© 2001 Kluwer Academic Publishers. Printed in the Netherlands. 67
Each species of
Glycine
collected in Taiwan has a unique seed protein
pattern
J.S. Hsieh
2, K.L. Hsieh
1, Y.C. Tsai
1& Y.I. Hsing
1∗1Institute of Botany, Academia Sinica, Taipei, Taiwan;2Department of Agronomy, National Taiwan University,
Taipei, Taiwan; (∗author for correspondence)
Received 8 September 1999; accepted 3 June 2000
Key words: Glycine species, GmPM proteins, heat soluble proteins, Western blot analysis
Summary
Several annual and perennial species in the genus Glycine Willd., including G. soja, long-pod G. tomentella, short-pod G. tomentella and G. tabacina, collected in Taiwan and nearby islands were studied for variations of their seed proteins. SDS-PAGE and Western blotting were used to analyze the total proteins, the heat soluble proteins, six seed maturation proteins (GmPMs) and one seed storage protein. The various species had different patterns of seed heat soluble proteins. In addition, each species of Glycine collected in Taiwan exhibited unique seed maturation protein patterns. They had several cross-reactive polypeptides recognized by specific antibodies against GmPM1, GmPM2 and GmPM8, but only one polypeptide recognized by antibodies against GmPM4, GmPM5 and MP130. The long pod G. tomentella, which has been suggested as a new species and renamed as G. dolichocarpa, could be distinct from the short pod G. tomentella on the basis of the analysis of these biochemical markers. It is also indicated that these GmPM antibodies may be used to distinguish between and within other Glycine species.
Abbreviations: GmPM – Glycine max physiological mature; LEA – late embryogenesis abundant
Introduction
The cultivated soybean, Glycine max (L.) Merr., is a major crop and has received a considerable atten-tion in the area of taxonomical studies. The genus
Glycine Willd. consists of many species belonging to
two subgenera, Soja and Glycine (Newell & Hymow-itz, 1983). The subgenus Soja includes the diploid (2n=40) cultivated soybean, G. max (L.) Merr., and its wild relative, G. soja Sieb. and Zucc. Both G.
max and G. soja are annual, and intercross freely
(Palmer et al., 1987). The subgenus Glycine includes about 15 described species, all of which are peren-nial (Hymowitz & Singh, 1987). G. soja is widely distributed including Russia, Korea, Japan, mainland China and Taiwan (Hymowitz & Singh, 1987), with Taiwan being the southernmost part of these areas. The two wild perennial Glycine species, G. tomentella Hayata and G. tabacina Benth., which have been sug-gested to be the probable ancestors of G. soja, are
found in Ryukyu islands, Taiwan, the Philippines, the South Pacific islands and Australia (Hymowitz et al., 1998), with Japan being the northernmost part of these areas. All other perennial Glycine species are found only in Australia (Tindale & Craven, 1988). There-fore, Taiwan and Japan are unusual because they have representatives of both subgenera. The collection and study of wild soybean and their relatives in Taiwan and adjacent islands thus provide information for a better understanding of the evolution and relation-ships between these two subgenera. Accordingly, we have analyzed the seed proteins of different Glycine species collected in Taiwan by SDS-PAGE followed by immunoblotting. These proteins included GmPM (Glycine max physiological mature) proteins, the seed storage proteins, and the soluble proteins after heat treatment. The general characteristics of these proteins are described in the following paragraphs.
