Molecular Ecology Notes (2003) 3, 595–597 doi: 10.1046/j.1471-8286.2003.00523.x
© 2003 Blackwell Publishing Ltd Blackwell Publishing Ltd.
P R I M E R N O T E
Isolation and characterization of microsatellite DNA
markers from the Lanyu scops owl (
Otus elegans botelensis
)
Y U - C H E N G H S U ,*† L U C I A L . S E V E R I N G H A U S ,‡ Y A O - S U N G L I N * and S H O U - H S I E N L I §
*Laboratory of Biodiversity, Institute of Zoology, National Taiwan University, Taipei 106 Taiwan, ROC; ‡Laboratory of Ornithology, Institute of Zoology, Academia Sinica, Taipei 115 Taiwan, ROC; §Laboratory of Genetic Diversity, Department of Biology, National Taiwan Normal University, Taipei 116 Taiwan, ROC
Abstract
From a genomic library enriched for GATA /CTAT and GAAA /CTTT repeats, 12 polymorphic
microsatellite markers were developed for the Lanyu scops owl (Otus elegans botelensis).
Polymorphism of these loci was evaluated in a sample of 58 adult individuals of unknown relationship. The allele numbers of each locus were from five to 25 and the observed heterozygosity of each locus ranges from 0.707 to 0.914. When using these loci in parentage assignment, the probability of false parent exclusion is greater than 0.999. All loci conformed to Hardy–Weinberg expectations.
Keywords: Otus elegans botelensis, Lanyu scops owl, microsatellites, strigidae, enrichment, genotyping
Received 18 June 2003; revision received 12 August 2003; accepted 12 August 2003
Lanyu scops owl (Otus elegans botelensis) is a subspecies of
elegant scops owl (O. elegans) and was listed as endangered
in the Red Data Book (King 1981). It is found only on Lanyu
Island (22° N, 121°5′-E), a 45.7-km2 island 60 km southeast
of Taiwan. A long-term study on the behaviour, ecology and population dynamics of this species has been conducted since 1985 (e.g. Severinghaus 2000; Severinghaus & Rothery 2001) making it one of the few tropical owl species intensively studied. Extra-pair copulations were observed occasionally during the breeding season (Severinghaus, personal observa-tion). To further investigate the population structure and genetic mating system of this species, highly polymorphic genetic markers, such as microsatellites, are needed. In this study, we constructed a microsatellite-enriched genomic DNA library of Lanyu scops owl to screen for highly poly-morphic microsatellite DNA loci.
Genomic DNA from an adult owl was used to construct
a library. After digestion with HaeIII, RsaI and AluI
restric-tion enzymes, the fragments were ligated with SNX linker
(Hamilton et al. 1999). These ligates were first amplified
using polymerase chain reaction (PCR) at 94 °C for 5 min,
30 cycles of 94 °C for 1 min, 55 °C for 1 min, and 70 °C for
2 min then 70 °C 5 min, followed by a solid phase
hybrid-ization with 3′-biotin-labelled (GAAA)10 or (GATA)10
oligo-nucleotides to select for microsatellite-containing DNA fragments. The biotin-labelled DNAs were eluted using Dynalbead streptavidin M280 (Dynal®) according to the manufacturer’s instruction. These microsatellite-enriched
DNAs were PCR-amplified (Li et al. 1997) then ligated into
vectors and transformed to JM109 competent cells, using pGEM®-T Easy vector system II (Promega). Transformed cells were grown on LB agar plates containing ampicil-lin, X-gal and IPTG for blue/white selection of positive
clones at 37 °C for 16 h. A total of 1500 positive clones were
lifted to Hybond-N+ membranes (Amersham Pharmacia
Biotech) and hybridized with [r32P] ATP end-labelled
(GAAA)8 or (GATA)8 oliginucleotides, then 192 positive
inserts were selected to extract their plasmid DNA, using the Montage Plasmid Miniprep kit (Millipore). These plasmid DNAs were sequenced using DYEnamic ET Dye Terminator Cycle Sequencing Kit for MegaBACE on MegaBACE 1000 autosequencer (Amersham Bioscience).
Sequences were edited in sequencher software (V4.0.5,
Gene Codes corporation) and 22 primer pairs were designed from 54 sequences containing tandem repeats and suitable
flanking sequences, using fastPCR 2.3.10 program which
is available at http://www.biocentre.helsinki.fi/bi/bare-1_html/download.htm.
