Received 30 May 2006 | Accepted 2 August 2006 | Published 4 August 2006
Myopia is the most common eye disorder worldwide. In the United States, the prevalence of myopia among blacks and whites is approximately 25% [1], whereas in Asian popula-tions, such as in Taiwan, prevalence may exceed 65% [2]. The prevalence of high myopia defined as myopia in excess of 6 D, has been estimated to be between 0.3% and 9.6% world-wide [3]. However, the prevalence has recently been shown to be as high as 21% in young Taiwanese populations, and evidence suggests that this number is increasing. The early onset and fast progression of myopia among children has been well documented [2].
Myopia is a complex disease involving multiple interact-ing genetic and environmental factors. Environmental factors, such as educational level, occupation, individual income, read-ing habits, and use of computers, may contribute to develop-ment of myopia [4,5]. Children of myopic parents are more likely to have myopia than those from nonmyopic parents [6]. In a study of twins, the ocular components (axial length, ante-rior chamber depth, and corneal curvature) and refractive er-rors of MZ twins were more closely aligned than were those of DZ twins, also indicating a hereditary component in the formation of myopia [7]. Population and family studies in China have suggested a genetic component of high myopia [8].
Previous studies have shown that scleral thinning in the highly myopic human eye is associated with a narrowing and dissociation of the collagen fiber bundles and a reduction in collagen fibril diameter [9]. Changes in the biochemical struc-ture of the sclera have also been reported in the sclera of highly myopic human eyes [9]. The proteoglycans, decorin and biglycan, members of the small leucine-rich repeat protein (SLRP) family, are major components of the scleral extracel-lular matrix. These small proteoglycans play an important role in regulating collagen fibril assembly and interaction [10]. Additionally, the human sclera has been shown to contain lumican (corneal keratan sulfate proteoglycan) core protein, where it is present in similar concentrations as in the cornea, and exists in a glycoprotein form with varying amounts of tyrosine sulfation [11]. An important property of SLRPs, par-ticularly decorin and lumican, is their role in the control of collagen fibrillogenesis [10,11]. Decorin, in particular, has been shown to decelerate fibril growth and increase fibril diameter [10]. The genetic locus for autosomal dominant high myopia has been identified on chromosome 12q21-23 (MYP 3) in which dermatan sulfate proteoglycan 3 (DSPG-3), decorin, and lumican are located [12,13].
The analysis of complex human diseases has been spurred by the number of genomic sequence variants discovered in the course of sequencing the human genome [14]. Single-nucleotide polymorphisms (SNPs), common variations among the DNA of individuals, are being uncovered and assembled into large SNP databases that promise to dissect the genetic basis of diseases and drug responses [15]. For example, Lam
The association of single nucleotide polymorphisms in the
5'-regulatory region of the lumican gene with susceptibility to
high myopia in Taiwan
I-Jong Wang,1 Ting-Hsuan Chiang,1 Yung-Feng Shih,1 Chuhsing Kate Hsiao,2 Shao-Chun Lu,3 Yi-Chih Hou,1
Luke Long-Kung Lin1
1Department of Ophthalmology, College of Medicine, 2Division of Biostatistics, Institute of Epidemiology, College of Public Health
and 3Department of Biochemistry and Molecular Biology, College of Medicine, National Taiwan University, Taipei, Taiwan
Purpose: To study the relationships between single nucleotide polymorphisms (SNPs) of lumican, decorin, and DSPG3
genes and high myopia.
Methods: One hundred and twenty adult patients with high myopia (< -10.0 D) and 137 controls were used to study the
relationships between the decorin, lumican, and DSPG genes and high myopia. All subjects were free of ocular diseases, other than myopia, as well as of other systemic genetic diseases. Genotyping was performed by direct sequencing after PCR amplification of chromosomal DNA. Allele frequencies were tested for Hardy-Weinberg disequilibrium. The χ2 or Fisher test was conducted to investigate the genotypic and allelic distribution between the high myopia and control groups.