Proteins synthesized during late maturation of seeds are called maturation proteins (MP)
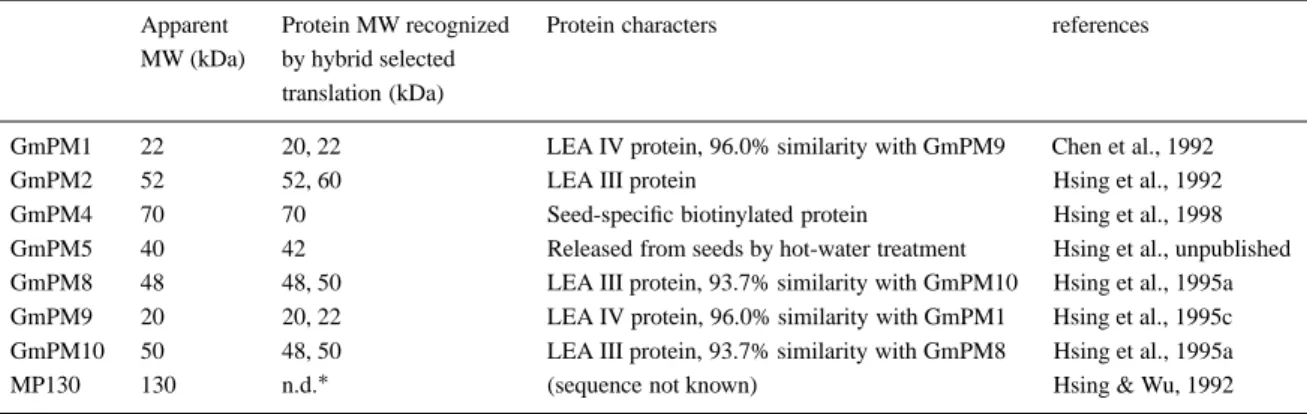
(Rosen-Table 1. Characteristics of the GmPM proteins in G. max seeds
Apparent Protein MW recognized Protein characters references MW (kDa) by hybrid selected
translation (kDa)
GmPM1 22 20, 22 LEA IV protein, 96.0% similarity with GmPM9 Chen et al., 1992
GmPM2 52 52, 60 LEA III protein Hsing et al., 1992
GmPM4 70 70 Seed-specific biotinylated protein Hsing et al., 1998 GmPM5 40 42 Released from seeds by hot-water treatment Hsing et al., unpublished GmPM8 48 48, 50 LEA III protein, 93.7% similarity with GmPM10 Hsing et al., 1995a GmPM9 20 20, 22 LEA IV protein, 96.0% similarity with GmPM1 Hsing et al., 1995c GmPM10 50 48, 50 LEA III protein, 93.7% similarity with GmPM8 Hsing et al., 1995a
MP130 130 n.d.∗ (sequence not known) Hsing & Wu, 1992
∗not determined.
berg & Rinne, 1986) or late embryogenesis abundant (LEA) proteins (Galau et al., 1986), which are cor-related with desiccation tolerance (Blackman et al., 1991), ABA content (Hughes & Galau, 1991), and transition to seedling growth (Rosenberg & Rinne, 1986). We have selected several cDNA clones encod-ing seed maturation proteins from a soybean (G. max cv. Shi-shi) pod-dried seed cDNA library by differen-tial screening (Hsing et al., 1990; Hsing & Wu, 1992). These clones are designated as GmPM. According to the results of genomic Southern analysis of several
Glycine species, most of the GmPM messages are
en-coded by single or low copy-number genes (Hsing et al., 1995b); therefore, it is likely that each GmPM antibody detects only one or a few protein bands. An-tibodies were raised against several of these GmPM proteins, whose characterization are listed in Table 1.
There are two major storage proteins in cultiv-ated soybean seeds. One is glycinin, which consists of acidic subunits and basic subunits (Kitamura et al., 1976). The other is conglycinin, which is a glycopro-tein composed of three (α’,α, and β) major subunits (Thanh & Shibasaki, 1977). These storage proteins are easily separated and recognized by SDS-PAGE and thus may be used as genotype markers.
Some seed proteins may remain in the supernatant after heat treatment of seed extracts. For instances, soybean seed heat soluble proteins are accumulated during desiccation period of seed development (Black-man et al., 1991). They can be obtained easily and rep-resent only a small fraction of the total seed proteins. We have also analyzed these heat soluble proteins in different Glycine species.
A wide variety of approaches, including cytogen-etics, morphology, isozyme, RFLP, RAPD and AFLP,
have been used to define the systematic relationships among species. These approaches necessitate large amounts of samples and/or efforts. However, the ap-proach using SDS-PAGE following by immunoblot-ting of seed proteins require only five seeds and two working days. The objectives of the current study were firstly to detect the above-mentioned proteins in
Glycine species collected in Taiwan and secondly to
test if they are polymorphic to reveal the systematic relationships among the Glycine species.
