede et al. [22], we submitted the amino acid sequence of P12 to the SWISS-MODEL (http://swissmodel.expasy.org/). Template se-lection, alignment, modeling process, and evaluation were completely au-tomated by the server. The auau-tomated modeling procedure started when at least one modeling template was available that had a sequence identity of more than 25% of the P12 sequence. These steps could be iteratively repeated until a satisfying model structure was achieved.
RESULTS
The Tertiary Structure of P12
The group of Kazal-type serine protease inhibitors is
characterized by a well-preserved amino acid sequence
containing three disulfide bridges [23]. Despite that the
strict coincidence of the polypeptide chain folding does not
occur throughout the inhibitor molecules, their extensive
sequence homology suggests a similarity of their overall
three-dimensional structures. In the absence of an x-ray
structure of P12, we extrapolated a comparative model of
its tertiary structure from the homologous modeling using
the SWISS-MODEL, which automatically selected the
known three-dimensional structures of related family
mem-bers, including the porcine trypsin inhibitor [24], human
pancreatic trypsin inhibitor [25], and pig intestine protease
inhibitor [26] as the templates (see Materials and Methods).
The high homology of target-template sequences increased
the model reliability. According to the molecular model
(Fig. 1), C
10, C
17, and C
25were, respectively, cross-linked
with C
39, C
36, and C
57in the formation of three disulfide
bonds. R
19, Y
21, D
22, R
43, K
44, R
45, R
52, and K
53were
dis-tributed over the entire protein surface. Around 40% of the
total amino acid residues formed secondary structures that
included one
a helix, which stretched from E
35to R
43, and
a small antiparallel
b sheet, which comprised the three
twisted strands of
23PVCG
26, 27TDGI
30, and
53KGGP
56. In
addition, there appeared to be a type I reverse turn centered
on G
26and T
27, an irregular reverse turn at the C-terminus
of the
a helix, where
43RKR
45allowed a sharp inversion
of the chain path, and a reverse turn at the C-terminal
re-gion, which were stabilized by a disulfide bond between
C
25and C
57. The remainder of the molecule may have
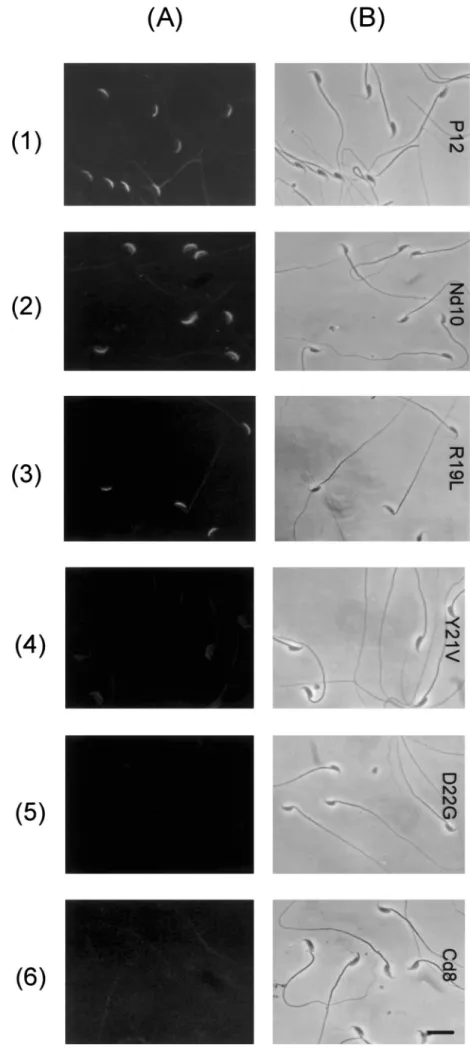
resi-FIG. 4. Cytological demonstration for the binding of a ligand to the acrosome of mouse spermatozoa. Mouse spermatozoa prepared from the caudal epididymis were devoid of P12. Freshly prepared cells were dried on glass slides. Slides were incubated with 15mM of each ligand in PBS for 30 min. The ligand-binding zone on the cell was immunolocalized by an indirect immu-nofluorescence method using rabbit antise-rum against P12 and fluorescein-conjugated anti-rabbit IgG. Slides were observed using a fluorescence microscope (A) or a light microscope (B). Bar5 10 mm.
