國 立 交 通 大 學
生化工程研究所
碩 士 論 文
減弱果蠅神經系統中酪氨酸亞硫酸基轉移酶
基因表現量對蛋白質亞硫酸化功能之探討
Functions of Protein Sulfation in Drosophila melanogaster by
Neuron-specific Knockdown of Tyrosylprotein Sulfotransferase
研究生: 吳咏馨
指導教授: 楊裕雄 教授
減弱果蠅神經系統中酪氨酸亞硫酸基轉移酶基因表現量 對蛋白質亞硫酸化功能之探討
Functions of Protein Sulfation in Drosophila melanogaster by Neuron-specific Knockdown of Tyrosylprotein Sulfotransferase
研究生: 吳咏馨 Student: Jennifer Yun-shin Wu 指導教授: 楊裕雄 教授 Advisor: Prof. Yuh-Shyong Yang
國立交通大學 生化工程研究所
碩士論文
A Thesis
Submitted to Institute of Biochemical Engineering College of Biological Science and Technology
National Chiao Tung University in Fulfillment of the Requirements
for the Degree of Master
in
Biochemical Engineering July 2009
Hsinchu, Taiwan, Republic of China
減弱果蠅神經系統中酪氨酸亞硫酸基轉移酶基因表現量 對蛋白質亞硫酸化功能之探討 學生: 吳咏馨 指導教授: 楊裕雄 教授 國立交通大學生化工程所碩士班 摘要 蛋白質的亞硫酸化是很重要的蛋白質後修飾之一,而酪氨酸亞硫酸基轉移酶 (tyrosylprotein sulfotransferase)負責將腺苷 3’-磷酸-5'-磷醯硫酸 (PAPS)的亞硫酸基轉移 到許多特定分泌性或膜上蛋白質的酪氨酸上。目前發現許多帶有亞硫酸化酪氨酸的蛋白 質參與了不同的生理過程,其中包含凝血反應、白血球貼附血管細胞、趨化素的訊息傳 遞、以及人類免疫缺陷病毒(HIV)感染細胞。至今對於亞硫酸化蛋白質體以及酪氨酸亞 硫酸機轉移酶本身所調控的生物功能都仍然不清楚。根據基因序列以及基因微陣列(gene microarray)的分析發現在果蠅體內僅存在單一個酪氨酸亞硫酸基轉移酶基因,並且此基 因的訊息核醣核酸(mRNA)在果蠅頭部及腦部有很高的表現量。利用了 GAL4-UAS 系統 將果蠅神經系統中的酪氨酸亞硫酸基轉移酶進行基因表現減弱(knockdown)。由二維蛋 白質電泳以及數位影像分析可以發現有四個蛋白質受到正調控(up-regulation),而有十八 個蛋白質受到負調控(down-regulation)。利用質譜儀針對這二十二個蛋白質做身分鑑定僅 鑑別出九個蛋白質,而他們大多與代謝以及氧化壓力(oxidative stress)有關。銅鋅超氧化 歧化酶(superoxide dismutase [Cu-Zn])是一種抗氧化的酵素,其主要負責清除體內的自由 基,而此酵素在神經系統中酪氨酸亞硫酸機轉移酶進行基因減弱的果蠅中有顯著的負調 控。利用巴拉刈(paraquat)進行氧化耐受度分析實驗,發現這些基因減弱的果蠅擁有顯著 較長的生存率。這些結果有助於進一步了解蛋白質上酪氨酸的亞硫酸化在神經系統中所 扮演的角色以及可能所參與的生理調控機制。 i
Functions of Protein Sulfation in Drosophila melanogaster by Neuron-specific Knockdown of Tyrosylprotein Sulfotransferase
Student: Jennifer Yun-shin Wu Advisor: Prof. Yuh-Shyong Yang Institute of Biochemical Engineering,
National Chiao Tung University
ABSTRACT
Protein tyrosine sulfation, catalyzed by tyrosylprotein sulfotransferase (TPST), is one of the most common post-translational modifications. TPST transfers the moiety of sulfuryl group from adenosine 3’-phosphate 5’-phosphosulfate (PAPS), to the hydroxyl group of specific tyrosine residues of various secreted and membrane-bound proteins overspread the eukaryotes. Tyrosine-sulfated proteins are known to mediate many physiological processes including coagulation, leukocyte adhesion, chemokine signaling, and HIV infection. At present, the sulfoproteomics and TPST-regulated biological functions remain largely unknown. A single TPST gene whose mRNA was highly expressed in the head and brain of
Drosophila melanogaster was identified following the analyses of genomic sequences and
gene microarray. TPST gene was knockdown specifically in the nervous system of D.