Correspondence: Shou-Hsien Li. †Present address: Department of Biology, National Taiwan Normal University, Taipei 116 Taiwan, ROC. Fax: 886-2-29312904; E-mail: t43028@cc.ntnu.edu.tw
596 PRIMER NOTE © 2003 Blackwell Publishing Ltd, Molecular Ecology Notes , 3, 595 – 597
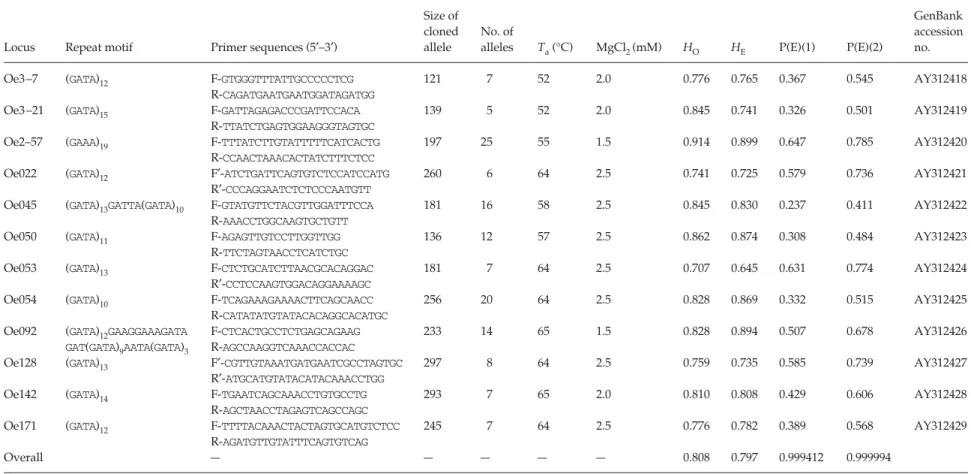
Table 1 Characterization of 12 microsatellite loci in the Lanyu scops owls
Locus Repeat motif Primer sequences (5′–3′)
Size of cloned allele
No. of
alleles Ta (°C) MgCl2 (mM) HO HE P(E)(1) P(E)(2)
GenBank accession no.
Oe3–7 (GATA)12 F-GTGGGTTTATTGCCCCCTCG 121 7 52 2.0 0.776 0.765 0.367 0.545 AY312418
R-CAGATGAATGAATGGATAGATGG
Oe3–21 (GATA)15 F-GATTAGAGACCCGATTCCACA 139 5 52 2.0 0.845 0.741 0.326 0.501 AY312419
R-TTATCTGAGTGGAAGGGTAGTGC
Oe2–57 (GAAA)19 F-TTTATCTTGTATTTTTCATCACTG 197 25 55 1.5 0.914 0.899 0.647 0.785 AY312420
R-CCAACTAAACACTATCTTTCTCC
Oe022 (GATA)12 F′-ATCTGATTCAGTGTCTCCATCCATG 260 6 64 2.5 0.741 0.725 0.579 0.736 AY312421
R′-CCCAGGAATCTCTCCCAATGTT
Oe045 (GATA)13GATTA(GATA)10 F-GTATGTTCTACGTTGGATTTCCA 181 16 58 2.5 0.845 0.830 0.237 0.411 AY312422
R-AAACCTGGCAAGTGCTGTT
Oe050 (GATA)11 F-AGAGTTGTCCTTGGTTGG 136 12 57 2.5 0.862 0.874 0.308 0.484 AY312423
R-TTCTAGTAACCTCATCTGC
Oe053 (GATA)13 F-CTCTGCATCTTAACGCACAGGAC 181 7 64 2.5 0.707 0.645 0.631 0.774 AY312424
R′-CCTCCAAGTGGACAGGAAAAGC
Oe054 (GATA)10 F-TCAGAAAGAAAACTTCAGCAACC 256 20 64 2.5 0.828 0.869 0.332 0.515 AY312425
R-CATATATGTATACACAGGCACATGC
Oe092 (GATA)12GAAGGAAAGATA F-CTCACTGCCTCTGAGCAGAAG 233 14 65 1.5 0.828 0.894 0.507 0.678 AY312426
GAT(GATA)9AATA(GATA)3 R-AGCCAAGGTCAAACCACCAC
Oe128 (GATA)13 F′-CGTTGTAAATGATGAATCGCCTAGTGC 297 8 64 2.5 0.759 0.735 0.585 0.739 AY312427
R′-ATGCATGTATACATACAAACCTGG
Oe142 (GATA)14 F-TGAATCAGCAAACCTGTGCCTG 293 7 65 2.0 0.810 0.808 0.429 0.606 AY312428
R-AGCTAACCTAGAGTCAGCCAGC
Oe171 (GATA)12 F-TTTTACAAACTACTAGTGCATGTCTCC 245 7 64 2.5 0.776 0.782 0.389 0.568 AY312429
R-AGATGTTGTATTTCAGTGTCAG
Overall — — — — — 0.808 0.797 0.999412 0.999994
Ta: annealing temperature; HO: observed heterozygosity; HE: expected heterozygosity; P(E)(1): exclusionary power of false parent when neither parent is unknown; P(E)(2): exclusionary
P R I M E R N O T E 597
© 2003 Blackwell Publishing Ltd, Molecular Ecology Notes, 3, 595–597
Nineteen of the primer pairs showed a PCR product with expected sizes and 12 of them were selected to screen for polymorphism. The forward primers of these 12 loci were labelled with HEX, FAM or TAMRA fluorescent dyes. For the characterization of polymorphism of these loci, 58 adult individuals with unknown relationship were genotyped. PCR were set up in 10 µL reaction volume