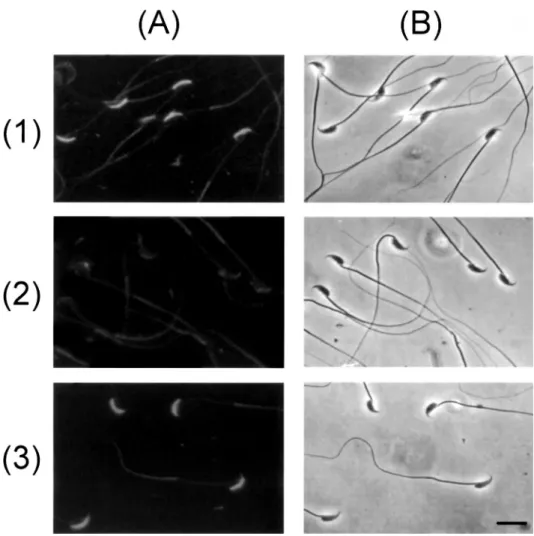
FIG. 5. Cytological examination of the in-hibitory effects of oligopeptides on P12-sperm binding. Freshly prepared P12- spermato-zoa on glass slides were preincubated with 2.0 mM of G-11HDAVAG16(row 1), G-18PRIYDPV24(row 2), or47EPVLIRKGGP56-G (row 3) in PBS for 30 min at room tempera-ture before the addition of P12 to a final concentration of 10.0mM in the cell incu-bation, which proceeded for another 30 min. The P12-binding zone on the cell was immunodetected as described in Figure 5. Slides were observed using a fluorescence microscope (A) or a light microscope (B). Bar5 10 mm.
dues 1–22 may have adopted an extended conformation that
stretches across the entire molecule. Homologous
align-ments of P12 to the known structures of other protease
inhibitors suggested that the reactive site for protease
in-hibition was at the peptide bond between R
19and I
20, and
43RKR
45was the regulatory site for temporary inhibition
[27]. It was noted that residues 1–10 of the N-terminal
re-gion,
43RKR
45, and residues 50–57 of the C-terminal region
were three-dimensionally distant from R
19, Y
21, and D
22in
the loop of residues 17–24.
Protein Conformation of P12 Variants
Based on the structural features of the molecular model,
we prepared eight P12 variants, which were purified to
ho-mogeneity (see Materials and Methods). Six were made
from single-site mutations, including R19L, Y21V, D22G,
R43G, K44S, and R45T, based on one-letter-code mutations
of amino acids. The other two, Nd10 and Cd8, were made
by, respectively, deleting 10 residues of the N-terminus and
8 residues of the C-terminus.
The CD profile of each P12 variant was compared with
that of P12 in PBS (Fig. 2). The spectra of P12 in the
wavelengths of 200–250 nm had two negative bands with
magnitudes of
28.1 3 10
3and
21.31 3 10
4deg·
cm
2·dmol
21around 220 nm (band I) and 205 nm (band II),
respectively, which suggested a considerable amount of
or-dered structures including a helix and a mixture of
b-forms
and
b-turns in the P12 molecule, based on the CD spectra
of the protein conformation [28–30]. The secondary
struc-ture shown in the molecular model of Figure 2 accounts
for the characteristic CD. R19L, Y21V, D22G, R43G,
K44S, R45T, and Nd10 shared very similar spectra with
P12. On the other hand, the protein conformation of Cd8
changed remarkably as evidenced by the disappearance of
bands I and II of the native protein and the appearance of
a strong negative band below 200 nm, which indicated that
Cd8 became unfolded. Apparently, the
b strand of
53
KGGP
56is important to maintaining the P12
conforma-tion.
The Reactive Site for Trypsin Inhibition
and the Sperm-Binding Site on P12
The inhibitory effect of each P12 variant on the
hydro-lysis of N-benzoyl-Phe-Val-Arg 7-amino-4 methyl
couma-rin ducouma-ring the course of trypsin digestion was compared
with its parent protein (Fig. 3). The K
iof each enzyme
inhibitor was determined from the kinetic data (Fig. 3, top).