Results: The genotyping success rate was 100%. Univariate analysis revealed significant differences between patients
and control subjects with respect to one of the SNPs (rs3759223, C->T) of the lumican gene, with a p value of 0.000283. There was no significant relationship between other SNPs of lumican, decorin, and DSPG genes and high myopia.
Conclusions: Our results indicate that an SNP (rs3759223), which is located in the promoter region of the lumican gene,
may be worth further investigation to determine its association with development of high myopia.
Correspondence to: Yung-Feng Shih, MD, Department of Ophthal-mology, National Taiwan University Hospital 7, Chung-Shan South Road, Taipei, Taiwan; Phone: 23123456; FAX: 886-2-23412875; email: yfshih@ha.mc.ntu.edu.tw
et al. [16] found that six SNPs in the coding exons of trans-forming growth factor (TGF)-beta-induced factor (TGIF) was a probable candidate gene for high myopia. However, Scavello et al. [17] found that the encoded TGIF gene did not identify sequence alterations associated with the myopia.
Therefore, it was of interest to examine whether individual differences of other SNPs in the extracellular matrix genes of the scleral coat may affect the response to the signal from retina-RPE complexes. Here, we report a case-control study
in which an attempt was made to identify SNPs of the decorin, lumican, and DSPG3 genes that are located in the MYP3 lo-cus by sequencing. Our results indicate a SNP at the 5'-regu-latory region of lumican may be susceptible to high myopia.
METHODS
Subjects: We recruited 120 unrelated Taiwanese subjects (50 males, 70 females; mean age of 34.4±15.2 years), who had
high myopia of -10.00 D or more negative refractive error in
TABLE 1.
Position
on DNA Length PCR conditions SNP ID contig Gene Primer sequence (bp) Locus (°C) --- --- --- --- --- ---rs1803344 15027690 Decorin F: AGAAGAGCTTCCTGCTTCCC 279 Exon 9 94/61/72 R: GAAAGGCATCCATGTGTGGT
rs3138268 15027704 Decorin F: AGAAGAGCTTCCTGCTTCCC 279 Exon 9 94/61/72 R: GAAAGGCATCCATGTGTGGT
rs2070985 15055254 Decorin F: GCTGCCTGAGTCATCGTCTA 294 Untranslated 94/58/72 R: CCAGGGGACACAGAAGAGAA
rs2070984 15055701 Decorin F: CCATCACTTTATTGACAAGACG 276 Untranslated 94/53/72 R: ACAAAATGCCCTGTTAGCAA
rs1802763 14980199 Lumican F: TGGATACTATGAAAACTGACACACA 277 Exon 3 94/53/72 R: TTGTTTTGAGCCAGTGTACTGAA
rs1802743 14980213 Lumican F: TGGATACTATGAAAACTGACACACA 277 Exon 3 94/53/72 R: TTGTTTTGAGCCAGTGTACTGAA
rs2300588 14982088 Lumican F: GGCCACATAGTGAATTTTCCA 287 Intron 94/55/72 R: GTTTTCTGGCCATGCTGAAT
rs7135740 14982193 Lumican F: GGCCACATAGTGAATTTTCCA 287 Intron 94/55/72 R: GTTTTCTGGCCATGCTGAAT
rs3741835 14987332 Lumican F: GCTCAGAACTGTGGATTTGCT 479 Untranslated 94/68-62 R: GAAGCAGGACTCAATTCTTGG (touchdown)/72 rs3741834 14987575 Lumican F: GCTCAGAACTGTGGATTTGCT 479 Putative 94/68-62 R: GAAGCAGGACTCAATTCTTGG promoter (touchdown)/72 rs3759223 14988974 Lumican F: GCTCTTGTTAGAAAAACTCCACC 379 Putative 94/57/72 R: TGAATGAGACTTTCTGACTCTCCA promoter
rs3759222 14989144 Lumican F: GCTCTTGTTAGAAAAACTCCACC 379 Putative 94/57/72 R: TGAATGAGACTTTCTGACTCTCCA promoter rs1135866 14847832 DSPG3 F: CAATGACAGCAAATAAAATTGGA 266 Exon 5 94/52/72 R: TGCTAGGTGATTTAAAAAGGATTG rs1920748 14881247 DSPG3 F: AAGCAGTAGGATCCAAATCACA 243 Promoter 94/52/72 R: TGAAGAAAGCAATTCAAGACTTC rs1920751 14876487 DSPG3 F: TCATGAGTTCATCACCAAGG 481 Intron 94/63-56 R: TGTTTGGGATCATGGCTTCT (touchdown)/72 rs1920752 14876173 DSPG3 F: TCATGAGTTCATCACCAAGG 481 Intron 94/63-56 R: TGTTTGGGATCATGGCTTCT (touchdown)/72
Primers designed for PCR amplifications of chosen SNPs. The names of corresponding genes are shown and their PCR primer sequences are shown in column 4. The length of PCR products (column 5) and corresponding positions of SNPs (column 1, 2 and 6) were also revealed. The denatured/annealing/extension temperatures are shown in the last column.
