Spatiotemporal modelling of ozone distribution in the State of California
全文
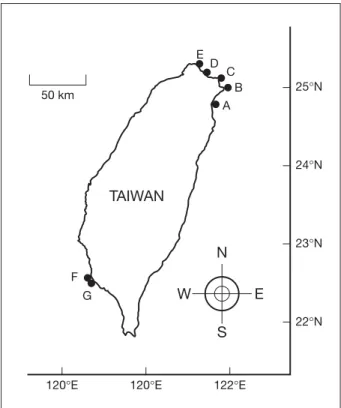
(2) Chiu et al. In the present study, 27 strains of marine LB were identified in water samples collected from the shallow coastal regions of northern and southern Taiwan. The isolation and characterization of these luminous bacterial strains are described.. E 50 km. D. C B A. 25°N. Methods 24°N. Culture media, collection of water samples and isolation of LB Culture media described by Shieh and Jean [11] were used for the present study. Modifications and additional media are described below. Peptone-yeast (PY) broth was modified to contain Bacto peptone at 3 g/L and Bacto yeast extract at 1 g/L, and pH was adjusted to 8.0. PY-II broth containing 3 g/L Bacto peptone and 1 g/L Bacto yeast extract was dissolved in 80% seawater, approximately 3.5% salinity, and was adjusted to pH 8.0. Bacto agar (Difco, Detroit, MI, USA) was added to PY broth and PY-II broth at 4 g/L and 15 g/L for the preparation of stab and plate media, respectively. PY-nitrate broth was prepared by adding potassium nitrate to PY broth at 2 g/L. Modified PY plate medium used for the lipase test was supplemented with calcium chloride (0.2 g/L) and Tween 80 (0.1%). PYcarbohydrate stab media differed from the original media in containing Bacto peptone, Bacto yeast extract, Bacto agar and bromothymol blue at concentrations of 3 g/L, 1 g/L, 8 g/L and 0.06 g/L, respectively. Modified Møller decarboxylase media contained 5 g/L of appropriate L -amino acid ( L -arginine, L -lysine or L -ornithine). Tryptone broth was adjusted to pH 8.0. Glucose-mineral medium differed from that described by Shieh et al [12] in containing Tris at 3 g/L. Nine water samples were collected at a depth of about 30 cm from seven stations set up in the shallow coastal regions of northern and southern Taiwan during the relatively warm seasons: 6 samplings from stations A, B, C and D in 2002, and 3 samplings from stations E, F and G in 2004 (Fig. 1). The water samples had original temperatures of 23-30°C, while the seawater temperature around the year in the sampling regions might vary from 16-30°C. Each water sample was serially diluted with sterile seawater. Plate counts of LB and heterotrophic bacteria were done by spreading 0.1 mL aliquots of each dilution onto PY-II plates in duplicate or triplicate. Incubation was carried out at 25°C in the dark for 7 days under aerobic conditions. Luminous colonies produced on the plates were examined and marked daily in a dark room © 2007 Journal of Microbiology, Immunology and Infection. TAIWAN 23°N. N F. W. G. E S. 120°E. 120°E. 22°N. 122°E. Fig. 1. Locations of water sampling stations (A, B, C, D, E, F, G).. during the incubation. All of the luminous colonies were picked off and purified by successive streaking on PYII plates. The isolates that again retained luminescence were maintained in PY-II stab medium and stored at 25°C.. Phenotypic characterization of luminous isolates Cells grown in PY plate medium were sampled with a straight needle for inoculation into each of the PYcarbohydrate stab media for tests of acid production from fermentation of various carbohydrates. The surface of the medium in each tube was overlaid with sterile liquid paraffin after inoculation. The cultures were examined for color changes daily for 7 days. Gas, if produced would be indicated by formation of gas bubbles or cracks in the medium, or by the separation of medium from the side or bottom of the tube. The cultures for tests of arginine dihydrolase and lysine and ornithine decarboxylases were also incubated under anaerobic conditions by overlaying the surface of the medium in each tube with sterile liquid paraffin after inoculation [13]. Cells grown for 1-3 days on PY plate medium were used to test Gram reaction by the potassium hydroxide lysis method [14]. They were also used to test luminescence, agarase, catalase, oxidase and swarming according to the procedures described by Shieh et al [15]. Amylase, caseinase, DNase, gelatinase and lipase 15.