Materials and methods
Plant materials
Soybean (Glycine max cv. Shi-shi) seeds were kindly provided by the Kaohsiung Agricultural Experimental Station, Pintung, Taiwan. The plants were grown to maturity at the Experimental Farm, Institute of Botany, Academia Sinica, Taiwan.
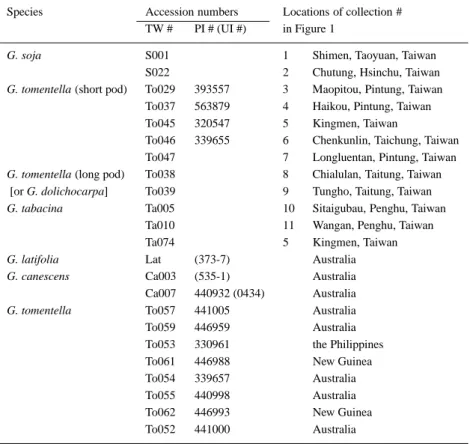
Seeds of wild soybeans and their relatives were collected from Taiwan and the nearby islands by Drs J.S. Hsieh and Y.C. Huang, Department of Agronomy, National Taiwan University. The accession numbers and locations of the collection are shown in Table 2 and Figure 1. The known permanent plant introduction (PI) numbers are also indicated. These wild soybeans were grown to maturity in a greenhouse at the Institute of Botany, Academia Sinica.
Protein extraction and analysis
Total seed soluble proteins were extracted by ho-mogenization of the seeds in an ice-cold grinding buffer consisting of 63 mM Tris-HCl (pH 7.8), 20
Table 2. Accessions of the wild soybean and their relatives used in this study and their places of collection
Species Accession numbers Locations of collection # TW # PI # (UI #) in Figure 1
G. soja S001 1 Shimen, Taoyuan, Taiwan
S022 2 Chutung, Hsinchu, Taiwan G. tomentella (short pod) To029 393557 3 Maopitou, Pintung, Taiwan
To037 563879 4 Haikou, Pintung, Taiwan To045 320547 5 Kingmen, Taiwan
To046 339655 6 Chenkunlin, Taichung, Taiwan To047 7 Longluentan, Pintung, Taiwan G. tomentella (long pod) To038 8 Chialulan, Taitung, Taiwan
[or G. dolichocarpa] To039 9 Tungho, Taitung, Taiwan G. tabacina Ta005 10 Sitaigubau, Penghu, Taiwan
Ta010 11 Wangan, Penghu, Taiwan
Ta074 5 Kingmen, Taiwan
G. latifolia Lat (373-7) Australia
G. canescens Ca003 (535-1) Australia Ca007 440932 (0434) Australia G. tomentella To057 441005 Australia To059 446959 Australia To053 330961 the Philippines
To061 446988 New Guinea
To054 339657 Australia To055 440998 Australia
To062 446993 New Guinea
To052 441000 Australia
mM MgCl2, 10 mM 2-mercaptoethanol and 1 mM
PMSF (phenylmethyl-sulphonyl fluoride). Following homogenization, an equal volume of Laemmli protein solubilization buffer (Laemmli, 1970) was added. The slurry was incubated at 100◦C for 10 min, and then subjected to SDS-PAGE. Electrophoresis (12.5% slab gel) was performed in the Laemmli system, and the gels were stained for proteins with Coomassie blue. About 100 µg proteins were loaded onto each lane.
Heat soluble proteins were extracted by homo-genization of soybean seeds in an ice-cold grinding buffer consisting of 60 mM Tris-HCl (pH 7.8) and 500 mM NaCl. The slurry was transferred to a centrifuge tube and incubated at 100◦C for 10 min and then at 4◦C for 10 min. Nonsoluble proteins including those coagulated by heat were removed by centrifugation. Proteins in the supernatant were concentrated by acet-one precipitation. Subsequently, the Laemmli protein solubilization buffer was added to the precipitate, and the slurry was incubated at 100◦C for 10 min, and then subjected to SDS-PAGE. Electrophoresis (12.5% slab
gel) was performed in the Laemmli system, and the gels were stained for proteins with Coomassie blue. About 60 µg proteins were loaded onto each lane.