melanogaster using GAL4-UAS system. 2-D electrophoresis and digital image analysis
indicated that 4 proteins were up-regulated and 18 proteins were down-regulated. Among them, 9 proteins were identified by MS spectrum and most of them were involved in the metabolism and oxidative stress. Superoxide dismutase [Cu-Zn], an antioxidant enzyme against reactive oxygen species, was mainly down-regulated in the neuron-specific TPST knockdown flies. Oxidative stress assay was examined with paraquat treatment, and surprisingly the result showed the knockdown flies had remarkably longer survival time (1.32-fold). These results are important to comprehend the biological roles and regulatory actions of protein tyrosine sulfation in the nervous system.
ii
Acknowledgment 碩士班的兩年真的是快得不可思議!回想這兩年間,有哭有笑,不知道接受過多少 人的幫助,真的要認真的打這個致謝說不定會比論文內文還厚!能拿到碩士的畢業證 書,真的是多虧了你們在不論在精神上或是實質上的支持。首先當然是我的指導教授楊 裕雄教授,從大學專題生就進入您的實驗室到現在已經四年了,感謝您為我提供這麼一 個良好的學習環境以及教導我許多身為一位研究生該具有的觀念。再來是 LEPE 的所有 成員:小米學姊,一個溫柔細心的正妹學姊,謝謝妳從大學以來就一直不斷的給我鼓勵。 普普學長,雖然你講話的 tempo 很快,但是我每次都有很努力要跟上你喔!政哲學長, 程允學長,淵仁學長,小志學長,雖然我們不同組沒有什麼聊天的機會,但是每次看到 你們就會讓覺得很安心,因為國科會計畫又有著落了。Sonia 和秀華,謝謝妳們不論在 行政上還是實驗上對我的幫助。康寧,謝謝你這幾次聚餐都充當我的司機,騎車的技術 不錯啦!文燦,每次去二樓打電話就會把你趕走,多謝包容囉! 然後是我們 TPST 組的成 員,陸宜學長,我們的頭兒,對你有太多太多的感謝無法用言語來表達,我很納悶究竟 有什麼事你不會的!小胖,全實驗室我好像就對你最兇,那就謝謝你容忍我的任性吧, 麥當勞真的不要吃太多啦!晨竹,我好像一直到去上海後才發現你是個好笑的人,以後 不要再隱藏自己了,放開你的心跟大家相處吧!接著是我的同袍們,音汝,跟妳一起共 患難,原本以為也會一起延畢的說,好險我們現在都一起畢業啦!欣怡,妳是我第一個 發現控制慾比我還強的人,這不是壞事喔,表示你以後一定是當主管的料啦!資翔,你 也算是個熱心的人,祝你結晶趕快長晶出來囉! 換了一個段落是因為我要感謝另外一批人,也就是小楊家的所有成員。淑萍,一直 覺得妳不碰四隻腳的動物真的特別,還有妳桌面的整潔度也讓人”驚豔”!旻秀,你這個三 八的學長,最喜歡聽你唱周杰倫的歌唱到破音。敏書,好像每次看到你都覺得你在賊賊 的笑,不過你每次自表真的都很好笑!淑貞,看到妳就想到妳的 WB 壓得很乾淨很漂亮, 就像妳給人的感覺一樣舒服。阿澎,聽說你大學的時候也叫公主是吧,以後不要仗著你 那俊俏的皮囊去欺騙良家婦女喔!佳真,從沒看過長個這麼老師臉的人,果然老師真的 是妳命中的職業。妍寧,妳認真的程度真是讓我羞愧阿,真的要好好向妳學習一番才是。 iii