both eyes, from the National Taiwan University Hospital. We also recruited 137 unrelated control subjects (77 males, 60 females; mean age of 41.9±8.5 years) who had refractive
er-rors of -1.5 D to 0.5 D in either eye. No participant had known ocular disease and insult that could predispose to myopia, such as a history of retinopathy or prematurity, or neonatal prob-lems, or a known genetic disease and connective tissue disor-der associated with myopia, such as Stickler and Marfan syn-drome [18,19]. All patients and control subjects involved in this study had similar social backgrounds and were from the local ethnic Han Chinese population, with no ethnic subdivi-sion. Informed consent was obtained from all subjects. The project had the approval of the Institutional Review Board (IRB)/Ethics Committees of National Taiwan University Hos-pital and was carried out in accordance with The World Medi-cal Association’s Declaration of Helsinki.
Every participant received a complete ocular examina-tion including retinoscopy, slit-lamp evaluaexamina-tion of the ante-rior segment, measurement of intraocular pressure, axial-length measurements (Sonomed Ultrasound A-1500, Lake Success, NY), keratometry measurements, and a fundus examination, with special notation as to the health and degree of cupping of the optic nerve head.
The mean axial length was 24.08±0.65 mm (range from
22.10 mm to 24.98 mm) for right eyes and 24.18±0.50 mm
(range from 22.71 mm to 24.96 mm) for left eyes of the con-trol group. The mean axial length was 29.47±1.84 mm (range
from 26.7 mm to 34.54 mm) for right eyes and 29.61±2.03
mm (range from 26.82 mm to 35.73 mm) for left eyes in myo-pic subjects. The mean keratometric readings were 43.56±1.22
D (40.12-46.75) for right eyes and 43.59±1.27 D (40.62-46.62)
for left eyes of the control group. The mean keratometric
read-ings were 43.70±2.07 D (40.62-49.87) for right eyes and
43.84±1.55 D (41.12-49.75) for left eyes of myopic group.
DNA extraction, amplification and mutation screening: Total genomic DNA was extracted from 10 to 15 ml of venous blood from all participants, after informed consent was ob-tained. DNA was purified from lymphocyte pellets according to standard procedures using a kit (Puregene kit; Gentra Sys-tems, Minneapolis, MN) or the phenol-chloroform extraction method [20].