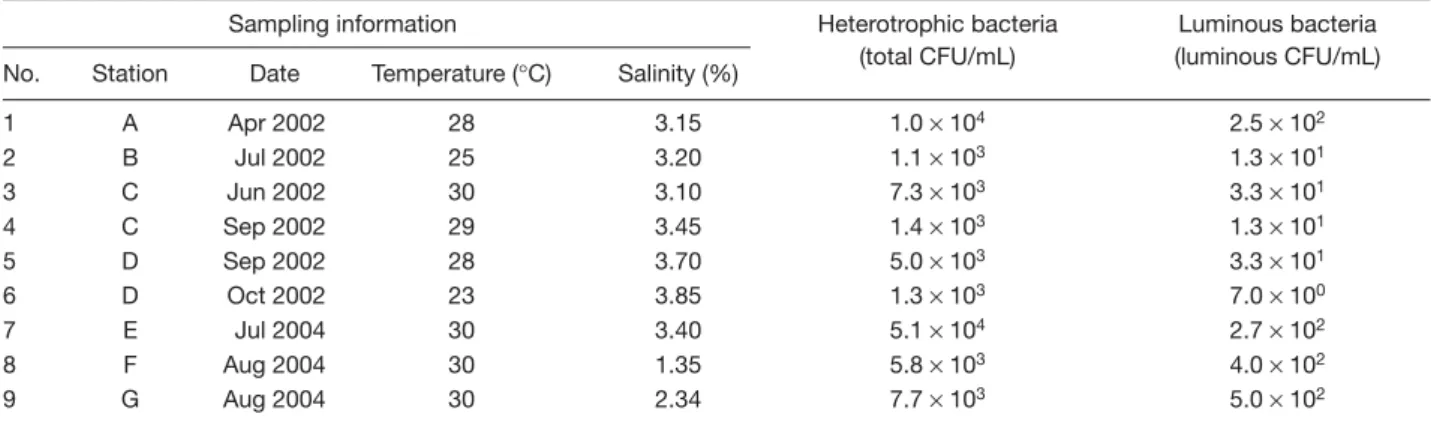
(3) Luminous marine bacteria. tests essentially followed the methods of Smibert and Krieg [13]. Cells grown in PY broth for 1-3 days were examined for shape and motility by phase-contrast microscopy. Cultures grown in PY-nitrate broth were examined for nitrate reduction [16] and denitrification [17] daily for up to 5 days. Indole production was determined by Kovac’s method [13] after incubating the tryptone broth cultures for 2-5 days. The ability to grow at different temperatures was determined in PY broth and recorded after incubating the cultures for 20 days at 4°C, and 3-7 days at 20-42°C. The ability to grow at various sodium chloride (NaCl) levels was determined in PY broth containing 0-8% NaCl. Utilization of various compounds as sole carbon and energy sources for growth was determined in glucose-mineral medium and its modifications containing test substrates used in place of glucose; sugars and sugar alcohols were both provided at 5 g/L, while organic acids were provided at 2 g/L. All of the test cultures were aerobically incubated at 25°C in the dark for 7 days unless stated otherwise.. Restriction patterns of 16S rRNA genes and polymerase chain reaction amplification PY broth cultures of the luminous isolates were aerobically incubated at 25°C in the dark for 24-36 h. Each culture was centrifuged to harvest the cells. Total genomic DNA was extracted and purified from the cells by using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA) in accordance with the instructions of the manufacturer. Hydration solutions of the purified DNA samples were prepared at concentrations in the range 200700 µg/mL. The DNA hydration solutions were used for polymerase chain reaction (PCR) amplification. PCR amplification of 16S rRNA genes was performed according to the procedures of Jean et al [18]. Aliquots (5 µL) of each of the PCR products of 16S rRNA genes were separately digested with restriction endonucleases ScaI (Takara, Japan), DdeI (Promega, Madison, WI, USA), HhaI (Takara) and RsaI (Promega) according to the manufacturer’s instructions. Restricted DNAs were analyzed by electrophoresis at 50 V for 60 min on 2% agarose gels in Tris-acetate-ethylenediamine tetra-acetic acid (TAE) buffer. After electrophoresis, the gels were stained with ethidium bromide (1 µg/mL) for 10 min in TAE buffer. DNA bands appearing on the gels were visualized and photographed under an image analyzing system consisting of an ultraviolet transilluminator (Spectroline, Westbury, NY, USA), a dark box (EDAS 290, Kodak, New Haven, CT, USA) and a room digital camera (DC290, Kodak). 16. A PCR method based on the 38-bp repetitive extragenic palindromic (REP) sequence in Enterobacteriaceae and other bacteria [19] was also applied to the typing of the luminous isolates.. Sequencing and phylogenetic analysis Sequencing reactions of 16S rRNA gene samples, alignment and comparison of the resulting sequences and reference sequences available in the GenBank database, calculation of the distance matrices for the aligned sequences and reconstruction of the phylogenetic dendrogram were as described by Shieh et al [20]. Sequences of gyrB were determined using the methods described by Ast and Dunlap [21]. Phylogenetic analysis of gyrB sequences essentially was the same as the analysis of the phylogeny for the 16S rRNA genes used in the present study.. DNA-DNA hybridization DNA-DNA hybridization experiments were performed among V. harveyi American Type Culture Collection (ATCC) 14126T, Vibrio campbellii ATCC 25920T and some of the luminous isolates. Bacterial DNA was isolated and extracted using the Puregene DNA isolation kit. The DNA was blotted on a Hybond-N+ membrane (Amersham Pharmacia Biotech, Arlington Heights, IL, USA) by use of a Bio-Dot microfiltration apparatus (BioRad Laboratories, Hercules, CA, USA), thereafter dotblot hybridization was done at 68°C [22]. Total DNA of either ATCC 14126T or ATCC 25920T labeled with a DIG DNA labeling kit (Roche Diagnostics, Basel, Switzerland) was used as a probe. Hybrid detection was performed by enzyme immunoassay and enzyme-catalyzed color reaction using a DIG nucleic acid detection kit (Boehringer Mannheim, Mannheim, Germany). DNA relatedness values among ATCC 14126T, ATCC 25920T and the luminous isolates were estimated according to the hybridization results examined using the 1D Image Analysis Software (Kodak).. Results and Discussion Enumeration and isolation of LB LB were found in all of the nine water samples taken from stations A-G. Plate counting values of these bacteria ranged from 7 to 500 colony-forming units (CFU) per mL, which accounted for 0.5-6.9% of the total CFU of heterotrophic bacteria (Table 1). Previous reports have shown that LB were of densities ranging from lower than 100 to 101 CFU per mL in nearshore © 2007 Journal of Microbiology, Immunology and Infection.
