Short communication
A novel phospholipase A
2
from the venom glands of Bungarus
candidus: cloning and sequence-comparison
q
Inn-Ho Tsai*, Hwa-Yao Hsu, Ying-Ming Wang
Institute of Biological Chemistry, Academia Sinica, and College of Sciences, National Taiwan University, P.O. Box 23-106, Taipei 106, Taiwan, ROC
Received 26 March 2002; accepted 14 May 2002
Abstract
The presence of phospholipase A2(PLA2) in the venom of Malayan krait (Bungarus candidus ) and its structure were studied.
The PLA2cDNAs from the venom gland of B. candidus (Indonesia origin) were amplified by the polymerase chain reactions
(PCR) and cloned. The primers used were based on the cDNA sequences of several homologous B. multicinctus venom PLA2s.
In addition to the A-chains of b-bungarotoxins, a novel B. candidus PLA2was cloned and its full amino acid sequence deduced.
Having totally 125 amino acid residues, the PLA2contains a pancreatic loop and is 61% identical to the acidic PLA2of king
cobra venom. However, the enzyme was not detected from the venom sample. Its structural relationships to other elapid venom PLA2s were analyzed with a phylogenetic tree and discussed. q 2002 Elsevier Science Ltd. All rights reserved.
Keywords: Malayan krait (Bungarus candidus ); Snake venom; cDNA cloning; Phospholipase A2; Phylogenetic analysis
Being a common component in snake venoms, the phospholipases A2(PLA2s, EC 3.1.1.4) are classified into
several structural groups. Amino acid sequences of more than 150 snake venoms PLA2 have been solved; the
molecules are 121 – 125 amino acids long with six or seven disulfide bonds. The group IA and IB PLA2s are found
in the venom of elapid and hydrophiid species and in the mammalian pancreases, while group II PLA2s are present in
the viperid snake venoms. Interestingly, PLA2s in venom
glands may undergo fast adaptive evolution to generate variants of diverse functions (Danse et al., 1997; Francis et al., 1998; Yu et al., 1999).
Snake of the genus Bungarus, commonly known as kraits, are distributed from Pakistan eastward through southern Asia and China. The Malayan krait Bungarus candidus and the Chinese/Taiwanese krait B. multicinctus are medically important, and are responsible for many
cases of lethal snakebites in Asian countries. The major symptom of the envenoming is severe neurotoxicity (Warrell, 1993). The venoms of both species show strong lethal potency toward mice and birds (Lee and Tseng, 1969; Tan and Ponnudurai, 1990) and a phylogenetic study on many kraits revealed close relationship between the two (Slowinski, 1994). In both venoms, the most lethal components are b-bungarotoxins, which exist as hetero-dimers with a PLA2 (A-chain) covalently linked with a
Kunitz type inhibitor (B-chain) (Tan et al., 1989). A monomeric PLA2has also been isolated from the venom
of B. multicinctus (Kondo et al., 1981), and it has been shown that the PLA2caused a sharp fall in arterial blood
pressure (Lee and Lee, 1979). The purpose of the present study is to identify and clone PLA2from the venom gland
tissue of B. candidus and examine its evolutionary relationships with other elapid venom PLA2s.
For cloning and sequencing of the PLA2, mRNA was
extracted and purified from the venom glands of B. candidus (Bali, Indonesia). The complement DNA was prepared using the cDNA synthesis kits according to the manufac-turer instructions (Stratagene, USA). Specific primers were designed based on the highly conserved cDNA sequences encoding the monomeric PLA2s and the A chains of
0041-0101/02/$ - see front matter q 2002 Elsevier Science Ltd. All rights reserved. PII: S 0 0 4 1 - 0 1 0 1 ( 0 2 ) 0 0 1 5 0 - 2
www.elsevier.com/locate/toxicon
q
The novel nucleotide sequences and deduced protein sequences were deposited in GenBank with the accession number AF492561. * Corresponding author. Tel.: 2362-0264; fax: þ886-2-2363-5038.