P12 produced a K
iof 0.15 nM. Except that R19L entirely
lost the ability to inhibit the protease, the other variants
remained active in protease inhibition. Apparently, R
19is
indispensable but the N-terminal 10 residues, Y
21, D
22, R
43,
K
44, R
45, and the C-terminal 8 residues are not essential for
the protease inhibition.
The sperm-binding ability of each P12 variant was
mea-sured. Mouse spermatozoa from the caudal epididymis
were incubated with each ligand, and its appearance on the
cell surface was examined using an indirect
immunofluo-rescence technique. The cells fixed on a glass slide were
incubated with P12 or the cells were incubated with P12
before their fixation on a slide, and no differences in the
FIG. 6. Inhibition of125I-P12-sperm binding by P12 variants. Spermato-zoa (2.53 106cells/ml) in PBS were incubated for 1 h in the presence of 100 nM125I-P12 and 0–10mM P12 or its variant at room temperature. Radioactivity associated with the cells was measured (see text for details). Results are expressed as percentages of counts measured in the absence of unlabeled ligands. Points are the mean of three determinations. The SD of each point is less than 5%. Symbols:●, P12; C, Nd10; ., Y21V; □, D22G; ¹, Cd8; m, R19L. Y21V showed very weak ability, while D22G and Cd8 failed to inhibit the binding of125I-P12 to sperm. The IC
50of each P12 variant was computed using a Cricket Graph and is listed at the top of the figure.
immunocytochemical patterns were examined. Figure 4
dis-plays the fluorescein fluorescence attributed to each ligand
on the cell surface. A crescent fluorescence zone on the
anterior region of the head of P12-treated sperm was seen,
indicating P12-binding sites on the acrosomal region (Fig.
4, row 1). Fluorescence intensity as strong as that of
P12-treated cells was visible on the cells preincubated with
Nd10 and R19L (Fig. 4, rows 2 and 3). Likewise,
immu-nochemical stainability was maintained on cells pretreated
with R43G, K44S, or R45T (data not shown). On the other
hand, very weak fluorescence appeared on Y21V-treated
cells (Fig. 4, row 4), and neither D22G nor Cd8
immuno-chemically stained the sperm acrosome after the cell was
preincubated with either variant (Fig. 4, rows 5 and 6). P12
barely stained cells after their preincubation with a
200-fold molar excess of G-
18PRIYDPV
24for 30 min (Fig. 5,
row 2). Substitution of this oligopeptide with either
G-11
HDAVAG
16or
47EPVLIRKGGP
56-G in the cell
preincu-bation did not hinder the binding of P12 to the cell surface
(Fig. 5, rows 1 and 3). This rules out the sperm-binding
site being within these two peptide regions. Meanwhile,
125
I-P12 was used for quantitative characterization of the
ligand-sperm binding. The sperm-binding ability of each
ligand was assayed by its inhibitory effect on the binding
of
125I-P12 to the cells. Figure 6 displays data of one
rep-resentative determination from cell incubation in the
pres-ence of 100 nM
125I-P12 and each unlabeled ligand. The
IC
50determined for each ligand is given at the top of Figure
6. The radiolabeled P12 bound to the cell surface was
com-pletely inhibited by unlabeled P12 with an IC
50of 98 nM,
but was not inhibited by D22G or Cd8 even when a large
excess of each ligand to
125I-P12 was present in the cell
incubation. Y21V seemed to have very weak
sperm-bind-ing ability, because the concentration of up to 10
mM in
the cell incubation inhibited less than 40% of
125I-P12-sperm binding. R43G was comparable with P12 in the
in-hibition of
125I-P12-sperm binding. Nd10, R19L, K44S, and
R45T were as strong as P12 in binding sperm. Furthermore,
the addition of G
18-PRIYDPV
24to the cell incubation in a
final concentration of 100
mM completely inhibited
125I-P12-sperm binding (data not shown).