Sunny,當了短暫兩個月的室友,發現妳真的是大好人一枚,謝謝每次都好心的帶飯回 宿舍給我吃,還有,妳那個怎麼裝都不會重的包包到底在哪裡買的?黃阿大,你這個大 傻妞,謝謝妳每次看到我都很興奮的對我大叫阿姨阿姨~~我都不知道自己什麼時候變得 這麼受歡迎了哩!個性好是好事,但是不要好到吃了虧被人欺負都默默吞下耶,學學人 家 Sunny 姨,當好人也是有底線的! 又分了一個段落是要特別特別感謝我兩年的室友,跟妳們從大學到現在認識了六年 耶!我的青春都獻給妳們了,看妳們要怎麼對我負責!小倩,首先要跟妳道歉,因為我好 像一直用高壓在欺負妳,可是這真的是一種很詭譎的生理反應喔!我也控制不了我自 己,所以只能繼續為未來的我先跟妳道歉啦!妳很專業,好像每次有什麼疑難雜症都可 以被妳解釋一番,但是請不要再把妳的推論說的跟真的一樣了。妳很單純,可惜那些單 純的舉動往往會引起共憤,但是或許大家內心其實跟妳是一樣的。妳很愛分享,不過好 像妳每次分享的東西我都不是很感興趣齁~哈哈!妳很瘦,所以不要每次都動不動就先開 衣服來刺激我們這些胖子了!最後,妳真的是個長得很古典的童顏正妹~開心了吧!阿毛, 從大一剛開始討厭妳到現在跟妳成為好朋友,真的是一整個很奇妙,而且其實漸漸的發 現其實我們倆的個性還真是越還越像,而且似乎都是像在不好的地方,例如欺負小倩等 等~哈哈!當了我兩年的司機真是讓妳辛苦了,感謝妳帶我進入代買板的世界中,認識妳 後好像讓我越花越多錢的感覺,而且我們兩個好像一湊在一起會有加乘的效果,真的很 恐怖!妳是最矮,但是也是我靠起來最舒服的一個。妳是最寬的,這應該也是靠起來會 這麼穩的原因。妳是最開心的,不管事情好不好笑妳都會先笑再說,這的確安慰了不少 人。妳是最棒的,因為能認識妳真的幸運,當然小倩也是一樣囉!既然說到這裡,那順 便感謝一下陰魂不散的楊 Stan 吧~謝謝你的白目讓我的生活多了這麼多的樂趣! 最後是我最愛的家人,辛苦工作的爸爸,謝謝你總是在我的人生中提供我正確的道 路。媽媽,謝謝妳無微不至的照顧以及容忍我的任性。姊姊,謝謝妳每次不管什麼情況 下都會第一時間打電話給我給予我鼓勵與關心。姊夫,謝謝你不斷的供給我食物讓我怎 麼想瘦都瘦不下來,我們一起繼續沉淪吧!蔡葳寶,妳喔~只要乖乖長大乖乖睡覺我就會 非常感謝妳了!致謝到這裡就告一段落了,希望沒有遺漏任何人阿~謝謝大家! iv
Flow chart
v
Contents Chinese abstract ... i English abstract ... ii Acknowledgment ... iii Flow chart ... v Contents ... vi
Contents of Tables ... viii
Contents of Figures ... ix Contents of Appendices ... x Abbreviations ... xi Chapter 1 Introduction ... 1 1.1 Post-translational modifications ... 1 1.2 Sulfotransferase ... 1 1.3 Tyrosylprotein sulfotransferase ... 2
1.4 Biological roles of tyrosine sulfation ... 3
1.4.1 Chemokine receptor ... 3
1.4.2 Leukocyte adhesion and inflammatory response ... 4