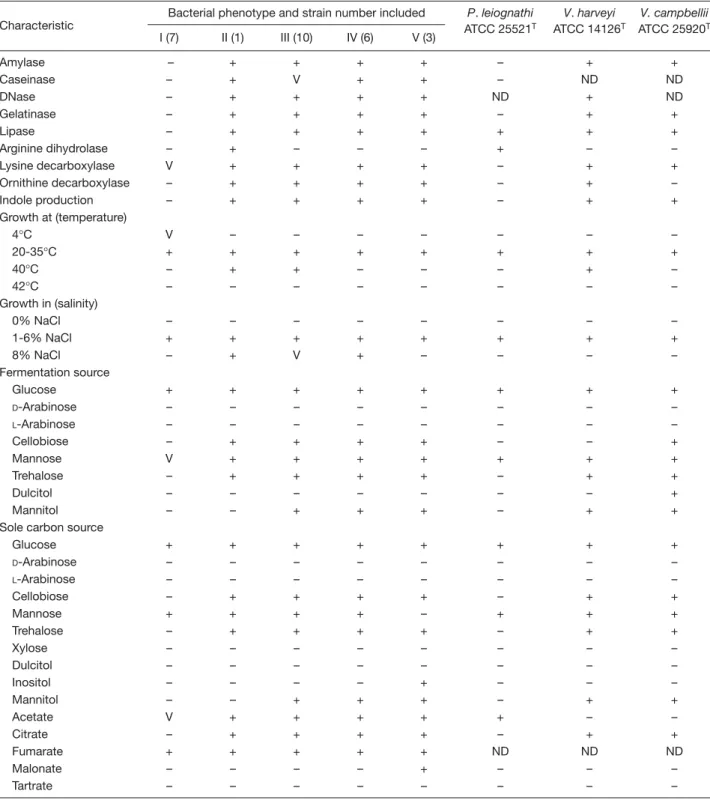
(4) Chiu et al. Table 1. Plate counts of luminous bacteria and heterotrophic bacteria in shallow coastal waters of Taiwan Sampling information No. 1 2 3 4 5 6 7 8 9. Station. Date. Temperature (°C). Salinity (%). A B C C D D E F G. Apr 2002 Jul 2002 Jun 2002 Sep 2002 Sep 2002 Oct 2002 Jul 2004 Aug 2004 Aug 2004. 28 25 30 29 28 23 30 30 30. 3.15 3.20 3.10 3.45 3.70 3.85 3.40 1.35 2.34. Heterotrophic bacteria (total CFU/mL). Luminous bacteria (luminous CFU/mL). 1.0 × 104 1.1 × 103 7.3 × 103 1.4 × 103 5.0 × 103 1.3 × 103 5.1 × 104 5.8 × 103 7.7 × 103. 2.5 × 102 1.3 × 101 3.3 × 101 1.3 × 101 3.3 × 101 7.0 × 100 2.7 × 102 4.0 × 102 5.0 × 102. Abbreviation: CFU = colony-forming units. waters [8,9,23-26]. The luminous CFU obtained from the water samples in the present study were generally greater than those obtained in the previous reports. This might be attributed to the different water temperature conditions. Eight out of the nine water samples counted here had original temperatures of 25-30°C, while in those previous reports, the original temperatures were rarely higher than 25°C. Differences in the water temperature have been correlated to the density of certain species of LB in seawater [24,26-28]. Twenty seven strains of LB were isolated from the counting plates. They were designated as LB1 to LB27, respectively. After subculturing, strains LB1-LB16 remained luminous, while the luminescence of strains LB17-LB27 was no longer visible. Losses of phenotypic features after subculture have also been observed in other bacteria, such as the loss of catalase activity [29], denitrifying activity [17,30], and the ability to produce gas vesicles [31], fascicle-like colonies [32] and square tablets of 8 to 64 cells [33]. Gene deletion was assumed to account for the occurrence of some of these phenomena [17,30]. Whether gene deletion could explain the loss of luminescence in our isolates remains to be further investigated.. Phenotypic characterization of luminous isolates All of the luminous isolates were motile, Gram-negative rods that produced flat and off-white colonies on PY plates. They were halophiles, unable to grow in the absence of NaCl. They were facultative anaerobes, capable of both respiratory and fermentative metabolism; acid but no gas was produced during fermentation of glucose. Oxidase and catalase tests were both positive. All of the isolates might be identified as members of the family Vibrionaceae according to the preliminary phenotypic characterization [34,35]. However, they were © 2007 Journal of Microbiology, Immunology and Infection. classified into five types based on more detailed phenotypic characteristics (Table 2). Phenotype I contained strains (LB1-LB3 and LB17LB20) that did not ferment cellobiose, trehalose and mannitol, and were unable to grow on these substrates as sole carbon and energy sources. The strains of this phenotype were negative in the tests for indole production and amylase, DNase, gelatinase, and lipase activities. Phenotypes II-V contained isolates that fermented cellobiose and trehalose and grew on these substrates as sole carbon and energy sources. Phenotypes II and III contained strains that grew at 40°C. The strain of phenotype II (LB4) differed from those of phenotype III (LB5-LB12 and LB21-LB22) in that it produced arginine dihydrolase and was unable to grow on mannitol as a sole carbon and energy source. Phenotypes IV (LB13-LB14 and LB23-LB26) and V (LB15-LB16 and LB27) contained strains that did not grow at 40°C. However, only the strains of phenotype IV grew in 8% NaCl and did not grow on inositol and malonate as sole carbon and energy sources.. Restriction pattern analysis of 16S rRNA genes and REP-PCR typing The 16S rRNA gene sequences of all luminous isolates were PCR-amplified with the primer pair mentioned above. All of the isolates produced a single band of about 1500 bp, corresponding to the predicted size of the 16S rRNA gene sequences amplified from this primer pair. Restriction patterns of the PCR-amplified 16S rRNA genes were analyzed by digestion with the restriction endonucleases DdeI, HhaI, RsaI and ScaI. Only one restriction pattern, designated pattern a, was obtained with DdeI, while two patterns, designated patterns a and b, respectively, were obtained with each of the other 17.