DISCUSSION
Our data strongly supported that D
22and/or Y
21but not
R
19were responsible for the sperm-binding site, while R
19but neither D
22nor Y
21was indispensable for trypsin
in-hibition. The sperm-binding site of P12 was not in its
C-terminal region, which was found by noting that
47
EPVLIRKGGP
56G was unable to inhibit P12-sperm
bind-ing. This is in line with the fact that Cd8 became unfolded
and was unable to bind sperm, which indicates that Y
21and
D
22should be in a conformation that fits them into the
P12-binding sites on the sperm head. As shown in the molecular
model (Fig. 1), the loop of residues 17–24 has a rather
certain architecture, such that the side chain of R
19faces
one direction to protrude into the active sites of a
trypsin-like protease, while the side chain of either Y
21or D
22faces
another direction. Such a steric restriction for Y
21/D
22was
maintained in R19L, R43G, K44S, and R45T, by noting
that all of these variants shared a very similar CD profile
with P12, and they were able to bind sperm. Apparently,
the sperm-binding site of P12 did not occur at R
19, R
43,
K
44, or K
45. The gross conformation of P12 was maintained
even when the N-terminal 10 residues were deleted to avoid
the formation of a disulfide bond between C
10and C
39. In
fact, Y
21/D
22in Nd10 retained active sperm binding. This
together with the illustration that G-
11HDAVAG
16was
un-able to inhibit P12-sperm binding ruled out the
sperm-bind-ing site besperm-bind-ing on the N-terminal 16 residues. On the
con-trary, the steric requirement for R
19in trypsin inhibition
was not so rigid that Cd8, which became unfolded,
re-mained active in the protease inhibition.
Winnica et al. [9] reported that excess rat caltrin I or P12
suppressed the proteolytic activity of individual rat or
mouse epididymal spermatozoa. This may be a
conse-quence of acrosome inhibition, blockage of
proacrosin/ac-rosin activation, or acproacrosin/ac-rosin release from acrosome. As
shown previously [15], P12 is exclusively secreted from
male accessory sexual glands and it binds to the plasma
membrane overlaying the acrosome. Because acrosin/
proacrosin should not be exposed on intact acrosomes, the
principal acrosomal protease is unlikely to be the
P12-bind-ing site on the spermatozoal head. More studies are needed
to clarify this aspect. Based on the molecular model shown
in Figure 1, the binding of D
22/Y
21on the P12 molecule to
its receptor on the sperm head from one direction in the
ejaculated semen would turn R
19to the other direction.
Such a structural feature allows for the binding of a
trypsin-like protease from another direction. Therefore, P12 may
dissociate from the sperm head when ejaculated
sperma-tozoa encounter a trypsin-like protease in the female
repro-duction tract under natural circumstance, considering that
P12 gave a K
ivalue of 0.15 nM for the trypsin inhibition
and a K
dvalue of 70 nM for the sperm binding.
Determin-ing the way in which P12 and the protease regulates the
activity of sperm during their transit in the female
repro-ductive tract is worthy of future study.
REFERENCES
1. Fink E, Fritz H. Proteinase inhibitors from guinea pig seminal vesi-cles. Methods Enzymol 1976; 45:825–833.
2. Fritz H, Tschesche H, Fink E. Proteinase inhibitors from boar seminal plasma. Methods Enzymol 1976; 45:834–847.
3. Meloun B, Cechova D, Jonakova V. Homologies in the structures of bull seminal plasma acrosin inhibitors and comparison with other ho-mologous proteinase inhibitors of the Kazal type 1. Hoppe Seylers Z Physiol Chem 1983; 364:1665–1670.
4. Tschesche H, Wittig B, Decker G, Muller-Esterl W, Fritz H. A new acrosin inhibitor from boar spermatozoa. Eur J Biochem 1982; 126: 99–104.
5. Huhtala ML. Demonstration of a new acrosin inhibitor in human sem-inal plasma. Hoppe Seylers Z Physiol Chem 1984; 365:819–825. 6. Cechova D, Jonakova V. Bull seminal plasma proteinase inhibitors.
Methods Enzymol 1981; 80:792–803.
7. Saling PM. Involvement of trypsin-like activity in binding of mouse spermatozoa to zonae pellucidae. Proc Natl Acad Sci U S A 1981; 78:6231–6235.
8. Coronel CE, Winnica DE, Novella ML, Lardy HA. Purification, struc-ture, and characterization of caltrin proteins from seminal vesicle of the rat and mouse. J Biol Chem 1992; 267:20909–20915.
9. Winnica DE, Novella ML, Dematteis A, Coronel CE. Trypsin/acrosin inhibitor activity of rat and guinea pig caltrin proteins. Structural and functional studies. Biol Reprod 2000; 63:42–48.