E-mail address: bc201@gate.sinica.edu.tw (I.H. Tsai). Abbreviations: Bc-PL, phospholipase of Bungarus candidus; PLA2or PLA, phospholipase A2.
b-bungarotoxins in the B. multicinctus venom glands (Chang et al., 1996). Primers 1 and 2 are 50 -CCA-GACGGCTTCATCATG-30 and 50 -AAAAGGAATRATC-CAGG-30, respectively. Primer 1 was in the sense orientation of the 50-end from untranslated region to the first two amino acids of signal peptide, whereas primer 2 was in the antisense direction of a conserved region at the 30 -end. The polymerase chain reaction (PCR) (Mullis and Faloona, 1987) was conducted to amplify the PLA2cDNAs,
using venom gland cDNA as templates as well as SuperTaq DNA polymerase (HT Biotech., UK).
DNA fragments of expected size (0.4 kb) were specifi-cally amplified as shown by 1.5% agarose gel electrophor-esis. After being treated with polynucleotide kinase, they were inserted into the pGEM-T vector and transformed into Escherichia coli strain JM109 (Maniatis et al., 1989). White transformants were picked up and sequenced on the DNA-Sequencing-System (model 373A, PE-Applied Biosystems, USA). About 20 PLA2clones were sequenced and most of
them were found to encode the b-bungarotoxins A-chains. Only two of the cDNA clones encode a novel PLA2which
we designated as Bc-PL. Its amino acid sequence was
deduced from the cDNA sequence (Fig. 1(A)). However, by gel filtration and high performance liquid chromatography (HPLC) fractionation of the B. candidus venom, we tried but failed to isolate Bc-PL or any other PLA2except the
b-bungarotoxins (data not shown). It would have been isolated if its content is $ 0.1% of the crude venom by mass. The highly conserved signal peptide of Bc-PL as compared with other kraits’ PLA2s (Fig. 1(B)) suggests that mRNA of
Bc-PL is originated from venom glands rather than other snake tissues.
Having totally 125 amino acid residues, Bc-PL belongs to group IB in the classification of the PLA2superfamily
(Danse et al., 1997). Its calculated molecular weight is 13982 assuming seven-disulfide bonds between the con-served Cys residues, and the theoretical isoelectric point is 5.1. Bc-PL contains a ‘pancreatic loop’ at 62 – 66, an acidic N-terminal half but a highly basic C-terminal region. In comparison to other venom PLA2s, Bc-PL has uncommon
substitutions including F2, N6, E10, N22, C25, and T31 and contains no Trp residue. Although an intact calcium binding Asp 49 and Gly-rich loop (residues 26 – 33) as well as other residues presumably essential for the enzymatic action (e.g.
Fig. 1. (A) Alignment of the deduced amino acid sequences of the venom PLA2s from B. candidus and other related elapids. Numbering follows
that ofRenetseder et al. (1988). Single letter codes for amino acids were used. Dots denote the amino acid residues identical to those in the first line and gaps are (2 ). The GenBank accession numbers are: O. hannah (KM, i.e. Kunming), AF297034; O. hannah (GX, i.e. Guangxi), AF302907; O. hannah OHV-A (of the venom from Fujian, China), AF302908; B. multicinctus, 350460, respectively; and the Swissprot numbers are: B. fasciatus PL-III, P14615; N. scutatus HTe, S65624, respectively. (B) Alignment of the signal peptide sequences. Non-conserved substitutions are bolded. References are: (1) the present study (residues used to design the primer at 50-end are underlined); (2) – (4) GenBank accession numbers CAA34782, CAD24466, and AAK62362, respectively; (5) see the legend of A, the signal sequences of the three O. hannah PLA2s are identical; (6) GenBank accession number X12605.
His48, Asp99 and Tyr52) are present in Bc-PL, the enzyme activity might be lowered as judged from previous chemical modification and mutagenesis studies at positions 2, 6, 10, 22 and 31 (Kuipers et al., 1990; Yang and Chang, 1989; Yuan and Tsai, 1999).
According to the Blast search of the sequence databank, Bc-PL is structurally most similar (about 61% identical) to the myotoxic/cardiotoxic PLA2from Ophiophagus hannah
(king cobra) venom from Kunming, China and the haemorrhagic/myotoxic PLA2(HTe) from Notechis
scuta-tus scutascuta-tus venom (Francis et al., 1995). Previous studies showed the possible presence of two acidic PLA2s in king
cobra venom (Huang et al., 1997), and geographic variations apparently exist such that the sequences of the king cobra PLA2s from the three Chinese provinces, Kunming,
Guangxi, and Fujian are not identical (Fig. 1(A)). The ‘pancreatic loop’ has been found in the venom PLA2s of
king cobra (Huang et al., 1997), several Australian elapids (Francis et al., 1995; Lambeau et al., 1995; Pearson et al., 1991), and New Guinea Micropechis ikaheka (Gao et al., 1999) but not in other kraits’ PLA2s. The sequences of the
pancreatic loops are rather variable (Fig. 1(A)).