(5) Luminous marine bacteria. Table 2. Phenotypic characteristics of luminous bacterial isolates in comparison with those of the type strains of Vibrio harveyi, Vibrio campbellii and Photobacterium leiognathia Bacterial phenotype and strain number included Characteristic Amylase Caseinase DNase Gelatinase Lipase Arginine dihydrolase Lysine decarboxylase Ornithine decarboxylase Indole production Growth at (temperature) 4°C 20-35°C 40°C 42°C Growth in (salinity) 0% NaCl 1-6% NaCl 8% NaCl Fermentation source Glucose D-Arabinose L-Arabinose Cellobiose Mannose Trehalose Dulcitol Mannitol Sole carbon source Glucose D-Arabinose L-Arabinose Cellobiose Mannose Trehalose Xylose Dulcitol Inositol Mannitol Acetate Citrate Fumarate Malonate Tartrate. P. leiognathi ATCC 25521T. V. harveyi ATCC 14126T. V. campbellii ATCC 25920T. I (7). II (1). III (10). IV (6). V (3). – – – – – – V – –. + + + + + + + + +. + V + + + – + + +. + + + + + – + + +. + + + + + – + + +. – – ND – + + – – –. + ND + + + – + + +. + ND ND + + – + – +. V + – –. – + + –. – + + –. – + – –. – + – –. – + – –. – + + –. – + – –. – + –. – + +. – + V. – + +. – + –. – + –. – + –. – + –. + – – – V – – –. + – – + + + – –. + – – + + + – +. + – – + + + – +. + – – + + + – +. + – – – + – – –. + – – – + + – +. + – – + + + + +. + – – – + – – – – – V – + – –. + – – + + + – – – – + + + – –. + – – + + + – – – + + + + – –. + – – + + + – – – + + + + – –. + – – + – + – – + + + + + + –. + – – – + – – – – – + – ND – –. + – – + + + – – – + – + ND – –. + – – + + + – – – + – + ND – –. Abbreviations: ATCC = American Type Culture Collection; NaCl = sodium chloride; + = positive; – = negative; V = variable between strains; ND = no data available aAll of the luminous isolates and the type strains are motile, Gram-negative rods that are positive for oxidase, catalase and nitrate reduction and are negative for swarming, agarase, denitrification and gas from glucose fermentation.. 18. © 2007 Journal of Microbiology, Immunology and Infection.
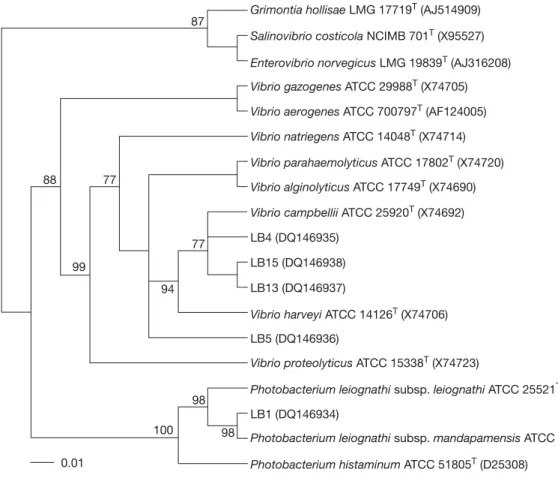
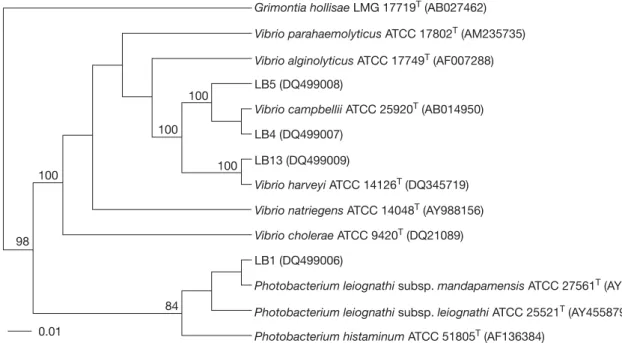
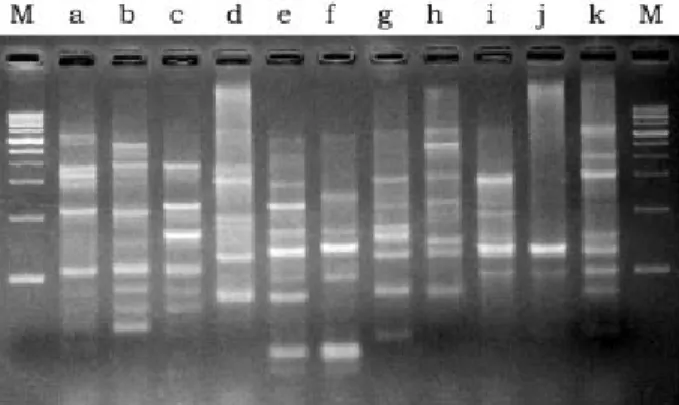
(6) Chiu et al. Table 3. Genotypes of luminous isolates derived from restriction patterns of 16S rRNA genes digested with various restriction enzymes Genotype I II. Phenotype. DdeI. HhaI. RsaI. ScaI. I II-V. a a. a b. a b. a b. restriction enzymes (Table 3). Two different genotypes of 16S rRNA genes were detected in the luminous isolates by analyzing the restriction patterns. Genotype I contained the seven isolates of phenotype I. Genotype II consisted of the twenty isolates of phenotypes II-V (Table 3). However, eleven REP-PCR typing patterns (3-15 amplified bands ranging in size from 250-3500 bp) were discernible in the luminous isolates LB1-LB16 (Fig. 2). This indicated that REP-PCR typing might be more effective than 16S rRNA genotyping in differentiation of the luminous isolates.. Phylogenetic analysis For the phylogenetic analysis of 16S rRNA gene and gyrB sequences, strains LB1, LB4, LB5, LB13, and LB15 were selected as representative strains of the phenotypes I-V, respectively. Almost complete 16S rRNA gene sequences of these strains (1433-1443 bp) were determined. The gyrB sequences of LB1, LB4, LB5 and LB13 were obtained at 