Preparation of antisera and immunoblotting
Eight specific soybean seed proteins were analyzed by SDS-PAGE followed by immunoblotting. The sera against glycinin, MP130 and GmPM5 were prepared by immunizing rabbits with each of these protein sep-arated by SDS-PAGE. The gel stripes containing the proteins were excised, grounded and then used for injection. The sera against seed maturation proteins GmPM1, GmPM2 and GmPM8 were prepared by im-munizing rabbits with purified recombinant proteins solubilized in 10 mM phosphate buffer (pH 7.0) and mixed with adjuvant before the injection. GmPM4 is a seed-specific biotin binding protein (Hsing et al., 1998), and alkaline phosphate-conjugated streptavidin (Boehringer Mannheim) was used for its detection.
Figure 1. Distribution of Glycine accessions collected in Taiwan used in the present study. The indicated accessions are described in Table 2.
Immunoblotting after SDS-PAGE was performed as described by Towbin et al. (1979). The dilution factors used for each primary antibody were: GmPM1, 50,000; GmPM2, 80,000; GmPM4, 2,000; GmPM5, 20,000; GmPM8, 160,000; MP130, 20,000 and gly-cinin, 10,000. For the secondary antiserum, goat anti-rabbit IgG conjugated to alkaline phosphatase was used, and nitroblue-tetrazolium was used as the chromogenic substrate.
Results and discussion
Total proteins and heat soluble proteins in seeds of Glycine species
SDS-PAGE analysis of total soluble seed proteins showed no significant difference in the banding pat-terns of glycinin and conglycinin in G. soja species and cultivated species. However, the polypeptide sizes of these proteins varied slightly in G. tomentella and
G. tabacina (Figure 2). Of the heat soluble protein
pro-files, there were prominent differences among those of G. max, G. soja, long-pod G. tomentella, short-pod G. tomentella and G. tabacina (Figure 3). There were several polypeptides present in G. soja but not
Figure 2. SDS-PAGE of total soluble seed proteins of various Gly-cine species. Proteins were prepared from (a) G. max Shi-shi; (b) G. soja S001, (c) S022; (d) G. tomentella (short pod) To029, (e) To037, (f) To045, (g) To046, (h) To047; (i) G. tomentella (long pod) To038, (j) To039;(k) G. tabacina Ta005, (l) Ta010, (m) Ta074. Arrows indicate molecular markers in kDa. The positions of α and β-conglycinin, and the acidic (aci.) and basic (bas.) polypeptides of glycinin, are indicated on the right.
Figure 3. SDS-PAGE of seed heat soluble proteins of various Gly-cine species. The accessions and their labelings are the same as those in Figure 2.
Figure 4. Immunostaining after SDS-PAGE of soybean seed proteins recognized by specific seed protein antibodies. The cross-reactive polypeptides recognized by antibodies against GmPM1 (Panel A), GmPM2 (Panel B), GmPM4 (Panel C), GmPM5 (Panel D), GmPM8 (Panel E), MP130 (Panel F) and glycinin (Panel G) are illustrated. The accessions and their labelings are the same as those in Figure 2. Numbers on the left indicate the apparent molecular weights of specific polypeptides.
in G. max. These polypeptides included those of 20 kDa, 30 kDa, and 50 - 75 kDa. The most obvious difference between the annual and perennial Glycine species was the presence of a 130-kDa protein in the former species and a 120-kDa protein in the latter species. The long-pod and short-pod G. tomentella gave very distinct patterns, especially in the proteins ranging of 25–35 kDa and 40–75 kDa. The three G.
tabacina accessions exhibited a similar protein pattern
among themselves, but this pattern was quite different from those of other species. Overall, the soybean seed heat soluble protein profile is a more useful parameter than the total seed protein profile for distinguishing the
Glycine species.
Seed proteins recognized by antibodies
Seven antibodies prepared against distinct soybean seed proteins were used to detect their presence in the Glycine species. The results of Western blot ana-lysis are shown in Figure 4. Two obviously different types of antibody reactivity were observed: 1. several polypeptides recognized by one antibody and 2. only one polypeptide specifically recognized by one anti-body. The antibodies against GmPM1, GmPM2 and GmPM8 belong to the first type, and the other four antibodies belong to the second type. These two types of antibody reactivity coincided well with the results of hybrid-select translation (Table 1).
