Segmentation of 3D MicroPET Images Using The Mixture Method
全文
(2) λ p* ( d ) and λ d* ( d ) respectively, the joint likelihood function of L (λ*p , λ*d ) is listed in Eq. (3). * p. * d. L (λ , λ ) =. D. ∏. (3). n *pd ( d ) ~ Poisson ( λ*d ( d )).. (5). The n ( b, d ) is the detected number of emissions * p. occur at bth pixel in dth tube and n *pd ( d ) is the detected number of accidental coincidence (AC) events in dth tube. It is assumed that n *p ( b , d ) and. n n. * p. ( d ) are statistically independent, then B. (d ) =. b =1. n. * p. ( b, d ) + n. * pd. (d ) .. where fk(Ixyz, k) refers to the probability density function in the kth cluster with parameter k. All parameters are collected to form a parameter vector Φ = ( π 1 ,..., π K , θ 1 ,..., θ K ). Since, the MLEs are usually difficult to solve directly. Alternatively, the EM algorithm can be applied to find the MLE iteratively [16-17]. Firstly, we introduce an index function as in Eq. (10). 1, if I xyz comes from k th normal distributi on. (10) = C xyzk. f ( I xyz | C xyz ) =. 1 2. ω xyzk =. p (b ' , d )λ. old. (d ). (b ') + λ. *old d. + n d* ( d ) ,. C xyzkπ k f k ( I xyz ;θ k ).. (11). K. π N. ( old ) j. f j ( I xyz ;θ. ( old ) j. (13). , ). µ. ( new ) k. =. ω xyzk ⋅ ( I xyz − µk( new) ) 2. xyz =1. N ⋅ π k( new) N. b' =1. For the study of a set of 3D images, it is denoted by Ixyz, where Ixyz represents the intensity at the xyzth voxel of one set of 3D images. In this study, xy in one slice has the size of 256x256 and the number of slices z has the size of 10. Since the variance scale in segments of 3D images may be different, we consider Gaussian mixture model with different mean vectors and covariance matrices [16-17]. Suppose that the probability of the data Ixyz comes from the kth cluster distribution is probability k, then, Eq. (8) holds.. (12). j =1. (d ). 2.2: Gaussian Mixtures Model on 3D Segmentation. ω xyzk ,. xyz =1. π k( old ) f k ( I xyz ;θ k( old ) ). (7). for all b = 1, 2,…, B, and d = 1, 2,…, D.. N. 1 N. π k( new) =. σ k2 ( new) =. n ( d )λ B. K. Similarly using EM approach finds MLE of parameters with the constraint of Eq. (8), the solutions of parameters for the local maximum are shown as in Eqs. (12) - (15).. b' =1. λ*dnew ( d ) =. 0, otherwise.. Let Cxyz = (Cxyz1, …, CxyzK) denote the unobserved index vector and {Ixyz, Cxyz} form the complete data for the EM algorithm. Given Cxyz, the conditional density of Ixyz becomes Eq. (11).. log likelihood function in complete data space. The optimal solution can be achieved directly by taking the first derivatives of the new parameters equal to zeros. The solutions are shown in Eqs. (6) and (7). D n *p (d ) p (b, d ) new old (6) λ (b) = λ (b) , B d =1 p (b' , d )λold (b') + λ*dold ( d ) *old d. (9). π k f k ( I xyz ; θ k ),. k =1. k =1. After applying EM approach, we can determine λ new and λ d* new as the solutions that maximize the expected. * p. K. f ( I xyz ; Φ) =. *. exp( − λ d* ( d )) λ d* ( d ) n d ( d ) . ∏ n d* ( d ) d =1 The EM algorithm can be applied to compute the maximum likelihood estimate iteratively [18-19]. Firstly, the observed data, n *p ( d ) and n d* ( d ) , are regarded as incomplete data. The EM algorithm needs to model the complete data. One possible model for PET is shown as in Eqs. (4) and (5). (4) n *p (b , d ) ~ Poisson ( p (b , d ) λ (b )),. * pd. , K . (8). Hence, the probability density function of Ixyz is as Eq. (9).. n *p ( d ). n *p ( d ). D. π k = 1, and 0 ≤ π k ≤ 1, for all k = 1,2,. k =1. exp( − λ p* ( d )) λ *p ( d ). d =1. K. ,. (14). ω xyzk ⋅ I xyz. xyz =1. N ⋅ π k( new). .. (15). Also, the Gaussian distribution for the kth cluster is listed as Eq. (16). f k ( x ;θ k ) = f k ( x ; µ k , σ k ) (16) 1 − 1 = ( 2πσ k2 ) 2 exp − ( x − µ k ) 2 / σ k2 . 2. 2.3: EM Algorithm for GMM The EM algorithm for GMM is described as follows. 1. Set the initial parameters (old). 2. Update the parameters by using Eqs. (12) - (15). 3. If log Lin( (new) ) –log Lin ( (old)) < tolerance, then the iteration stops. Otherwise, go to Step 2 with the old values of parameters replaced by the new values.. - 1315 -.