1169-1180 bp, while that of LB15 was not determined successfully. Gene sequences that had been determined were aligned and compared with all bacterial sequences available in the GenBank database. The GenBank accession numbers for the 16S rRNA gene sequences of LB1, LB4, LB5, LB13,. Fig. 2. Diagram of amplification patterns of selective luminous isolates with repetitive extragenic palindromic sequencepolymerase chain reaction method. Patterns: a, LB1; b, LB2; c, LB3; d, LB4, LB7, LB9, LB10 and LB11; e, LB5; f, LB6; g, LB8; h, LB12; i, LB13 and LB16; j, LB14; k, LB15. M, molecular size markers (from top to bottom: 10,000, 8000, 6000, 4000, 3000, 2500, 2000, 1500, 1000, and 500 bp).. © 2007 Journal of Microbiology, Immunology and Infection. and LB15 were DQ14934-DQ14938 and those for the gyrB sequences of LB1, LB4, LB5 and LB13 were DQ499006-DQ499009. Our phylogenetic analysis confirmed that all of the representative strains were members of the family Vibrionaceae in the gammaProteobacteria. Fig. 3 shows the 16S rRNA gene-based phylogenetic positions of these strains within the radiation of their related taxa in the family Vibrionaceae. Strain LB1 clustered with species of the genus Photobacterium. Its closest relatives were P. leiognathi subsp. mandapamensis (99.3% sequence similarity) and P. leiognathi subsp. leiognathi (98.6% sequence similarity) [21]. No other Photobacterium species shared more than 97% sequence similarity with the strain. Strains LB4, LB5, LB13 and LB15 had sequence similarity levels of 97.6-99.4%. They formed a cluster at sequence similarity levels of 97.6-99.8% with V. harveyi and other Vibrio species, including V. campbellii, Vibrio natriegens, Vibrio alginolyticus, Vibrio parahaemolyticus and Vibrio proteolyticus. Fig. 4 shows the gyrB-based phylogenetic relationships between strains LB1, LB4, LB5 and LB13 and other related members in the family Vibrionaceae. Again, strain LB1 clustered with species of the genus Photobacterium, with P. leiognathi subsp. mandapamensis (98.7% sequence similarity) and P. leiognathi subsp. leiognathi (97.6% sequence similarity) as its closest relatives. However, LB4, LB5 and LB13 fell into two separate clusters. LB4 and LB5 clustered with V. campbellii at sequence similarities of 97.1-98.2%, and LB13 clustered with V. harveyi at a sequence similarity of 98.6%. The sequence similarities between strains of the two clusters were only 93.094.4%. A prokaryote whose 16S rRNA gene sequence differs by more than 3% from that of all other organisms is generally considered as a new species, and sequence differences of greater than 5-7% (93-95% identity) have been taken as support for a new genus [36]. Strain LB1 could be identified as P. leiognathi, since the two had rather similar sequences (similarity > 98.5%) and shared high similarity (93.3-97.8%) in their phenotypic characteristics (Table 2). Such identification was also supported by high levels of gyrB sequence similarity between LB1 and P. leiognathi. The high 16S rRNA sequence similarity levels of strains LB4, LB5, LB13 and LB15 with V. harveyi, V. campbellii, V. natriegens, V. alginolyticus, V. parahaemolyticus and V. proteolyticus indicated the phylogenetic impracticability for differentiation of V. harveyi, 19.