10. Mills JS, Needham M, Parker MG. A secretory protease inhibitor requires androgens for its expression in male sex accessory tissues but is expressed constitutively in pancreas. EMBO J 1987; 6:3711– 3717.
11. Needham M, Mills JS, Parker MG. Organization and upstream DNA sequence of the mouse protease inhibitor gene. Nucleic Acids Res 1988; 16:6229.
12. Guerin SL, Pothier F, Robidoux S, Gosselin P, Parker MG. Identifi-cation of a DNA-binding site for the transcription factor GC2 in the promoter region of the p12 gene and repression of its positive activity
by upstream negative regulatory elements. J Biol Chem 1990; 265: 22035–22043.
13. Lai ML, Chen SW, Chen YH. Purification and characterization of a trypsin inhibitor from mouse seminal vesicle secretion. Arch Biochem Biophys 1991; 290:265–271.
14. Lai ML, Li SH, Chen YH. Purification and biochemical characteriza-tion of a recombinant mouse seminal vesicle trypsin inhibitor pro-duced in Escherichia coli. Protein Expr Purif 1994; 5:22–26. 15. Chen LY, Lin YH, Lai ML, Chen YH. Developmental profile of a
caltrin-like protease inhibitor, P12, in mouse seminal vesicle and char-acterization of its binding sites on sperm surface. Biol Reprod 1998; 59:1498–1505.
16. Luo CW, Lin HJ, Chen YH. A novel heat-labile phospholipid-binding protein, SVS VII, in mouse seminal vesicle as a sperm motility en-hancer. J Biol Chem 2001; 276:6913–6921.
17. Huang YH, Chu ST, Chen YH. Seminal vesicle autoantigen, a novel phospholipid-binding protein secreted from luminal epithelium of mouse seminal vesicle, exhibits the ability to suppress mouse sperm motility. Biochem J 1999; 343:241–248.
18. Huang YH, Chu ST, Chen YH. A seminal vesicle autoantigen of mouse is able to suppress sperm capacitation-related events stimulated by serum albumin. Biol Reprod 2000; 63:1562–1566.
19. Markwell MA. A new solid-state reagent to iodinate proteins. I. Con-ditions for the efficient labeling of antiserum. Anal Biochem 1982; 125:427–432.
20. Aarons D, Boettger-Tong H, Holt G, Poirier GR. Acrosome reaction induced by immunoaggregation of a proteinase inhibitor bound to the murine sperm head. Mol Reprod Dev 1991; 30:258–264.
21. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680–685.
22. Schwede T, Kopp J, Guex N, Peitsch MC. Swiss-model: an automated protein homology-modeling server. Nucleic Acids Res 2003; 31: 3381–3385.
23. Laskowski M Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem 1980; 49:593–626.
24. Bolognesi M, Gatti G, Menagatti E, Guarneri M, Marquart M, Papa-mokos E, Huber R. Three-dimensional structure of the complex be-tween pancreatic secretory trypsin inhibitor (Kazal type) and trypsin-ogen at 1.8 A resolution. Structure solution, crystallographic refine-ment and preliminary structural interpretation. J Mol Biol 1982; 162: 839–868.
25. Hecht HJ, Szardenings M, Collins J, Schomburg D. Three-dimension-al structure of a recombinant variant of human pancreatic secretory trypsin inhibitor (Kazal type). J Mol Biol 1992; 225:1095–1103. 26. Liepinsh E, Berndt KD, Sillard R, Mutt V, Otting G. Solution structure
and dynamics of PEC-60, a protein of the Kazal type inhibitor family, determined by nuclear magnetic resonance spectroscopy. J Mol Biol 1994; 239:137–153.
27. Kikuchi N, Nagata K, Shin M, Mitsushima K, Teraoka H, Yoshida N. Site-directed mutagenesis of human pancreatic secretory trypsin in-hibitor. J Biochem (Tokyo) 1989; 106:1059–1063.
28. Chen YH, Yang JT, Martinez HM. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dis-persion. Biochemistry 1972; 11:4120–4131.
29. Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochem-istry 1974; 13:3350–3359.
30. Chang CT, Wu CS, Yang JT. Circular dichroic analysis of protein conformation: inclusion of the beta-turns. Anal Biochem 1978; 91: 13–31.