行政院國家科學委員會專題研究計畫 成果報告
探討慢性骨髓單核球白血病之純化單核球的整體基因表達
研究成果報告(精簡版)
計 畫 類 別 : 個別型
計 畫 編 號 : NSC 95-2314-B-002-261-
執 行 期 間 : 95 年 08 月 01 日至 96 年 07 月 31 日
執 行 單 位 : 國立臺灣大學醫學院檢驗醫學科
計 畫 主 持 人 : 周文堅
共 同 主 持 人 : 唐季祿、田蕙芬
計畫參與人員: 計畫主持人:黃彥寧、周文堅
處 理 方 式 : 本計畫可公開查詢
中 華 民 國 96 年 05 月 22 日
執行報告 執行報告執行報告 執行報告
Introduction
Myelodysplastic syndrome (MDS) is a heterogeneous group of disease characterized with dysplasia, ineffective hematopoiesis, and a potential of transformation to acute myeloid leukemia. It frequently occurs in elderly population and about 50% of patients harbor a chromosomal abnormality, with monosomy 7 and trisomy 8 most common 1,2. The etiologies of MDS still remain to be investigated. For more than 20 years since the proposal of French-American-British (FAB) classification of this disease, the names coined for various subtypes of MDS are still morphology-oriented, without any definitive breakthrough in understanding the underlying causes of this enigmatic disease 3.
Previous studies about MDS had revealed several genes relevant to the pathogenesis of this disease. Loss of one copy of nucleophosmine (NPM) gene, which is located at chromosome 5q, can cause symptoms mimic human MDS in mice 4. Interestingly, chromosome 5q is one of the most frequently affected regions in MDS patients. Another gene, death inducer and obliterator (Dido), which was mapped at chromosome 20q, another hot spot of abnormality in MDS patients, had abnormal expression in MDS patients, and targeted deletion of this gene in mice resulted in MDS/MPD-like disease 5. Mutation in Ras gene was found in 20% of MDS patients, and acquisition of FLT3 and N-Ras mutation is a frequent event in transformation to acute leukemia 1,6. Another gene, Delta-like (Dlk), expressed specifically in MDS rather than AML patients 7. Overall, the evidence supporting any genes definitively leading to MDS in human is still weak and remains to be explored.
One way to search for the genetic etiologies of MDS is microarray study. By comparing the genetic profiles of normal control and MDS patients, the specific genetic aberrations can be identified and explored. However, several characteristics of MDS make this approach limiting. For example, the disease is heterogeneous in clinical manifestation; there are 8 and 4 sub-categories in MDS and MDS/MPD, respectively, according to WHO classification 8. Moreover, patients of MDS present dysplasia in different numbers of lineage; some have only erythroid dysplasia while others might have trilineage dysplasia. To make things more complicated, there are different degrees of dysplasia within one single lineage in one single patient. Thus, to get convincing results from microarray, pure and homogeneous samples are mandatory.
pathogenesis 7,9-13. These studies used purified neutrophils or stem cells (expressing CD34 or AC133) from MDS patients and normal persons as materials. They presented data by clustering analysis. One pitfall of these studies is the uncertainty of the disease status of the purified cells. No data definitively support that neutrophils or stem cells are real target cells for MDS research. In those studies there was no evidence that the purified cells were unanimously from the MDS clones.
To overcome these intrinsic problems, we decide to focus on chronic myelomonocytic leukemia (CMML), a specific group of disease previously classified in MDS according to FAB system, but now diverted to MDS/MPD by WHO 8. This disease has dysplastic features and increased monocyte number in the peripheral blood (PB). The advantages of selecting this group of patients are multifaceted in that the monocytes are abnormal based on the persistently high counts, the cells are readily available from PB, and most important, the high acceptance of patients to donate PB rather than bone marrow. We have tried magnetic bead-based purification method to purify monocytes from some of the patients and normal controls. We plan to perform microarray analysis on these purified cells. We believe this approach can add insights of the pathogenesis of MDS.