(7) Luminous marine bacteria. Grimontia hollisae LMG 17719T (AJ514909) 87 Salinovibrio costicola NCIMB 701T (X95527) Enterovibrio norvegicus LMG 19839T (AJ316208) Vibrio gazogenes ATCC 29988T (X74705) Vibrio aerogenes ATCC 700797T (AF124005) Vibrio natriegens ATCC 14048T (X74714) Vibrio parahaemolyticus ATCC 17802T (X74720) 77. 88. Vibrio alginolyticus ATCC 17749T (X74690) Vibrio campbellii ATCC 25920T (X74692) LB4 (DQ146935). 77. LB15 (DQ146938). 99. LB13 (DQ146937). 94. Vibrio harveyi ATCC 14126T (X74706) LB5 (DQ146936) Vibrio proteolyticus ATCC 15338T (X74723) Photobacterium leiognathi subsp. leiognathi ATCC 25521T (X74686). 98. LB1 (DQ146934) 100 0.01. 98. Photobacterium leiognathi subsp. mandapamensis ATCC 27561T (AY341441) Photobacterium histaminum ATCC 51805T (D25308). Fig. 3. Unrooted phylogenetic tree deduced from neighbor-joining analysis of the 16S rRNA gene sequences of selective luminous isolates and other related taxa in the family Vibrionaceae. Bootstrap confidence values (percentages) obtained with 100 resamplings are given at the branch points; values below 75 are not shown. Bar = one nucleotide substitution per 100 nucleotides. LMG = Laboratorium voor Microbiologie, Universiteit Gent (UGent); NCIMB = National Collections of Industrial Food and Marine Bacteria; ATCC = American Type Culture Collection; LB = luminous bacteria.. V. campbellii, V. natriegens, V. alginolyticus, V. parahaemolyticus and V. proteolyticus and for identification of the four luminous isolates at the species level. However, the four luminous isolates might be classified as V. harveyi considering that only V. harveyi was defined to include LB in these Vibrio spp. [37]. Such classification was supported by high DNA-DNA relatedness values (68.2-88.8%) between the four strains and V. harveyi ATCC 14126T. However, both LB4 and LB5 had higher DNA-DNA relatedness values with V. campbellii ATCC 14126T than with V. harveyi ATCC 14126T (LB4, 88.8% vs 80.7%; LB5, 81.0% vs 74.3%). This together with phylogeny based on gyrB sequences (Fig. 4) indicated that LB4 and LB5 are best classified as V. campbellii rather than V. harveyi. Our work provides the first evidence that V. campbellii may include LB. It also suggests that the gyrB-based phylogeny could easily differentiate V. harveyi from V. campbellii, despite the phenotypical and genomical similarity of the two species [37,38]. 20. The Microtox® test, based on the inhibition of light emission of V. fischeri, is a practical method for monitoring the toxicity of aquatic samples. This test is considered one of the most sensitive, rapid and reliable microbial bioassays applicable for detection of water quality [39]. Microbiosensor B17 677F, a biotest based on the luminescence quenching of P. phosphoreum, is also useful for assessing the toxicity of sewage and river water [40]. The application of the present luminous isolates in tests of water quality and toxicity remains to be evaluated. Previous studies based on phenotypic characterization have shown that V. harveyi and V. fischeri are generally the most frequently encountered species of LB in temperate nearshore seawaters. The two species accounted for more than 90% of the LB isolated from Sargasso Sea waters at depths of 160-320 m, where P. leiognathi and P. phosphoreum constituted the remainder of the isolates [7]. They even occupied 99% of the luminous isolates from California coastal surface © 2007 Journal of Microbiology, Immunology and Infection.