1.4.3 Hemostasis and anticoagulation ... 4
1.4.4 TPST knockout animal model ... 5
1.5 Bottlenecks of protein sulfation researches ... 6
1.6 Tyrosylprotein sulfotransferase in Drosophila melanogaster ... 7
1.7 Tyrosylprotein sulfotransferase in nervous system ... 7
Chapter 2 Materials and methods ... 9
2.1 Materials ... 9
2.1.1 Fly Strains ... 9
2.1.2 Oligonucleotide primers ... 9
vi
2.1.3 Proteomics ... 9
2.2 Methods ... 10
2.2.1 RT-PCR ... 10
2.2.2 Sample Preparation ... 10
2.2.3 Protein Quantitation ... 11
2.2.4 Two Dimensional Electrophoresis ... 11
2.2.5 Protein Staining and analysis of 2-DE gels ... 12
2.2.6 In-gel Digestion of Protein Spots in 2-DE gels ... 13
2.2.7 Protein Identification by MALI-TOF Mass Spectrometry ... 13
2.2.8 Oxidation Stress Assay of Flies from APPL-GAL4>UAS-TPSTRNAi, APPL-GAL4/+, and UAS-TPSTRNAi /+ ... 13
Chapter 3 Results ... 15
3.1 TPST mRNA distribution of wild type D. melanogaster ... 15
3.2 TPST mRNA expression in neuron-specific knockdown D. melanogaster ... 15
3.3 Proteome analysis of neuron-specific TPST knockdown D. melanogaster ... 16
3.4 Identification of proteins changes in neuron-specific TPST knockdown of D. melanogaster ... 16
3.5 Oxidative stress assay of neuron-specific TPST knockdown in D. melanogaster ... 17
3.6 mRNA expression of superoxide dismutase [Cu-Zn] in neuron-specific knockdown D. melanogaster ... 17 Chapter 4 Discussions ... 18 Chapter 5 Conclusions ... 23 References ... 24 Tables ... 28 Figures ... 32 Appendices ... 41 vii
Contents of Tables
Table 1. The tyrosylprotein sulfotransferase gene expression analysis of adult D.
melanogaster ... 28