Materials and Methods
Patients: Patients of CMML were diagnosed if they fit any one of the following criteria (WHO classification):
1. Persistent peripheral blood monocytosis > 1x109 /L
2. Absence of Philadelphia chromosome or BCR/ABL fusion gene 3. Less than 20% blasts in the blood or marrow
4. Dysplasia in one or more myeloid lineages. If myelodysplasia is minimal or absent, then patients have to have an acquired and clonal cytogenetic abnormality, or the monocytosis has to be persistent for more than 3 months without obvious causes.
Sample collection: After diagnosis is confirmed and informed consents are available, patients’ peripheral blood (20 mL) is collected with EDTA and heparin-anticoagulated tubes. The former blood is for smear preparation for morphological assessment, and the latter is subjected to Ficoll-Hypaque centrifugation. The mononuclear cells are collected, washed, and then for purification of monocytes.
Purification of monocytes by immunomagnetic methods: Mononuclear cells are re-suspended in 107/80 µL of MACS buffer (1xPBS buffer containing
0.5% bovine serum albumin and 2 mM EDTA with pH 7.2. Ten µL of anti-CD14 microbeads (Miltenyi Biotec, Auburn, CA) is added and the sample is incubated on ice with gentle tapping every 5 minutes for 15 minutes. The cells are washed with 5 mL of MACS buffer and centrifuged at 200g for 10 minutes at 4°C. The pellet is then re-suspended with 500 µL of MACS buffer and transferred to LS columns (Miltenyi Biotec), which has been rinsed with 3 mL of MACS buffer and attached to miniMACS magnetic field (Miltenyi Biotec), but about 2 µL of cells are spared and not subjected to column purification. These cells are labeled as “before purification” for subsequent flow cytometrical analysis. After 3 times of wash with 3 mL of MACS buffer, the column is removed from the magnetic field and the CD14(+) cells are flushed out with 5 mL of MACS buffer. About 10 µL of these purified cells is collected and labeled as “after purification” for subsequent flow cytometrical analysis. The other purified cells are put on ice for RNA extraction.
Flow cytometry: The purity of the cells labeled as “before purification” and “after purification” is assessed by a flow cytometer (Epics XL/MCL, Beckman Coulter, Miami, FL) by staining anti-CD45 and anti-CD14 antibodies (Immunotech, Marseille, France).
Microarray analysis: After verification of the purity of the cells, the RNA is extracted with Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA of 10 µg in 10 µL of DEPC-treated water is sent for bioassay to confirm the RNA quality and microarray analysis (Affymetrix, Santa Clara, CA), according to the manufacturer’s instruction. Briefly, ribosomal RNA is removed from the total RNA, and then the first strand cDNA is generated by reverse transcriptase (RT) reaction. Complementary RNA (cRNA) is generated by adding T7 promoter-tagged random hexamers and RNA polymerase. Finally, the second round of RT reaction is performed to generate amplified, biotin-labeled and fragmented cDNA for subsequent hybridization onto Affymetrix gene chip. The signals are read and analyzed by bioinformatics.
Real-time PCR: Quantitative real-time PCR (qPCR) was performed with universal PCR master mix (Applied Biosystems, Foster City, CA). In every 20 µL of reaction mixture, there were 10 µL of 2x SYBR master mix, 250 nM each primer, and cDNA generated from 200 ng total RNA. The reaction was performed on 7300 sequence detection system (Applied Biosystems),
comprising 50°C for 2 minutes, 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. The primers for JAK2 are
JAK2-3528F: 5'-GAACCTGGTGAAAGTCCCATATTC-3' and
JAK2-3599R: 5'-TGAGGCCACAGAAAACTTGCT-3'. The primers for ANXA1 are ANXA1 213F: 5'-GATGTCGCTGCCTTGCATAA-3' and
ANXA1 310R: 5'-TGTTGACGCTGTGCATTGTTT-3'. The control gene for equal loading is RPLP0 (NM_053275.3). The quantity of expression of each gene was corrected according to expression of RPLP0.