Sixteen SNPs and their primers were identified for decorin (4), lumican (8), and DSPG3 (4), using GenBank and a data-base of Japanese single nucleotide polymorphisms (Table 1). They were designed to amplify the sequence of the identified SNPs with 250-500 bp extensions beyond the SNPs. PCR was performed on 50 ng of genomic DNA with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Foster City, CA). Temperatures for PCR reactions are shown in Table 1. The PCR cycling conditions of SNPs included an initial dena-turation for 5 min at 94 °C, followed by 30 cycles of
denatur-ation for 45 s at 94 °C, annealing for 45 s at a different
tem-perature, extension for 45 s at 72 °C, and a final extension for
7 min at 72 °C. The Touchdown PCR cycling conditions of
SNPs rs3741835 and rs3741834 were performed as follows: initial preheating step for 11 min at 95 °C to achieve a hot
start, followed by 12 cycles of 94 °C for 20 s, 68 °C for 20 s,
and 72 °C for 45 s. The annealing temperature was decreased
0.5 °C per cycle until 62 °C and was followed by 30 cycles of
94 °C for 20 s, 62 °C for 20 s, and 72 °C for 45 s with a final
elongation at 72 °C for 10 min. For SNPs rs14876487 and
rs14876183, the touchdown annealing temperature decreased 0.5 °C per cycle from 63 °C to 56 °C, followed by 30 cycles
of 94 °C for 20 s, 56 °C for 20 s, and 72 °C for 45 s with a final
TABLE 2.
Genotype Frequency (%)
Patients (n=120) Control (n=137)
Sequence --- --- Genotype Gene alternation 1/1 1/2 2/2 HWD 1/1 1/2 2/2 HWD p value --- --- --- --- --- --- --- --- ---Decorin rs1803344 (C->G) 120 0 0 0 137 0 0 0 1.0000 rs3138268 (C->T) 120 0 0 0 137 0 0 0 1.0000 rs2070985 (C->G) 89 26 5 2.7118 107 25 5 4.4938* 0.7594 rs2070984 (A->G) 120 0 0 0 137 0 0 0 1.0000 Lumican rs1802763 (T->C) 120 0 0 0 137 0 0 0 1.0000 rs1802743 (G->T) 120 0 0 0 137 0 0 0 1.0000 rs2300588 (G->T) 11 56 53 0.4858 21 69 47 0.2766 0.1550 rs7135740 (A->T) 64 48 8 0.0620 54 68 15 0.8821 0.0698 rs3741835 (C->T) 65 47 8 0.0161 56 66 15 0.4654 0.0867 rs3741834 (C->T) 64 49 7 0.3579 56 65 16 0.1910 0.0743 rs3759223 (C->T) 61 32 27 21.1796* 55 68 14 1.1185 0.000283 rs3759222 (A->C) 18 55 47 0.0841 17 67 53 0.3515 0.8000 DSPG3 rs1135866 (A->G) 120 0 0 0 137 0 0 0 1.0000 rs1920748 (A->G) 4 13 103 12.4105* 1 14 122 0.6855 0.3124 rs1920751 (A->C) 8 32 80 3.3333 10 37 90 4.4571* 0.9761 rs1920752 (A->C) 7 33 80 1.9321 10 37 90 4.4571* 0.8946
Genotype frequencies and probabilities of decorin, lumican, and DSPG3 sequence alterations in patients with myopia < -10 D and control subjects. In the table, 1/1 indicates a genotype with homozygous normal allele; 1/2 indicates a genotype with heterozygous sequence alter-ations; 2/2 indicates genotype with homozygous sequence alteralter-ations; and HWD indicates a χ2 statistic for Hardy-Weinberg disequilibrium. The asterisk indicates a p<0.05 for the Hardy-Weinberg equilibrium. The genotype p value is based on a χ2 test.
elongation at 72 °C for 10 min. Amplified PCR products were
separated by agarose gel electrophoresis and visualized by staining with ethidium bromide. They then were purified in purification columns (QIAquick; Qiagen, Valencia, CA) and sequenced using dye terminator chemistry (BigDye Termina-tor version 3.1 on a model 3100 Genetic Analyzer; Applied Biosystems). Sequences were trimmed for quality and aligned using BioEdit software (version 5.0.6). Normal and affected individual DNA sequences were aligned to the known refer-ence genomic sequrefer-ence (NT_019546), available via the Na-tional Center for Biotechnology Information (NCBI) database and compared for sequence variations.