Highly similar PLA2s from Asian elapid venoms have
been purified and characterized in the past three decades. Recently, the cDNAs encoding many PLA2isoforms in the
venoms of Naja sputatix (Armugam et al., 1997), Naja kaouthia (Chuman et al., 2000), and Naja atra (Pan et al., 1994) were cloned and sequenced. As many as eight PLA2
isoforms have been isolated and sequenced from the pooled
venom of golden krait Bungarus fasciatus (Liu et al., 1989; Liu and Lo, 1994), including four catalytically active PLA2s
(III, Vb-1, X-1, X-2), three abundant but less-active PLA2s
(Va, Vb-2 and VI), and one inactive Ala49 PLA2. A
phylogenetic tree has been constructed based on the amino acid sequences of the group IA and group IB PLA2s from the
venoms. The bioinformatics tool used was the neighbor-joining methodology using PHYLIP (Felsenstein, 1992). The bovine pancreatic PLA2 sequence was used as the
outgroup.
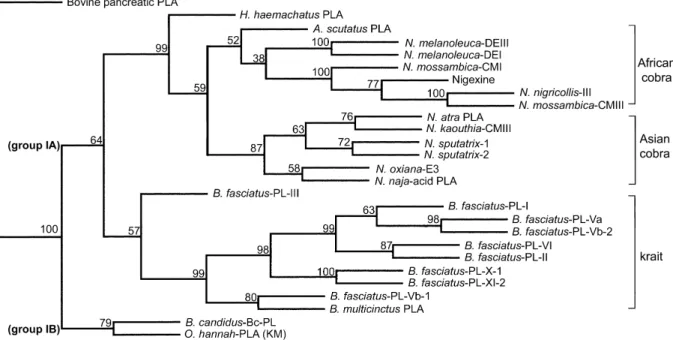
The dendrogram (Fig. 2) has focused on the structural relationships between the cobra and the krait venom PLA2s.
It shows linkage of the group IB members, i.e. Bc-PL and the king cobra venom PLA2, independent of other group IA
PLA2s. All the B. fasciatus PLA2s are closely related to each
other and to the B. multicinctus PLA2. Moreover, cobras
appear to be evolved under two phylogeographic groups, i. e. Asian and African, respectively. The topology of the tree (Fig. 2) showed taxonomic affinity between cobras and kraits with the robustness at most of the major nodes supported by bootstrap analysis. We did not consider the PLA2s of hydrophiid and Australian/New Guinea elapid
venoms since previous phylogenetic analysis of the group I PLA2s showed that the venom PLA2s of Hydrophiinae and
Australian Elapidae evolved independently of those of the Asian and African Elapidae (Tsai, 1997).
This is the first case that a group IB PLA2is found in
krait venom glands although the protein is not expressed in significant amount. The presence of variable surface loop at
Fig. 2. Dendrogram showing structural relationships among the venom PLA2s of Asian and African Elapidae. The tree was constructed using
the amino acid sequences of the elapid venom PLA2s (Armugam et al., 1997; Chuman et al., 2000; Danse et al., 1997; Liu et al., 1989; Liu and
Lo, 1994; Pan et al., 1994), and of bovine pancreatic PLA2(as the outgroup). Abbreviations are: N., Naja; O., Ophiophagus; and B., Bungarus,
and the GenBank accession numbers for B. fasciatus PL-I and PL-II and O. hannah PLA are AAK62362, AAK62361 and AF297034, respectively.
residues 62 – 66 of a PLA2supposedly would not affect its
over-all structure (White et al., 1990) but may decrease the hydrolytic activity (Kuipers et al., 1989; Thunnissen et al., 1990) and possibly regulates its function (Gao et al., 1999). The phylogenetic analysis (Fig. 2) represents an attempt to study the snake taxonomy based on the sequences of their venom PLA2s, this has been demonstrated to be rather
successful in another case (Tsai et al., 2001). Nevertheless, it is known that B. candidus and B. multicinctus are very similar but not so to king cobra (Slowinski, 1994, and our unpublished result). The expression of distinct monomeric PLA2s in both krait venoms, as either group IA or group IB,
probably resulted from natural selection between the venom PLA2paralogs through evolution, and both groups of PLA2
may be present in their ancestrial species.