Results
Totally we collected 4 patients who fit the strict criteria of CMML as described above. We collected the purified monocytes by magnetic beads from 20 mL of PB. We have obtained every patient’s informed consent before collection of the PB. As shown in Fig. 1, the purity of the collection is satisfactory, reaching 90%.
Legend: The purity of monocytes of a patient of CMML after immunomagnetic
100 101 102 103 104 SS LOG 1 0 0 1 0 1 1 0 2 1 0 3 1 0 4 C D 4 5 -P C 5 R1 100 101 102 103 104 CD14-FITC 1 0 0 1 0 1 1 0 2 1 0 3 1 0 4 C D 4 5 -P C 5 47.37% 52.63% 0.00% 0.00% R2 100 101 102 103 104 SS LOG 1 0 0 1 0 1 1 0 2 1 0 3 1 0 4 C D 4 5 -P C 5 R1 100 101 102 103 104 CD14-FITC 1 0 0 1 0 1 1 0 2 1 0 3 1 0 4 C D 4 5 -P C 5 2.03% 97.97% 0.00% 0.00% R2
bead purification. Upper left and right: Cells after Ficoll-Hypaque centrifugation but before bead purification. Most of the neutrophils are deleted, and the monocytes account for 52% of the total mononuclear cells, as shown in the upper right CD14+ fraction. Lower left and right: after bead purification, CD14+ monocytes reached to 98% of the mononuclear cells.
We also included monocytes purified from 6 normal control volunteers and pooled them together as a control specimen. The 4 patient samples plus the normal pool were sent for microarray study.
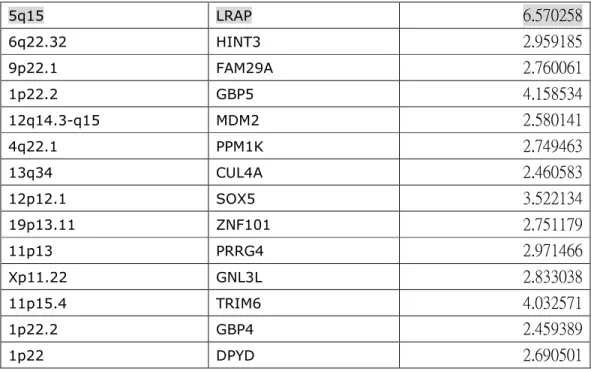
Out of 30,000 open reading frame of human genome, there were only 65 genes (Table 1) up-regulated and 7 genes (Table 2) down-regulated in all four patient monocytes compared with the normal pooled sample. Among them, we were particularly interested in JAK2 (Janus kinase 2, NM_004972) and ANXA1 (Annexin 1, NM_000700), which were up-regulated and down-regulated in CMML patients’ monocytes, respectively. JAK2 is a cytoplasmic kinase, which phosphorylates intracellular targets, including signaltransducers and activators of transcription (STATs), upon ligand binding to the hematopoietic receptors 14. Mutation causing a phenylalanine to valine at position 617 (JAK2V617F) is present in almost all
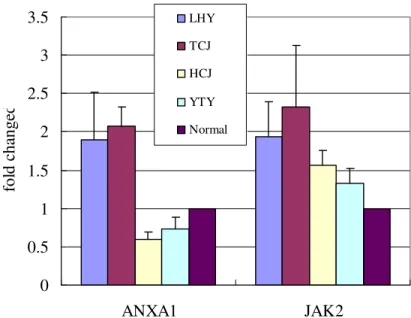
polycythemia vera and about 50% of essential thrombocythemia and idiopathic myelofibrosis 151617181920212223. The up-regulation of this gene in CMML, a myeloproliferative condition, is provocative. ANXA1 is a member of annexin superfamily. It has anti-inflammation and anti-proliferation activities, probably related to suppression of cytosolic phospholipase and arachidonic acid 2425. The down-regulation of this gene in CMML is also worth of exploration. But a big problem is that the difference of each gene between the normal pool and the patients was quite small, at most 4 fold difference. We designed the primers for these two genes and performed real-time PCR using SYBR. We used the RNA from the patients and normal pool control as materials. But the difference was small for both ANXA1 and JAK2. Moreover, the ANXA1 expression patter is not consistent with the microarray data (Fig. 1a and 1b). The main reason is the small difference in
expression level of these genes.