Statistical Analysis: To examine the association for each of the 16 SNPs, we used the χ2 test to compare the alteration
of genotypes between 120 patients and 137 control subjects. This test evaluates the difference between the observed geno-type frequency and the expected frequency under the null hy-pothesis of no association. However, when the expected fre-quency is small, the Fisher exact test is conducted instead [21]. After each test, the Bonferroni correction was applied for multiple tests by comparing the p value to the significance level divided by the total number of tests (16). Next, all de-tected SNPs were assessed for Hardy-Weinberg disequilib-rium using the χ2 test [21]. Again, Bonferroni correction was
applied for the multiple tests [22]. To evaluate the effects of SNPs on the risk of high myopia, we conducted logistic re-gression analysis with a stepwise approach [23]. The depen-dent variable was the disease status (patients, 1; control sub-jects, 0), and the independent variables were values of SNPs (homozygotes, 2; heterozygotes,1; wild type, 0). The covariates of interactions between SNPs were also included in the model. The final optimal model contains the statistically significant
variables (SNPs) that were selected by the stepwise proce-dure [23]. No Bonferron correction is considered in this analy-sis. These statistical analyses were performed on computer (SPSS software, version 10.1; SPSS Science, Chicago, IL).
RESULTS
For the lumican gene, rs3759222 (A->C), rs3759223 (C->T), and rs3741834 (C->T) are located in the putative promoter region of the gene (-4176, -4006 bp and -2607 bp, respec-tively), while rs1802763 (A->G, Ser->Ser), rs3741834 (C->T, untranslated), and rs1802743 (G->T, Gly->Cys) are located in exon 3 of the lumican gene. Also, rs2300588 (G->T), rs7135740 (A->T), and rs3741835 (C->T) are in the intron of THE lumican gene (Table 2). For the lumican gene, rs1802763, rs1802743, rs2300588, rs7135740, rs3741835, rs3741834, and rs3759222, there were no significant differences between the high myopic group and normal subjects with respect to geno-types. However, there was a statistically significant difference between high myopic group and normal subjects (genotype
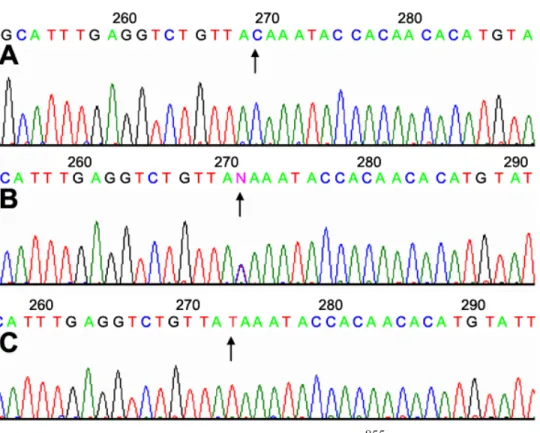
χ2, p=0.000283) for rs3759223 (C->T; Figure 1). After
Bonferroni correction by comparing each p value with the adjusted significance level, α/16=0.05/16=0.003125, it is still
significant. In the logistic regression mode, the SNP (rs3759223) also showed a significant result with the odds ratio 8.178 and the 95% confidence interval (2.424-29.834, p=0.001).
In DSPG3, rs1920748 (A->G; -2714 bp) is located in the promoter region of the gene and rs1135866 (C->T, Ser->Leu) is in exon 5, while rs1920751 (A->C) and rs1920752 (A->C) are in the intron of the DSPG3 gene. For the DSPG3 gene, there was no significant difference for rs1135866, rs1920748, rs1920751, or rs1920752. For the decorin gene, rs1803344
Figure 1. Direct sequence of SNPs at rs3759223 (C->T) from the high myopia group. A: The arrow points to a normal sequence (C). B: The arrow shows a heterozygote. C: The arrow marks a homozygote for the SNP of rs3759223 (T).
(C->G, Gln->Glu, exon 9), rs3138268 (C->T, Thr->Met, exon 9), rs2070985 (C->G, untranslated), and rs2070984 (A->G, untranslated), there were no significant differences between the highly myopic group and normal subjects with respect to genotypes (Table 2).