(8) Chiu et al. Grimontia hollisae LMG 17719T (AB027462) Vibrio parahaemolyticus ATCC 17802T (AM235735) Vibrio alginolyticus ATCC 17749T (AF007288) LB5 (DQ499008) 100 Vibrio campbellii ATCC 25920T (AB014950) 100. LB4 (DQ499007) LB13 (DQ499009). 100. 100. Vibrio harveyi ATCC 14126T (DQ345719) Vibrio natriegens ATCC 14048T (AY988156) Vibrio cholerae ATCC 9420T (DQ21089). 98. LB1 (DQ499006) Photobacterium leiognathi subsp. mandapamensis ATCC 27561T (AY455883) 84. Photobacterium leiognathi subsp. leiognathi ATCC 25521T (AY455879). 0.01. Photobacterium histaminum ATCC 51805T (AF136384). Fig. 4. Unrooted phylogenetic tree deduced from neighbor-joining analysis of the gyrB sequences of selective luminous isolates and other related taxa in the family Vibrionaceae. Bootstrap confidence values (percentages) obtained with 100 resamplings are given at the branch points; values below 75 are not shown. Bar = one nucleotide substitution per 100 nucleotides. LMG = Laboratorium voor Microbiologie, Universiteit Gent (UGent); ATCC = American Type Culture Collection; LB = luminous bacteria.. waters, where V. harveyi predominated (60-70%) in the summer, but was almost completely replaced by V. fischeri during the winter [8]. Fifteen strains of LB isolated from Tyrrhenian Sea coastal waters off northeastern Sicily were characterized by a combination of phenotypic and molecular tests. All of the strains were identified to be members of V. harveyi [5]. The present work showed that culturable LB in the shallow coastal waters of Taiwan during the relatively warm sampling seasons would be dominated by V. harveyi/campbellii and P. leiognathi, and that the Table 4. Distribution of luminous strains of various species in the sampling stations No. of luminous strainsa. Sampling No.. Station. 1 2 3 4 5 6 7 8 9. A B C C D D E F G. Vibrio harveyi/ campbellii 5 1 1 1 0 2 3 5 2. (3) (1) (1) (0) (0) (0) (1) (1). Photobacterium leiognathi 0 1 0 0 3 0 1 0 2. former species (V. harveyi/campbellii) appeared to be more prevalent and numerous than the latter species in general (Table 4). Culturable LB distributed in temperate and tropical nearshore waters may comprise few predominant species other than V. harveyi/campbellii, V. fischeri and P. leiognathi. The species distribution patterns of these bacteria, however, may vary greatly due to geographical and seasonal differences.. Acknowledgments We thank Dr. J.S. Chen, School of Medicine, China Medical University for advice and critical reading of the manuscript. This work was supported by grant NSC93-2313-B-002-060 from the National Science Council.. References (1). (2) (1) (0). aNumbers. in parentheses show the strains that lost visible luminescence after subculture.. © 2007 Journal of Microbiology, Immunology and Infection. 1. Lee KH, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565-71. 2. Leisman G, Cohn DH, Nealson KH. Bacterial origin of luminescence in marine animals. Science. 1980;208:1271-3. 3. McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491-4. 4. Nealson K, Hastings JW. The luminous bacteria. In: Balows 21.
(9) Luminous marine bacteria. A, Trüper HG, Dworkin M, Harder W, Schleifer KH, eds. The prokaryotes, a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. New York: Springer; 1991:625-39. 5. Caccamo D, Di Cello F, Fani R, Gugliandolo C, Maugeri TL. Polyphasic approach to the characterisation of marine luminous bacteria. Res Microbiol. 1999;150:221-30. 6. Makemson JC, Fulayfil N, Landry W, Van Ert LM, Wimpee CF, Widder EA, et al. Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int J Sys Bacteriol. 1997;47:1034-9. 7. Orndorff SA, Colwell RR. Distribution and identification of luminous bacteria from the Sargasso Sea. Appl Environ Microbiol. 1980;39:983-7. 8. Ruby EG, Nealson KH. Seasonal changes in the species composition of luminous bacteria in the nearshore seawater. Limnol Oceanogr. 1978;23:530-3. 9. Yetinson T, Shilo M. Seasonal and geographic distribution of luminous bacteria in the eastern Mediterranean Sea and the Gulf of Elat. Appl Environ Microbiol. 1979;37:1230-8. 10. Liu PC, Lee KK, Chen SN. Pathogenicity of different isolates of Vibrio harveyi in tiger prawn, Penaeus monodon. Lett Appl Microbiol. 1996;22:413-6. 11. Shieh WY, Jean WD. Alterococcus agarolyticus, gen. nov., sp. nov., a halophilic thermophilic bacterium capable of agar degradation. Can J Microbiol. 2003;44:637-45. 12. Shieh WY, Chen AL, Chiu HH. Vibrio aerogenes sp. nov., a facultatively anaerobic marine bacterium that ferments glucose with gas production. Int J Syst Evol Microbiol. 2000;50: 321-9. 13. Smibert RM, Krieg NR. Phenotypic characterization. In: Gerhardt P, Murray RG, Wood WA, Krieg NR, eds. Manual of methods for general and molecular bacteriology. Washington, DC: American Society for Microbiology; 1994:607-54. 14. Buck JD. Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol. 1982; 44:992-3. 15. Shieh WY, Simidu U, Maruyama Y. Enumeration and characterization of nitrogen-fixing bacteria in an eelgrass (Zostera marina) bed. Microb Ecol. 1989;18:249-59. 16. Collins CH, Lyne PM, Grange JM, eds. Identification methods: Collins and Lyne’s microbiological methods. 7th ed. London: Butterworth-Heinemann; 1995:102-20. 17. Shieh WY, Yang JT. Denitrification in the rhizosphere of the two seagrasses Thalassia hemprichii (Ehrenb.) and Halodule uninervis (Forsk.) Aschers. J Exp Mar Biol Ecol. 1997;218: 229-41. 