Anti-GmPM1 antibody recognized two identical polypeptides in G. max and G. soja seeds. It re-cognized two polypeptides in the five accessions of short-pod G. tomentella, one polypeptide in two long-pod G. tementella, and three polypeptides in three
G. tabacina; the apparent molecular mass of these
recognized polypeptides were quite different. Anti-GmPM2 antibody detected two polypeptides of differ-ent sizes in G. max and G. soja. It detected two poly-peptides in short-pod G. tomentella, four polypoly-peptides in long-pod G. tomentella, and three polypeptides in
G. tabacina; again, these polypeptides were different
in sizes. One of the accessions of short-pod G.
to-mentella, To047, gave a pattern which was slightly
different from that of other short-pod G. tomentella (lane h, Figure 4B). Anti-GmPM8 antibody reacted with two polypeptides in G. max and only one poly-peptide in G. soja. It reacted with three polypoly-peptides in short-pod G. tomentella, long-pod G. tomentella and G. tabacina; these three polypeptides were dif-ferent in sizes. One accession of the short-pod G.
tomentella, To037, gave a pattern which was slightly
different from that of other short-pod G. tomentella (lane e, Figure 4E). Hybrid select translation prepared from G. max seeds indicated that there was only one polypeptide of GmPM4 or GmPM5. Anti-GmPM4 or anti-GmPM5 each detected only one polypeptide of an identical size in all the accessions of Glycine tested. Also, there was one polypeptide detected by anti-MP130 in all Glycine species, but the size of the polypeptide in the perennial Glycine species was smal-ler, about 120 kDa. Anti-glycinin recognized one or two polypeptides of similar sizes in all the Glycine species tested (Figure 4G).
Based on Western blot analysis, several conclu-sions can be drawn (Figure 4). There are two types of soybean seed maturation proteins, one including
GmPM1, GmPM2 and GmPM8, and the other in-cluding GmPM4, GmPM5 and MP130. For each species, the former type contains several different cross-reactive polypeptides, and gives distinct patterns for each accession. There are one or two polypeptides in G. max and G. soja while are many polypeptides in G. tomentella and G. tabacina. These three GmPM (GmPM1, GmPM2 and GmPM8) proteins belong to group 3 or 4 LEA proteins (Chen et al., 1992; Hsing et al., 1992; Hsing et al., 1995a). The latter type gives only one cross-reactive polypeptide with an identical or slightly different molecular weight for each acces-sion. None of these three GmPM (GmPM4, GmPM5 and MP130) proteins belongs to any known LEA pro-tein. G. max, G. soja, short-pod G. tomentella, long-pod G. tomentella and G. tabacina each has a unique seed maturation protein pattern. These results indicate that the antibodies against soybean seed maturation proteins may provide a powerful tool for genetic stud-ies. For instance, the antibody against GmPM2 may detect 52-kDa and 60-kDa polypeptides in G. max but detect 55-kDa and 60-kDa polypeptides in G. soja. F1 hybrids resulted from a cross between these two
species may thus be confirmed by using this antibody.
Are long and short podded G. tomentella different species?
The morphological characteristics, chromosome num-bers, and interspecific crosses of several Glycine spe-cies collected in Taiwan were studied intensively by Tang and his colleagues (Tang & Chen, 1959; Tang & Lin, 1962; Tang & Tai, 1962). At the time of study, the long-pod G. tomentella was called G. tomentosa, and the short-pod G. tomentella was called G. tomentella. Even though both long and short podded G. tomentella were tetraploid (2n = 80), their morphological features were quite different.