DISCUSSION
High myopia is caused by excessive axial elongation that pri-marily involves the ora-equatorial area and the posterior pole. The changes in scleral components (e.g., collagen fibrils and proteoglycans) are associated with myopia in human and ex-perimental animals. Also, significant changes in proteoglycan synthesis have been shown to be correlated with changes in the rate of axial elongation during postnatal ocular growth and during the development of myopia in a variety of animal mod-els, suggesting that proteoglycans play a critical role in deter-mining the biomechanical properties of the sclera [24]. Fibrillogenesis of the sclera may be affected by mutations in these candidate proteins, as has been demonstrated in other connective-tissue disorders that manifest with myopia, such as Sticker syndrome and Marfan syndrome [18,25]. Moreover, two previous genetic loci associated with familial high myo-pia (MYP1 and MYP3) have been mapped to Xq28 and 12q21-23 [13,26] which include the loci for biglycan (Xq27ter) [27], decorin (12q21-q22) [28], and lumican (12q21.3-q22) genes [29], suggesting that mutations in these extracellular matrix components may be involved in some forms of human myo-pia.
A large genetic locus for autosomal dominant high myo-pia has been identified at 12q21-23 (MYP 3), which includes several SLRP genes, including DSPG-3, decorin, and lumican [12,13]. However, there was no significant difference for decorin and DSPG genes in our study. Decorin and lumican are members of the small interstitial proteoglycan family of proteins that are expressed in the extracellular matrix of vari-ous tissues [30,31]. Both interact with collagen and limit the growth of fibril diameter [10,11]. Decorin and lumican are present in corneal stroma and in the interstitial matrices of the heart, aorta, skeletal muscle, skin, and intervertebral disks [32]. Furthermore, lumican has also been demonstrated in both mouse and human sclera [33,34]. DSPG3, another small in-terstitial proteoglycan, is expressed in cartilage as well as in ligament and placental tissues [35].
Lumican is a member of the proteoglycan family. Al-though heteroduplex and sequence analysis excluded lumican as the causative gene involved in the family with 12q21-23-linked high myopia [12], it is possible that a mutation in one or both alleles of the lumican gene may cause significant de-fects in the scleral extracellular matrix, which, in turn, could result in alterations in ocular shape and size. The defects ob-served in sclera collagen fibril diameter and organization in lumican-deficient mice were expected to lead to severe de-fects in ocular shape and size [11]. Volumetric estimations of eye size in wild type and lumican-deficient mice suggest that eyes are larger in these mice [36]. Another mouse study also implicated the proteoglycans, lumican and fibromodulin, as functional candidate genes for high myopia [37]. However,
Paluru et al. [38] suggested that this is not the case. They con-sidered the possibility of false-positive results of lumican and fibromodulin knockout mice due to the “hitchhiker” effect, in which adjacent altered genes (hitchhiking genes) may influ-ence the phenotype, such as eye size. The current results indi-cate that an SNP of the lumican gene may confer susceptibil-ity to high myopia. SNP (rs3759223) of the lumican gene is located 4,406 bp upstream from exon 1, does not account for any codon change and is considered a putative regulatory ele-ment of the lumican gene. Our results showed that certain varia-tions of SNPs in the lumican regulatory region that influence the promoter activities of lumican and affect fibrillogenesis is different in myopic and normal eyes. This finding is compat-ible with the results obtained from lumican-deficient mice [11]. We surmise that rs3759223 (C->T) can regulate the promoter activity of the lumican gene, and affect the formation of col-lagen fibrils of the scleral coat during the development of myopia.
All 16 SNPs were tested for Hardy-Weinberg equilibrium (HWE; Table 2). Deviations from HWE may indicate popula-tion stratificapopula-tion or genotyping errors [39]. However, the tests were not significant in the control group and hence may rule out the possibility of population admixture. To examine the genotyping error and to validate our findings, we repeated the genotyping analysis several times, and obtained consistent results. Our sequences were mostly clean at baseline. Most important, the mutated peaks in the heterozygous conditions were much higher than the normal peaks, which clearly indi-cated the presence of heterozygous mutation. Therefore, oc-currence of genotyping errors in this study was kept to a mini-mum. In the high myopia group, however, two out of the 16 SNPs considered in our study were not in HWE (Table 2). The deviations from HWE may indicate possible association with the myopia status. This disequilibrium in the patients seems to provide further support for the association of rs3759223 (C->T) and rs1920748 (A->G) with the myopia status. The other SNPs, rs2070985 (C->G), rs1920751 (A->C), and rs1920752 (A->C) although found not in HWE in the pa-tients, were not found associated with myopia. This disequi-librium may be due to the imbalanced observed genotypes and hence down weights its implication of association unless further studies are conducted.