con-taining 30 ng genomic DNA, 0.3 µm each primer, 0.5 mm
dNTP, 10 mm Tris-HCL, pH 9.0, 50 mm KCL, 0.01% (w/V) gelatin, 0.1% Triton X-100, 0.4 U Pro Taq DNA polymerase
(Protech) and 1.5–2.5 mm MgCl2 (see Table 1). The PCR
condition was as follows: Firstly, denaturation at 94 °C for
3 min, then 34 cycles of 94 °C 30 s, 30 s at the optimal
annealing temperature of each primer pair (Table 1) and 72 °C for 30 s, followed by a final extension at 72 °C for 3 min. All of the PCR in this study were conducted using iCycler thermal cycler (Bio-Rad). The PCR products were electrophoresed in megabace 1000 autosequencer (Amer-sham Biosciences). Sizing of alleles was analysed with the software genetic proflier 2.0 (Amersham Biosciences).
Table 1 summarized the polymorphism of the 12 micro-satellite loci. The observed and expected heterozygosities, probability of false parent exclusion and conformation to Hardy–Weinberg expectations were calculated using pro-gram cervus 2.0 (Marshall et al. 1998). No significant devi-ation from Hardy–Weinberg equilibrium was detected across all loci. Overall, the mean number of alleles per locus was 11.17 and the mean expected heterozygosity was 0.797. When applying these loci to paternity analysis, more
than 99.9% of false putative parents can be excluded. The highly polymorphic nature of these microsatellite loci makes it an ideal genetic marker system to study the population genetics and mating system of the Lanyu scops owl.
Acknowledgements
This work was funded by the National Science Council to LLS (NSC 90–2313-B-001–036) and by the Council of Agriculture to SHL [COA 90AT-1.3.3-F-R1(5)]. We thank Dr Y. J. Huang and members of Genetic Diversity Laboratory for the help in lab work and all assistants of Ornithology Laboratory for collecting blood samples.
References
Hamilton MB, Pincus EL, Fiore AD, Fleischer RC (1999) Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques, 27, 500–507. King WB (1981) Endangered Birds of the World. The ICBP Bird Red
Data Book. Smithsonian Institute Press, Washington, DC. Li SH, Huang YJ, Brown JL (1997) Isolation of tetranucleotide
microsatellites from the Mexican Jay Aphelocoma ultramarina. Molecular Ecology, 6, 499–501.
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology, 7, 639–655.
Severinghaus LL (2000) Territoriality and the significance of call-ing in the Lanyu scops owl. Otus Elegans Botelensis. IBIS, 142, 297–304.
Severinghaus LL, Rothery P (2001) The survival rate of Lanyu scops owls. Otus Elegans Botelensis. IBIS, 143, 540–546.