Acknowledgements
We are grateful for the gift of a live specimen of B. candidus from Mr Duncan MacRae. This research was supported by grants from National Science Council, Taiwan.
References
Armugam, A., Earnest, L., Chung, M.C., Gopalakrishnakone, P., Tan, C.H., Tan, N.H., Jeyaseelan, K., 1997. Cloning and characterization of cDNAs encoding three isoforms of phos-pholipase A2in Malayan spitting cobra (Naja naja sputatrix )
venom. Toxicon 35, 27 – 37.
Chang, L.S., Wu, P.F., Chang, C.C., 1996. cDNA sequence analysis and expression of the A chain of b-bungarotoxin from Bungarus multicinctus (Taiwan banded krait). Biochem. Biophys. Res. Commun. 221, 328 – 332.
Chuman, Y., Nobuhisa, I., Ogawa, T., Deshimaru, M., Chijiwa, T., Tan, N.H., Fukumaki, Y., Shimohigashi, Y., Ducancel, F., Boulain, J.C., Menez, A., Ohno, M., 2000. Regional and accelerated molecular evolution in group I snake venom gland phospholipase A2isozymes. Toxicon 38, 449 – 462.
Danse, J.M., Gasparini, S., Menez, A., 1997. Molecular biology of snake venom phospholipases A2. In: Kini, R.M., (Ed.), Venom
Phospholipase A2Enzyme: Structure, Function and Mechanism,
Wiley, UK, pp. 29 – 71.
Felsenstein, J., 1992. PHYLIP: the PHYLogeny Inference Package, 3.573, Computer program distributed by the Department of Genetics, University of Washington, Seattle.
Francis, B., Coffield, J.A., Simpson, L.L., Kaiser, I.I., 1995. Amino acid sequence of a new type of toxic phospholipase A2from the
venom of the Australian tiger snake (Notechis scutatus scutatus ). Arch. Biochem. Biophys. 318, 481 – 488.
Francis, B.R., Meng, J., Kaiser, I.I., 1998. Classification of snake venom Group II Phospholipases A2 according to amino acid
sequence. In: Bailey, G.S., (Ed.), Enzymes from Snake Venom, Alaken, Colorado, pp. 503 – 544.
Gao, R., Kini, R.M., Gopalakrishnakone, P., 1999. Purification, properties, and amino acid sequence of a
hemoglobinuria-inducing phospholipase A2, MiPLA-1, from Micropechis
ikaheka venom. Arch. Biochem. Biophys. 369, 181 – 192. Huang, M.Z., Gopalakrishnakone, P., Chung, M.C., Kini, R.M.,
1997. Complete amino acid sequence of an acidic, cardiotoxic phospholipase A2 from the venom of Ophiophagus hannah
(King Cobra): a novel cobra venom enzyme with pancreatic loop. Arch. Biochem. Biophys. 338, 150 – 156.
Kondo, K., Toda, H., Narita, K., 1981. Amino acid sequence of phospholipase A from Bungarus multicinctus venom. J. Biochem. 89, 37 – 47.
Kuipers, O.P., Thunnissen, M.M., de Geus, P., Dijkstra, B.W., Drenth, J., Verheij, H.M., de Haas, G.H., 1989. Enhanced activity and altered specificity of phospholipase A2by deletion
of a surface loop. Science 244, 82 – 85.
Kuipers, O.P., Kerver, J., van Meersbergen, J., Vis, R., Dijkman, R., Verheij, H.M., de Haas, G.H., 1990. Influence of size and polarity of residue 31 in porcine pancreatic phospholipase A2on
catalytic properties. Protein Eng. 3, 599 – 603.
Lambeau, G., Ancian, P., Nicolas, J.P., Beiboer, S.H., Moinier, D., Verheij, H., Lazdunski, M., 1995. Structural elements of secretory phospholipases A2 involved in the binding to
M-type receptors. J. Biol. Chem. 270, 5534 – 5540.