Table 1. Genes Upregulated in CMML Patients
Chromosomes Gene Name Fold Increase
12p12.3 EMP1 4.517188 11p11.2 NR1H3 2.962982 19p12 LOC115648 2.611855 4q21.1 SEPT11 3.505876 19p13.3-p13.2 EPOR 2.292148 5q13.2 LOC153561 2.696943 4q21.22 PLAC8 3.078228 1p13.3 GSTM1 2.456706 3p21 CCR5 4.662811 2q37.1 SP100 2.767005 15q25.1 WDR61 5.188797 16q12 SIAH1 2.45428 8p22 MTSS1 2.541579 2p22.3 YIPF4 2.742896 9p21 CDKN2B 3.100337 16p13.2 PRO0149 2.331688 11p14.1 METT5D1 2.886908 19q13.3 ZNF83 3.582893 10q21.2 ARID5B 2.503072 2p16-p15 VRK2 3.698076 17p13.1 TMEM107 3.60344 3q13.2 ZBTB20 5.381376 12q24.3 RSN 2.735776 22q11.23 CABIN1 2.766765 3q28 KIAA0804 3.663062 5q23.1 COMMD10 3.208947 9p24 JAK2 2.478556 13q12-q13 PFAAP5 5.037734 9p22.2 C9orf39 2.933555 7q34-q35 CASP2 2.53463 12q23.1 KIAA0701 3.191479 2q31.1 DHRS9 2.890913 17p13.3 TSR1 3.384458 5q15 LRAP 5.927094 7p11 IGF2BP3 4.452298 11p15.4 ADM 2.723855 6p21.2 CDKN1A 3.077854
5q15 LRAP 6.570258 6q22.32 HINT3 2.959185 9p22.1 FAM29A 2.760061 1p22.2 GBP5 4.158534 12q14.3-q15 MDM2 2.580141 4q22.1 PPM1K 2.749463 13q34 CUL4A 2.460583 12p12.1 SOX5 3.522134 19p13.11 ZNF101 2.751179 11p13 PRRG4 2.971466 Xp11.22 GNL3L 2.833038 11p15.4 TRIM6 4.032571 1p22.2 GBP4 2.459389 1p22 DPYD 2.690501
Table 2. Downregulated genes in CMML patients
Chromosomes Gene Name Fold Decrease
1q23 FCER1A 0.250295 20q13.32 RAB22A 0.320251 6p21.3 HLA-DRB6 0.215809 9q12-q21.2 ANXA1 0.389623 22q13 FAM118A 0.236789 12q24.2 CAMKK2 0.435426 14q32.33 TDRD9 0.341853
0 0.5 1 1.5 2 2.5 3 3.5 AN XA 1 AN XA 1 AN XA 1 JAK 2 JAK 2 JAK 2 fo ld c ha ng e LHY TCJ HCJ YTY Normal
Fig. 1a. The three independent experiments of ANXA1 and JAK2 expression in the four patients and the normal pool. The fold change was calculated by normalization with a house keeping gene, RPLP0.
0 0.5 1 1.5 2 2.5 3 3.5 ANXA1 JAK2 fo ld c h an g ed LHY TCJ HCJ YTY Normal
Fig. 1b. The averaged fold change of ANXA1 and JAK2 in the four patients and the normal pool. The fold change was calculated by normalization with a house keeping gene, RPLP0.
References:
clinical characteristics in primary myelodysplastic syndrome. A study on 68 Chinese patients in Taiwan. Cancer Genet Cytogenet. 1994;74:40-49.