In a taxonomic study of the Leguminosae in Taiwan, Ohashi et al. (1991) observed a distinct mor-phological form of G. tomentella and described it as a new species, Glycine dolichocarpa Tateishi et Ohashi. They pointed out that G. dolichocarpa was clearly distinguishable from G. tomentella on the basis of the pods, seeds, leaflets, flowers and hairiness of the stems and petioles (Ohashi et al., 1991; Tateishi & Ohashi, 1992). However, this taxonomic report has not been generally accepted. The genomic diversity of tet-raploid G. tomentella has been studied by cytogenetic analysis, seed protein profiles, protease inhibitor activ-ity band profiles, Western blot analysis of anti-Kunitz
Figure 5. Immunostaining after SDS-PAGE of soybean seed pro-teins recognized by seed maturation protein antibodies. The cross-reactive polypeptides recognized by antibodies against GmPM1 (Panel A), GmPM2 (Panel B), and GmPM8 (Panel C) are illustrated. Proteins were prepared from (a) G. max Shi-shi; (b) G. soja S001; (c) G. latifolia Lat; (d) G. tabacina Ta010; (e) G. canescens Ca003; (f) G. canescens Ca007; (g) G. tomentella To039; (h) G. tomentella To057; (i) G. tomentella To059; (j) G. tomentella To047; (k) G. tomentella To053; (l) G. tomentella To061; (m) G. to-mentella To054; (n) G. toto-mentella To055; (o) G. toto-mentella To062; (p) G. tomentella To052. Numbers on the left indicate the apparent molecular weights of specific polypeptides.
trypsin inhibitor, RFLP analysis and synthetic allotet-raploids (Kollipara et al., 1994). On the basis of this research, G. tomentella was divided into seven groups. The short-pod G. tomentella collected in Taiwan was placed in group T4; the long-pod G. tomentella was not included in the above study.
In this paper, we have shown that there are differ-ences in the seed heat soluble protein profiles and the antibody cross-reactive seed maturation protein pat-terns between short-pod and long-pod G. tomentella collected in Taiwan. By the analysis of nucleotide sequences of the internal transcribed spacer, we’ve also demonstrated that short-pod G. tomentella and long-pod G. tomentella indeed are different (Hsing et al., submitted). Therefore, we suggest that G.
do-lichocarpa (the long-pod G. tomentella) should be
separated from G. tomentella, which generally have short pod.
Can GmPM antibodies be used to distinguish between and within other Glycine species?
In order to test if the immunoblot analysis may be used to assess the degree of genetic variability between and within different Glycine species, we employed the antibodies against GmPM1, GmPM2 and GmPM8. Several accessions (listed in Table 2) from G. max, G.
soja, G. latifolia, G. tabacina, G. canescens and G. tomentella collected in Taiwan, the Philippines, New
Guinea, and Australia were used for the Western blot analyses (Figure 5). The results indicate that these three antibodies indeed can be used to distinguish between and within different Glycine species.
Acknowledgements
The authors are grateful to Kaohsiung Agricultural Experimental Station in Pintung, Taiwan for providing soybean cultivar Shi-shi seeds. We also express our deep thanks to Drs Anthony Huang and Alan Warneke for critical reading of the manuscript. This research was supported, in part, by grants from the National Science Council, ROC and Academia Sinica, ROC to Y.I.H. and J.S.H.
References
Blackman, S.A., S.H. Wettlaufer, R.L. Obendorf & A.C. Leopold, 1991. Maturation proteins associated with desiccation tolerance in soybean. Plant Physiol 96: 868–874.
Chen, Z.Y., Y.I. Hsing, P.F. Lee & T.Y. Chow, 1992. Nucleotide sequences of a soybean cDNA encoding an 18 kilodalton late embryogenesis abundant protein. Plant Physiol 99: 773–774. Galau, G.A., D.W. Hughes & L. Dure III, 1986. Abscisic acid
induction of cloned cotton late embryogenesis-abundant (Lea) mRNAs. Plant Mol Biol 7: 155–170.
Hsing, Y.I., R.W. Rinne, A.G. Hepburn & R.E. Zielinski, 1990. Expression of maturation-specific genes in soybean seeds. Crop Sci 30: 1343–1350.
Hsing, Y.I., Z.Y. Chen & T.Y. Chow, 1992. Nucleotide sequences of a soybean complementary DNA encoding a 50-kilodalton late embryogenesis abundant protein. Plant Physiol 99: 354–355. Hsing, Y.I. & S.J. Wu, 1992. Cloning and characterization of cDNA
clones encoding soybean seed maturation polypeptides. Bot Bull Acad Sin 33: 191–199.