In conclusion, our study demonstrated that SNP (rs3759223) may be associated with high myopia in Taiwan.
ACKNOWLEDGEMENTS
The studies were in part supported by NTUH95-000292 and NSC grants: NSC91-2314-B002-294, NSC92-2314-B002-154, NSC93-2314-B002-019, and NSC94-2314-B002-257.
REFERENCES
1. Sperduto RD, Seigel D, Roberts J, Rowland M. Prevalence of myopia in the United States. Arch Ophthalmol 1983; 101:405-7.
2. Lin LL, Shih YF, Hsiao CK, Chen CJ, Lee LA, Hung PT. Epide-miologic study of the prevalence and severity of myopia among schoolchildren in Taiwan in 2000. J Formos Med Assoc 2001; 100:684-91.
3. Curtin BJ. Adult myopia. Acta Ophthalmol Suppl 1988; 185:78-9. 4. Wong TY, Foster PJ, Hee J, Ng TP, Tielsch JM, Chew SJ, Johnson GJ, Seah SK. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci 2000; 41:2486-94.
5. Whitmore WG. Congenital and developmental myopia. Eye 1992; 6:361-5.
6. Goldschmidt E. The importance of heredity and environment in the etiology of low myopia. Acta Ophthalmol (Copenh) 1981; 59:759-62.
7. Edmund J, Goldschmidt E. [Concordance for myopia and discor-dance for optic disk cupping in a pair of monozygotic twins]. Klin Monatsbl Augenheilkd 1988; 193:645-6.
8. Chu R, Ni P, Ni M, Shen F. Genetic epidemiology study of patho-logical myopia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2000; 17:178-80.
9. Curtin BJ, Iwamoto T, Renaldo DP. Normal and staphylomatous sclera of high myopia. An electron microscopic study. Arch Ophthalmol 1979; 97:912-5.
10. Kuc IM, Scott PG. Increased diameters of collagen fibrils pre-cipitated in vitro in the presence of decorin from various con-nective tissues. Connect Tissue Res 1997; 36:287-96. 11. Austin BA, Coulon C, Liu CY, Kao WW, Rada JA. Altered
col-lagen fibril formation in the sclera of lumican-deficient mice. Invest Ophthalmol Vis Sci 2002; 43:1695-701.
12. Young TL, Roughley PJ, Ronan SM, Alvear AB, Fryer JP, King RA. Lumican candidate gene analysis in chromosome 12q linked high myopia. Invest Ophthalmol Vis Sci 1999; 40:S594. 13. Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS,
Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet 1998; 63:1419-24.
14. Nowotny P, Kwon JM, Goate AM. SNP analysis to dissect hu-man traits. Curr Opin Neurobiol 2001; 11:637-41.
15. Pirmohamed M, Park BK. Genetic susceptibility to adverse drug reactions. Trends Pharmacol Sci 2001; 22:298-305.
16. Lam DS, Lee WS, Leung YF, Tam PO, Fan DS, Fan BJ, Pang CP. TGFbeta-induced factor: a candidate gene for high myopia. In-vest Ophthalmol Vis Sci 2003; 44:1012-5.
17. Scavello GS, Paluru PC, Ganter WR, Young TL. Sequence vari-ants in the transforming growth beta-induced factor (TGIF) gene are not associated with high myopia. Invest Ophthalmol Vis Sci 2004; 45:2091-7.