(3) 2.4: Slice Normalization The intensity heterogeneity in slices of microPET images comes from differences in emission activities and reconstruction processes. The heterogeneity between slices affects the accuracy of the segmentation. Hence, slice normalization is needed before 3D segmentation to adjust image levels between slices. The slice normalization is defined in Eq (17). new I xyz =a. where. new I xyz. normalization, voxel,. is. I xyz − Min z Max z − Min z the. new. ,. intensity. (17) after. slice. I xyz is the original intensity at xyzth. Minz is the minimum value of the zth slice,. Max z is the maximum value of the zth slice, and a is a. none zero constant for adjusted image level, like a = 32768.. 2.5: Determination of the Cluster Number for GMM The kernel density estimation (KDE) [20] is adopted to determinate the numbers of cluster used in GMM. Those high peaks could be regarded as mean of the groups and low peaks were used to decide standard deviation of the groups. It is based on the empirical rule the range of µ ± 3σ covers the most of observations from a normal distribution. Hence, the initialized values of GMM are determined by applying this empirical rule. Let the value of high peak be hp and value of the right low peak be lp, then this empirical rule is used to estimate by (hp-lp)/3. Therefore, hp and (hp-lp)/3 will be used as the starting values of and for one cluster. This approach can automatically decide the cluster number and starting values of each cluster in EM algorithm of GMM.. 3: Results 3.1: Simulation Studies The simulated phantom study with 457932 total counts is displayed in Figure 1. This simulated study is focused on testing and evaluating the performance of GMM. Figure 1A shows target image with five ROIs. Figure 1B displays target image with 50% noise added. Figure 1C presents the clustering results by GMM. The number of clusters is decided by the KDE shown in Figure 1D. There are four local high peaks regarded as the means of four clusters. For accuracy comparison, Figure 2 shows the indices of ROIs by GMM. Figure 3 presents the accurate comparison between the results by K-means and GMM. It is observed that the GMM has a clearer segmentation result than the K-means method. has. Detail results of ROIs are shown in Table 1. The total accuracy of GMM is 92.1% and that of K-means is 66.6%. Another simulated volume data based on the modified Shepp-Logan' s head phantom image is shown as in Figure 4A and 4B. Fifty percentage of noise ratio to phantom images are added. In order to compare the effects of variation between slices, different image levels and shapes of ROIs are considered in slice 1 and 2. First, we use the MLE-EM reconstruction, the result is shown in Figure 4C and 4D. Meanwhile, the GMM is also applied to segment two images without slice normalization as shown in Figure 4E and 4F. It is observed that the boundaries of ROIs are difficult to distinguish. Therefore, slice normalization is applied to the volume data and then GMM is used to segment images as shown in Figure 4G and 4H. The boundaries of these segmentations become clearer after slice normalization. Figure 4I plotted estimated kernel density curve of volume data for finding the number of clusters and initialized values. For these simulation cases, the performance and accuracy using GMM is better than those of using K-means. The KDE is adopted to decide the number of clusters and the starting values of parameters in the EM algorithm. The slice normalization is necessary when GMM is applied to segment volume data in this study.. 3.2: Empirical Studies The empirical data of one rat (i.e., one big mouse) injected by F18 isotope scanning is collected from the microPET R4 system. The acquired configurations are listed as below. Scanner energy is between 350 and 750 Kev with total scanning 3600 seconds. There are 32 rings in microPET R4 system. File format of histogram data is stored by 2 bytes for each voxel. Ten slices (from the 51st to the 60th slice) of the volume data are used for investigation and evaluation. Figure 5 shows estimated kernel density curve of volume data. Based on this KDE, four groups are determinate by local high peaks and their starting values are obtained for EM algorithm. Figure 6 shows the reconstructed rat images by MLE-EM from the 51st to the 60th slice. Besides, Figure 7 and 8 show the segmentation results by GMM and K-means respectively. The detail segmentation from GMM are shown with the comparison to Figure 6. The details uptake areas can be segmented by GMM from 59th and 60th images, in addition, it can segment small areas with high gene expression than those by K-means. On the contrary, the K-means method segments big areas and ignores small uptake areas. For this real rat microPET study, the GMM leads to more detail segmentation results than the K-means method does. The GMM also has better performance than K-means. The full width half maximum (FWHM) is usually used to evaluate performance of segmented results. The horizontal line profile near the center of the 60th slice is used to investigate the performance between. - 1316 -.