Hsing, Y.I., Z.Y. Chen, M.D. Shih, J.S. Hsieh & T.Y. Chow, 1995a. Unusual sequences of group 3 LEA mRNA inducible by maturation or drying in soybean seeds. Plant Mol Biol 29: 863–868.
Hsing, Y.I., K.L. Hsieh, Y.C. Huang & J.S. Hsieh, 1995b. The rela-tionships of cultivated soybeans and their wild relatives collected from Taiwan: revealed by seed proteins. Bot Bull Acad Sin 36: 65–72.
Hsing, Y.I., P.F. Lee, Z.Y. Chen & T.Y. Chow, 1995c. A soy-bean cDNA (X63565) encoding a late embryogenesis abundant protein. Plant Physiol 109: 1125.
Hsing, Y.I., C.H. Tsou, T.F. Hsu, Z.Y. Chen, K.L. Hsieh, J.S. Hsieh & T.Y. Chow, 1998. Tissue- and stage-specific expression of a soybean (Glycine max L.) seed-maturation, biotinylated. Plant Mol Biol 38: 481–490.
Hughes, D.W. & G.A. Galau, 1991. Developmental and envir-onmental induction of Lea and LeaA mRNAs and the post-abscission program during embryo culture. Plant Cell 3: 605– 618.
Hymowitz, T. & R.J. Singh, 1987. Taxonomy and speciation. In J.R. Wilcox, Soybeans: Improvements, Production, and Uses, 2nd ed. Agronomy Monograph No. 16, pp. 23–48.
Hymowitz, T., R.J. Singh & K.P. Kollipara, 1998. The genomes of the Glycine. Plant Breeding Rev 16: 289–317.
Kitamura, K., T. Takagi & K. Shibasaki, 1976. Subunit structure of soybean 11S globulin. Agric Biol Chem 40: 1837–1844. Kollipara, K.P., R.J. Singh & T. Hymowitz, 1994. Genomic
di-versity and multiple origins of tetraploid (2n = 78, 80) Glycine tomentella. Genome 37: 448–459.
Laemmli, U.K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. Newell, C. & T. Hymowitz, 1983. Hybridization in genus Glycine subgenus Glycine Willd. (Leguminosae, Papilionoideae). Am Jour Bot 70: 334–348.
Ohashi, H., Y. Tateishi, T. Nemoto & H. Hoshi, 1991. Taxonomic studies on the Leguminosae of Taiwan IV. Sci Rep Tohoku Univ 4th Ser (Biol) 40: 1–37.
Palmer, R., K. Newhouse, R. Graybosch & X. Delannay, 1987. Chromosome structure of the wild soybean. J Hered 78: 243– 247.
Rosenberg, L.A. & R.W. Rinne, 1986. Moisture loss as a prerequis-ite for seedling growth in soybean seeds (Glycine max L. Merr.). Jour Exper Bot 37: 1663–1674.
Tang, W.T. & C.H. Chen, 1959. Preliminary studies on the hy-bridization of cultivated and wild soybean (Glycine max× G. formosana). Jour Agr Assoc China NS 28: 17–23.
Tang, W.T. & C.C. Lin, 1962. Studies on the characteristics of some Glycine species found in Taiwan. Jour Agr Assoc China NS 37: 15–22.
Tang, W.T. & G. Tai, 1962. Studies on the qualitative and quantitat-ive inheritance of an interspecific cross of soybean Glycine max and G. formosana. Bot Bull Acad Sin 3: 39–60.
Tateishi, Y. & H. Ohashi, 1992. Taxonomic studies on Glycine of Taiwan. J Jpn Bot 67: 127–147.
Thanh, V.H. & K. Shibasaki, 1977. Beta-conglycinin from soy-bean proteins. Isolation and immunological and physicochemical properties of the monomeric forms. Biochem Biophys Acta 490: 370–384.
Tindale, M.D. & L.A. Craven, 1988. Three new species of Glycine (Fabaceae: Phaseolae) from north-western Australia, with notes on amphicarpy in the genus. Aust Syst Bot 1: 399–410. Towbin, H., T. Staehlin & J. Gordon, 1979. Electrophoretic
trans-fer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354.