18. Nollen GJ, Mulder BJ. What is new in the Marfan syndrome? Int J Cardiol 2004; 97:S103-8.
19. Logan NS, Gilmartin B, Marr JE, Stevenson MR, Ainsworth JR. Community-based study of the association of high myopia in children with ocular and systemic disease. Optom Vis Sci 2004; 81:11-3.
20. Mann W, Jeffery J. Isolation of DNA from yeasts. Anal Biochem 1989; 178:82-7.
21. Falconer DS, Mackay TFC. Introduction to quantitative genet-ics. 4th ed. Essex (UK): Longman; 1996.
22. Boehringer S, Epplen JT, Krawczak M. Genetic association stud-ies of bronchial asthma—a need for Bonferroni correction? Hum Genet 2000; 107:197.
23. Ott J, Hoh J. Statistical multilocus methods for disequilibrium analysis in complex traits. Hum Mutat 2001; 17:285-8. 24. Rada JA, McFarland AL, Cornuet PK, Hassell JR. Proteoglycan
synthesis by scleral chondrocytes is modulated by a vision de-pendent mechanism. Curr Eye Res 1992; 11:767-82.
25. Snead MP, Yates JR. Clinical and Molecular genetics of Stickler syndrome. J Med Genet 1999; 36:353-9.
26. Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet 1990; 38:281-6.
27. Fisher LW, Heegaard AM, Vetter U, Vogel W, Just W, Termine JD, Young MF. Human biglycan gene. Putative promoter, in-tron-exon junctions, and chromosomal localization. J Biol Chem 1991; 266:14371-7.
28. Pulkkinen L, Alitalo T, Krusius T, Peltonen L. Expression of decorin in human tissues and cell lines and defined chromo-somal assignment of the gene locus (DCN). Cytogenet Cell Genet 1992; 60:107-11.
29. Grover J, Chen XN, Korenberg JR, Roughley PJ. The human lumican gene. Organization, chromosomal location, and expres-sion in articular cartilage. J Biol Chem 1995; 270:21942-9. 30. Scott JE. Proteoglycan: collagen interactions in connective
tis-sues. Ultrastructural, biochemical, functional and evolutionary aspects. Int J Biol Macromol 1991; 13:157-61.
31. Iozzo RV. The biology of the small leucine-rich proteoglycans. Functional network of interactive proteins. J Biol Chem 1999; 274:18843-6.
32. Chakravarti S, Petroll WM, Hassell JR, Jester JV, Lass JH, Paul J, Birk DE. Corneal opacity in lumican-null mice: defects in collagen fibril structure and packing in the posterior stroma. Invest Ophthalmol Vis Sci 2000; 41:3365-73.
33. Ying S, Shiraishi A, Kao CW, Converse RL, Funderburgh JL, Swiergiel J, Roth MR, Conrad GW, Kao WW. Characterization and expression of the mouse lumican gene. J Biol Chem 1997; 272:30306-13.
34. Dunlevy JR, Rada JA. Interaction of lumican with aggrecan in the aging human sclera. Invest Ophthalmol Vis Sci 2004; 45:3849-56.
35. Simionescu C. [The invasion of neighboring structures in malig-nant choroid melanoma]. Oftalmologia 1992; 36:283-8. 36. Lea GS, Amann J, Chakravarti S, Lass J, Edelhauser HF.
Intraocu-lar volumes and surface area measurements of the mouse eye: wildtype vs. lumican-deficient. Invest Ophthalmol Vis Sci 2000; 41:S739.
37. Chakravarti S, Paul J, Roberts L, Chervoneva I, Oldberg A, Birk DE. Ocular and scleral alterations in gene-targeted lumican-fibromodulin double-null mice. Invest Ophthalmol Vis Sci 2003; 44:2422-32.
38. Paluru PC, Scavello GS, Ganter WR, Young TL. Exclusion of lumican and fibromodulin as candidate genes in MYP3 linked high grade myopia. Mol Vis 2004; 10:917-22.
39. Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 2005; 76:887-93.
857
The print version of this article was created on 4 Aug 2006. This reflects all typographical corrections and errata to the article through that date. Details of any changes may be found in the online version of the article. α