(4) GMM and K-means. Figure 9 is plotted with four regions in this line profile and their FWHMs for Fig. 6. Table 2 shows that the FWHMs of segmented results by GMM are closer to target FWHMs than those by K-means. Meanwhile, the signal to noise ratio (SNR) defined by the ratio of mean value to standard deviation is used to compare the segmentation performance between GMM and K-means. The SNRs of four regions of GMM are higher than those of K-means.. 4: Conclusion and Discussion We have proposed the GMM to segment 3D microPET images with the MLE-EM reconstruction. With fewer artifacts, the MLE-EM reconstruction is more precise than the FBP reconstruction is. For the seeding region growing methods proposed in literature for the segmentation of PET images, initial seeds were crucial to perform images segmentation. The number of clusters was determined by a subjective choice or sequentially searching. On the other hand, the GMM proposed in this study can perform the segmentation automatically through the KDE method. The KDE approach can automatically estimate the number of clusters and provide the initial values for the EM algorithm of GMM. If the activities of various clusters have different temporary patterns, the slice normalization approach incorporated with GMM is useful to segment 3D images. For further investigation, it will be of great interest to evaluate the qualitative and quantitative performance by more phantom and empirical studies with the comparisons to judgments from medical experts in the future.. Acknowledgement The authors thank Dr. Meei-Ling Jan and her lab for supporting microPET rat data in Institute of Nuclear Energy Research. This research was supported by the grants of NSC in Taiwan.. REFERENCES [1] Cunningham VJ, Jones T. Spectral analysis of dynamic PET studies. J. Cereb. Blood Flow Metab., 1993, 13, 15-23. [2] Shepp LA, Vardi Y. Maximum likelihood reconstruction for emission tomography. IEEE Trans. Med. Image, 1982, MI-1, 99, 113-122. [3] Vardi Y, Shepp LA, Kaufman L. A statistical model for positron emission tomography. Journal of the American Statistical Association, 1985, 80, 389, 8-20. [4] Politte DG., Snyder DL. Corrections for accidental coincidences and attenuation in maximum-likelihood image reconstruction for positron-emission tomography.. IEEE Transactions on Medical Image, 1991, 10, 1, 99, 82-89. [5] Ollinger JM, Fessler JA. Positron-emission tomography. IEEE Signal Processing Magazine, 1997, 41, 1, 43-55. [6] Lu HHS, Tseng WJ. On Accelerated Cross-Reference Maximum Likelihood Estimates for Positron Emission Tomography. Proceedings of the IEEE Nuclear Science Symposium, 1997, 2, 1484 -1488. [7] Lu HHS, Chen CM, Yang IH. Cross-Reference Weighted Least Square Estimates for Positron Emission Tomography. IEEE Transactions on Medical Imaging, 1998, 17, 1, 1-8. [8] Chen JC, Tu KY, Lu HHS, Chen TB, Chou KL, Liu RS. Statistical Image Reconstruction for PET Studies. The Biomedical Engineering Society 1998 Annual Symposium. 1998, 71-72. [9] Tu KY, Chen JC, Lu HHS, Chen TB, Chou KL, Liu RS. Random Correction Using Iterative Reconstruction for PET. European Association of Nuclear Medicine Annual Congress. 2000. [10] Chen JC, Liu R., Tu KY, Lu HHS, Chen TB, Chou KL. Iterative Image Reconstruction with Random Correction for PET studies. Proceedings of the Society of Photo-Optical Instrumentation Engineers, 2000, 3979, 1218-1229. [11] Tu KY, Chen, TB, Lu, HHS, Liu RS, Chen KL, Chen CM, Chen JC. Empirical Studies of Cross-Reference Maximum Likelihood Estimate Reconstruction for Positron Emission Tomography. Biomed. Eng. Appl. Basis Comm. 2001, 13, 1, 1-7. [12] Chen CM, Lu HHS, Hsu YP. Cross-Reference Maximum Likelihood Estimate Reconstruction for Positron Emission Tomography. Biomed. Eng. Appl. Basis Comm., 2001, 13, 4, 190-198. [13] Akaike H. A new look at the statistical model identification. IEEE Trans. Automat. Ontr., 1974, AC-19, 716-723. [14] Wong KP, Feng D, Meikle SR, Fulham MJ. Simultaneous estimation of physiological parameters and input function: In vivo PET data. IEEE Trans. Inform. Technol. Biomed., 2001, 5, 67-76. [15] Wong KP, Feng D, Meikle SR, Fulham MJ. Segmentation of Dynamic PET Images Using Cluster Analysis. IEEE Transactions on Nuclear Science, 2002, 49, 1. [16] McLachlan G.J, Basford KE. Mixture Models. Inference and Applications to Clustering. Dekker. 1988. [17] Hsiao IT, Rangarajan A, Gindi G. Joint-Map Reconstruction/Segmentation for transmission Tomography Using Mixture-Model as Priors. Proc. IEEE Nuclear Science Symposium and Medical Image Conference, 1998, 3, 1689-1693. [18] Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm (with discussion). J. R. Statist. Soc. B, 1977, 39, 1-38. [19] Wu CFJ. On the convergence properties of the EM algorithm. Annals of Statistics, 1983, 11, 95-103. [20] Sheather, S. J. and Jones M. C. A reliable data-based bandwidth selection method for kernel density estimation. J. Roy. Statist. Soc. B, 1991, 683-690.. - 1317 -.