18. Jean WD, Shieh WY, Chiu HH. Pseudidiomarina taiwanensis gen. nov., sp. nov., a marine bacterium isolated from shallow coastal water of An-Ping Harbour, Taiwan, and emended 22. description of the family Idiomarinaceae. Int J Syst Evol Microbiol. 2006;56:899-905. 19. Wong HC, Lin CH. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J Clin Microbiol. 2001;39:4233-40. 20. Shieh WY, Lin YT, Jean WD. Pseudovibrio denitrificans gen. nov., sp. nov., a marine, facultatively anaerobic, fermentative bacterium capable of denitrification. Int J Syst Evol Microbiol. 2004;54:2307-12. 21. Ast JC, Dunlap PV. Phylogenetic analysis of the lux operon distinguishes two evolutionarily distinct clades of Photobacterium leiognathi. Arch Microbiol. 2004;181: 352-61. 22. Fesefeldt A, Kloos K, Bothe H, Lemmer H, Gliesche CG. Distribution of denitrification and nitrogen fixation genes in Hyphomicrobium spp. and other budding bacteria. Can J Microbiol. 1998;44:181-6. 23. Feldman KA, Buck JD. Distribution and characterization of luminescent bacteria in a temperate estuary. Estuaries. 1984;7: 93-7. 24. O’Brien CH, Sizemore RK. Distribution of the luminous bacterium Beneckea harveyi in a semitropical estuarine environment. Appl Environ Microbiol. 1979;38:928-33. 25. Makemson JC, Fulayfil N, Basson P. Association of luminous bacteria with artificial and natural surfaces in Arabian Gulf seawater. Appl Environ Microbiol. 1992;58:2341-3. 26. Venkateswaran K, Nakano H, Hashimoto H. Occurrence and distribution of bioluminescent bacteria in the Seto inland sea. Nippon Suisan Gakkaishi. 1989;55:467-73. 27. Ruby EG, Greenberg EP, Hastings JW. Planktonic marine luminous bacteria: species distribution in the water column. Appl Environ Microbiol. 1980;39:302-6. 28. Shilo M, Yetinson T. Physiological characteristics underlying the distribution patterns of luminous bacteria in the Mediterranean Sea and the Gulf of Elat. Appl Environ Microbiol. 1979;38:577-84. 29. Rurangirwa FR, Teitzel CA, Cui J, French DM, McDonough PL, Besser T. Streptococcus didelphis sp. nov., a streptococcus with marked catalase activity isolated from opossums (Didelphis virginiana) with suppurative dermatitis and liver fibrosis. Int J Syst Evol Microbiol. 2000;50:759-65. 30. Gamble TN, Betlach MR, Tiedje JM. Numerically dominant denitrifying bacteria from world soils. Appl Envion Microbiol. 1977;33:926-39. 31. Lehtimäki J, Lyra C, Suomalainen S, Sundman P, Rouhiainen L, Paulin L, et al. Characterization of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int J Syst Evol Microbiol. 2000;50:1043-53. 32. Gugger M, Lyra C, Henriksen P, Couté A, Humbert JF, Sivonen K. Phylogenetic comparison of the cyanobacterial genera © 2007 Journal of Microbiology, Immunology and Infection.
(10) Chiu et al. Anabaena and Aphanizomenon. Int J Syst Evol Microbiol. 2002; 52:1867-80. 33. Lee N, Cellamare CM, Bastianutti C, Rosselló-Mora R, Kämpfer P, Ludwig W, et al. Emended description of the species Lampropedia hyalina. Int J Syst Evol Microbiol. 2004; 54:1709-15. 34. Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68:403-31. 35. Farmer III JJ, Janda JM. Family 1. Vibrionaceae Véron 1965, 5245AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, eds. Bergey’s manual of systematic bacteriology. 2nd ed. Vol 2. New York: Springer-Verlag; 2005:491-555. 36. Stackerbrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846-9.. © 2007 Journal of Microbiology, Immunology and Infection. 37. Farmer III JJ, Janda JM, Brenner FW, Cameron DN, Birkhead KM. Genus I. Vibrio Pacini 1854, 411AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, eds. Bergey’s manual of systematic bacteriology. 2nd ed. Vol 2. New York: Springer-Verlag; 2005: 494-546. 38. Baumann P, Furniss AL, Lee, JV. Genus I Vibrio Pacini 1854, 411AL. In: Krieg NR, Holt JG, eds. Bergey’s manual of systematic bacteriology. 1st ed. Vol 1. Baltimore: Williams & Wilkins; 1984:518-38. 39. Magi E, Righetti C, Di Carro M, Sanguinetti MS, Ferri M. Study of the water quality close to urban sewers in eastern Liguarian coast by means of bioluminescence tests and conventional analyses. Chem Ecol. 2005;21:47-60. 40. Kuznetsov AM, Rodicheva EK, Medvedeva SE. Biotesting of effluent and river water by lyophilized luminous bacteria biotest. Field Anal Chem Technol. 1998;2:267-75.. 23.
(11)
數據
相關文件
The first row shows the eyespot with white inner ring, black middle ring, and yellow outer ring in Bicyclus anynana.. The second row provides the eyespot with black inner ring
You are given the wavelength and total energy of a light pulse and asked to find the number of photons it
Robinson Crusoe is an Englishman from the 1) t_______ of York in the seventeenth century, the youngest son of a merchant of German origin. This trip is financially successful,
fostering independent application of reading strategies Strategy 7: Provide opportunities for students to track, reflect on, and share their learning progress (destination). •
Strategy 3: Offer descriptive feedback during the learning process (enabling strategy). Where the
Now, nearly all of the current flows through wire S since it has a much lower resistance than the light bulb. The light bulb does not glow because the current flowing through it
volume suppressed mass: (TeV) 2 /M P ∼ 10 −4 eV → mm range can be experimentally tested for any number of extra dimensions - Light U(1) gauge bosons: no derivative couplings. =>
• Strange metal state are generic non-Fermi liquid properties in correlated electron systems near quantum phase transitions. • Kondo in competition with RVB spin-liquid provides