2. Maserati E, Aprili F, Vinante F, et al. Trisomy 8 in myelodysplasia and acute leukemia is constitutional in 15-20% of cases. Genes Chromosomes Cancer. 2002;33:93-97.
3. Bennett JM, Catovsky D, Daniel MT, et al. Criteria for the diagnosis of acute leukemia of megakaryocyte lineage (M7). A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:460-462.
4. Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147-153.
5. Futterer A, Campanero MR, Leonardo E, et al. Dido gene expression alterations are implicated in the induction of hematological myeloid neoplasms. J Clin Invest. 2005;115:2351-2362.
6. Shih LY, Huang CF, Wang PN, et al. Acquisition of FLT3 or N-ras mutations is frequently associated with progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia. 2004;18:466-475.
7. Miyazato A, Ueno S, Ohmine K, et al. Identification of myelodysplastic syndrome-specific genes by DNA microarray analysis with purified hematopoietic stem cell fraction. Blood. 2001;98:422-427.
8. Elaine S. Jaffe NLH, Harald Stein, James W. Vardiman. World Health Organization Classification of Tumours. Pathology & Genetics. Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC press; 2001.
9. Chen G, Zeng W, Miyazato A, et al. Distinctive gene expression profiles of CD34 cells from patients with myelodysplastic syndrome characterized by specific
chromosomal abnormalities. Blood. 2004;104:4210-4218.
10. Schmelz K, Sattler N, Wagner M, Lubbert M, Dorken B, Tamm I. Induction of gene expression by 5-Aza-2'-deoxycytidine in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) but not epithelial cells by
DNA-methylation-dependent and -independent mechanisms. Leukemia. 2005;19:103-111.
11. Hofmann WK, de Vos S, Komor M, Hoelzer D, Wachsman W, Koeffler HP. Characterization of gene expression of CD34+ cells from normal and myelodysplastic bone marrow. Blood. 2002;100:3553-3560.
12. Ueda M, Ota J, Yamashita Y, et al. DNA microarray analysis of stage progression mechanism in myelodysplastic syndrome. Br J Haematol. 2003;123:288-296.
13. Pellagatti A, Esoof N, Watkins F, et al. Gene expression profiling in the myelodysplastic syndromes using cDNA microarray technology. Br J Haematol. 2004;125:576-583.
14. Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296:1653-1655.
15. Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054-1061. 16. James C, Ugo V, Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144-1148. 17. Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387-397.
18. Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779-1790.
19. Antonioli E, Guglielmelli P, Pancrazzi A, et al. Clinical implications of the JAK2 V617F mutation in essential thrombocythemia. Leukemia. 2005;19:1847-1849. 20. Campbell PJ, Scott LM, Buck G, et al. Definition of subtypes of essential thrombocythaemia and relation to polycythaemia vera based on JAK2 V617F mutation status: a prospective study. Lancet. 2005;366:1945-1953.
21. Wolanskyj AP, Lasho TL, Schwager SM, et al. JAK2 mutation in essential thrombocythaemia: clinical associations and long-term prognostic relevance. Br J Haematol. 2005;131:208-213.
22. Campbell PJ, Griesshammer M, Dohner K, et al. V617F mutation in JAK2 is associated with poorer survival in idiopathic myelofibrosis. Blood.
2006;107:2098-2100.
23. Tefferi A, Lasho TL, Schwager SM, et al. The JAK2(V617F) tyrosine kinase mutation in myelofibrosis with myeloid metaplasia: lineage specificity and clinical correlates. Br J Haematol. 2005;131:320-328.
24. Flower RJ, Rothwell NJ. Lipocortin-1: cellular mechanisms and clinical relevance. Trends Pharmacol Sci. 1994;15:71-76.
25. Alldridge LC, Bryant CE. Annexin 1 regulates cell proliferation by disruption of cell morphology and inhibition of cyclin D1 expression through sustained activation of the ERK1/2 MAPK signal. Exp Cell Res. 2003;290:93-107.