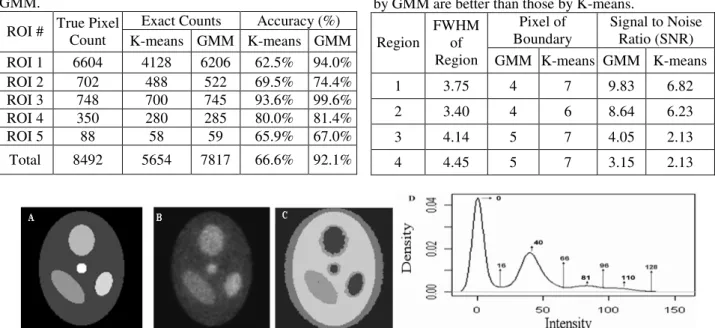
(5) Table 1: Compared clustering results by K-means and GMM. Accuracy (%) True Pixel Exact Counts ROI # Count K-means GMM K-means GMM ROI 1 6604 4128 6206 62.5% 94.0% ROI 2 702 488 522 69.5% 74.4% ROI 3 748 700 745 93.6% 99.6% ROI 4 350 280 285 80.0% 81.4% ROI 5 88 58 59 65.9% 67.0% Total. 8492. 5654. 7817. 66.6%. Table 2: The FWHMs and SNRs of segmented results by GMM are better than those by K-means. Pixel of Signal to Noise FWHM Boundary Ratio (SNR) Region of Region GMM K-means GMM K-means. 92.1%. 1. 3.75. 4. 7. 9.83. 6.82. 2. 3.40. 4. 6. 8.64. 6.23. 3. 4.14. 5. 7. 4.05. 2.13. 4. 4.45. 5. 7. 3.15. 2.13. Fig.1. A) Simulation image of five clusters is displayed. B) Adding 50% noise into Fig. A. C) Clustering results using GMM. D) Kernel density curve using C with values of high and low peaks. It identifies four peaks. Hence, the number of groups is set as four.. Fig.2. Target ROIs are marked.. Fig.3. The results of A) by K-means and B) by GMM clustering are shown.. Fig.4. Simulated volume data including slice 1 and 2 are marked as A and B. C and D are reconstructed images after added 50% noise ratio to A and B. E and F are segmented results without slice normalization. G and H are segmented results with slice normalization. I is the estimated kernel density curve of simulated volume data after slice normalization.. Fig.5. Kernel density curve using rat volume data (10 slices) with values of high and low peaks. There are four peaks. Hence, the number of groups is set as four. Values of peak are applied to compute starting values in EM algorithm.. - 1318 -.
(6) Fig.6. The reconstructed rat images are shown from the 51st (top-left) to the 60th (bottom -right) slice.. Fig.7. The results of segmentation by the GMM are shown from the 51st (top-left) to the 60th (bottom -right) slice.. Fig.8. The results of segmentation by the K-means method are shown from the 51st (top-left) to the 60th (bottom -right) slice.. Fig.9. The horizontal line profile of the 60th slice of Fig. 6 is shown with FWHMs. The FWHMs of region 1, 2, 3 and 4 are 3.75, 3.40, 4.14 and 4.45 pixels respectively. The top part shows the location of this line profile in the MLE-EM reconstruction image and the segmented results by GMM and K-means.. - 1319 -.
(7)
數據
相關文件
Valor acrescentado bruto : Receitas do jogo e dos serviços relacionados menos compras de bens e serviços para venda, menos comissões pagas menos despesas de ofertas a clientes
A cylindrical glass of radius r and height L is filled with water and then tilted until the water remaining in the glass exactly covers its base.. (a) Determine a way to “slice”
Promote project learning, mathematical modeling, and problem-based learning to strengthen the ability to integrate and apply knowledge and skills, and make. calculated
Microphone and 600 ohm line conduits shall be mechanically and electrically connected to receptacle boxes and electrically grounded to the audio system ground point.. Lines in
The remaining positions contain //the rest of the original array elements //the rest of the original array elements.
– S+U can also preserve annotations of synthetic images – Refined images really help improving the testing result – Generate > 1 images for each synthetic
A digital color image which contains guide-tile and non-guide-tile areas is used as the input of the proposed system.. In RGB model, color images are very sensitive
Finally, the Delphi method is used to verify and finalize the assessing framework.. Furthermore, the AHP method is used to determine the relative weights of factors in the