Lee, C.Y., Tseng, L.F., 1969. Species differences in susceptibility to elapid venoms. Toxicon 7, 89 – 93.
Lee, C.Y., Lee, S.Y., 1979. Cardiovascular effect of snake venoms. In: Lee, C.Y., (Ed.), Snake Venoms: Handbook of Experimental Pharmacology, Vol. 52, Springer, Berlin, pp. 546 – 569. Liu, C.S., Lo, T.B., 1994. Chemical studies of Bungarus fasciatus
venom. J. Chin. Biochem. Soc. (ROC) 23, 69 – 75.
Liu, C.S., Hsiao, P.W., Chang, C.S., Tzeng, M.C., Lo, T.B., 1989. Unusual amino acid sequence of fasciatoxin, a weak reversibly acting neurotoxin in the venom of the banded krait, Bungarus fasciatus. Biochem. J. 259, 153 – 158.
Maniatis, T., Fritsch, E.F., Sambrook, J., 1989. Molecular Cloning, a Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
Mullis, K.B., Faloona, F.A., 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Meth. Enzy-mol. 155, 335 – 350.
Pan, F.M., Yeh, M.S., Chang, W.C., Hung, C.C., Chiou, S.H., 1994. Sequence analysis and expression of phospholipase A2 from
Taiwan cobra. Biochem. Biophys. Res. Commun. 199, 969 – 976.
Pearson, J.A., Tyler, M.I., Retson, K.V., Howden, M.E., 1991. Studies on the subunit structure of textilotoxin, a potent presynaptic neurotoxin from the venom of the Australian common brown snake (Pseudonaja textilis ). 2. The amino acid sequence and toxicity studies of subunit D. Biochim. Biophys. Acta 1077, 147 – 150.
Renetseder, R., Dijkstra, B.W., Huizinga, K., Kalk, K.H., Drenth, J., 1988. Crystal structure of bovine pancreatic phospholipase A2
covalently inhibited by p-bromophenacyl bromide. J. Mol. Biol. 200, 181 – 188.
Slowinski, J.B., 1994. A phylogenetic analysis of Bungarus (Elapidae ) based on morphological characters. J. Herpetol. 28, 440 – 446.
Tan, N.H., Ponnudurai, G., 1990. A comparative study of the biological properties of krait (genus Bungarus ) venoms. Comp. Biochem. Physiol. 95, 105 – 109.
properties of Bungarus candidus (Malayan krait) venom and venom fractions. Toxicon 27, 1065 – 1070.
Thunnissen, M.M., Kalk, K.H., Drenth, J., Dijkstra, B.W., 1990. Structure of an engineered porcine phospholipase A2 with
enhanced activity at 2.1 A˚ resolution. Comparison with the wild-type porcine and Crotalus atrox phospholipase A2. J. Mol.
Biol. 216, 425 – 439.
Tsai, I.H., 1997. Phospholipase A2 from Asian snake venom.
J. Toxicol., Toxin Rev. 16, 79 – 113.
Tsai, I.H., Chen, Y.H., Wang, Y.M., Tu, M.C., Tu, A.T., 2001. Purification, sequencing, and phylogenetic analyses of novel Lys-49 phospholipases A2from the venoms of rattlesnakes and
other pit vipers. Arch. Biochem. Biophys. 394, 236 – 244. Warrell, D.A., 1993. Severe neurotoxic envenoming by the
Malayan krait (B. candidus ). Br. Med. Bull. 286, 678 – 680.
White, S.P., Scott, D.L., Otwinowski, Z., Gelb, M.H., Sigler, P.B., 1990. Crystal structure of cobra-venom phospholipase A2in a
complex with a transition-state analogue. Science 250, 1560 – 1563.
Yang, C.C., Chang, L.S., 1989. Studies on the status of lysine residues in phospholipase A2 from Naja naja atra (Taiwan
cobra) snake venom. Biochem. J. 262, 855 – 860.
Yu, B.Z., Rogers, J., Tsai, M.D., Pidgeon, C., Jain, M.K., 1999. Contributions of residues of pancreatic phospholipase A2 to
interfacial binding, catalysis, and activation. Biochemistry 38, 4875 – 4884.
Yuan, C., Tsai, M.D., 1999. Pancreatic phospholipase A2: new