Table 2. Criteria of selecting spots in 2-D gel for protein analysis ... 29 Table 3. Protein changes in neuron-specific TPST knockdown Drosophila melanogaster ... 30 Table 4. The superoxide dismutase [Cu-Zn] gene expression analysis of adult D.
melanogaster ... 31
viii
Contents of Figures
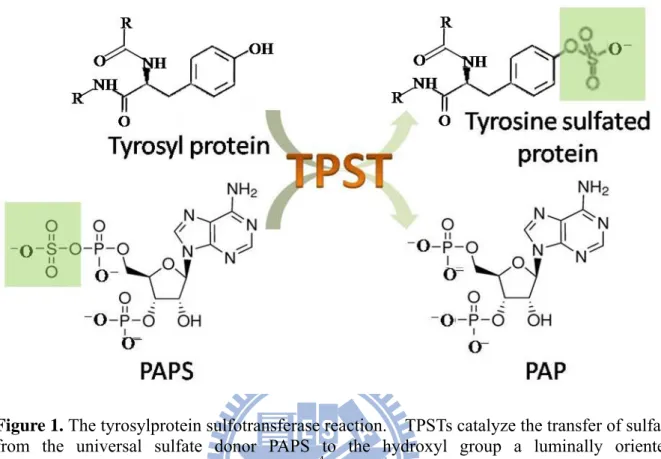
Figure 1. The tyrosylprotein sulfotransferase reaction ... 32
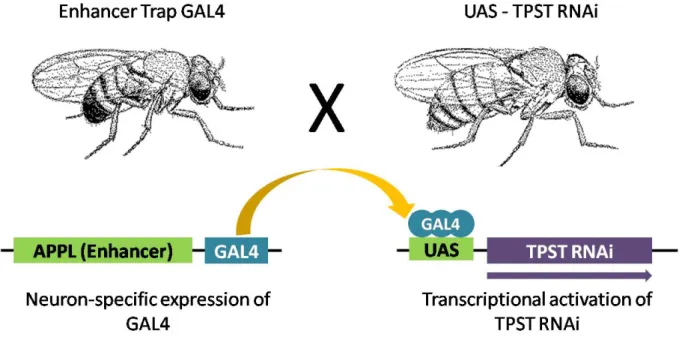
Figure 2. Directed gene expression in D. melanogaster... 33
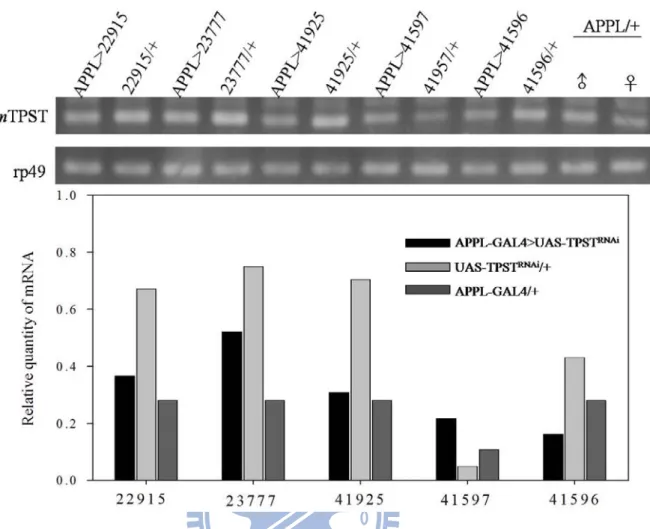
Figure 3. TPST mRNA expression of five neuron-specific TPST knockdown lines detected by RT-PCR ... 34
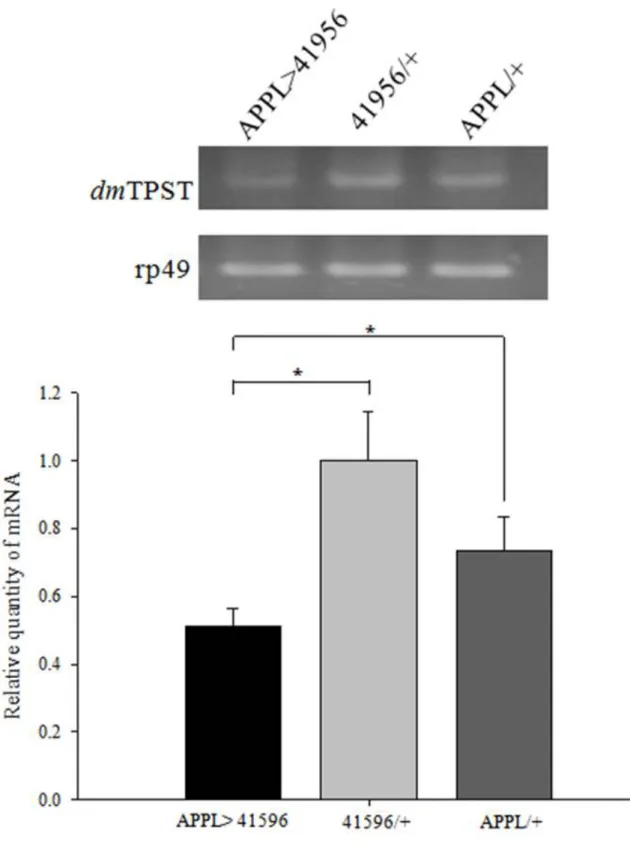
Figure 4. RNAi knockdown of TPST in the fly detected by RT-PCR ... 35
Figure 5. 2-D electrophoresis of neuron-specific TPST knockdown fly and two controls ... 36
Figure 6. Oxidative stress assay of neuron-specific TPST knockdown flies ... 37
Figure 7. The mRNA of superoxide dismutase [Cu-Zn] ... 38
Figure 8. Sequence alignment of human TPST1 (hTPST1), human TPST2 (hTPST2), and D. melanogaster TPST (dmTPST) ... 39
Figure 9. Sequence alignment of D. melanogaster TPST ... 40
ix
Contents of Appendices
Appendix 1. Some common and important post-translation modifications ... 41 Appendix 2. General ST-catalyzed reaction with PAPS as the cosubstrate ... 42 Appendix 3. Sulfate activation and tyrosine O-sulfation ... 43 Appendix 4. Comparison of the expression level of TPST-1 and TPST-2 in 20 human
tissues ... 44 Appendix 5. Schematic representation of cell entry by HIV-1 following sulfonation of CCR5
by a tyrosylprotein sulfotransferase ... 45 Appendix 6. Tyrosine sulfation plays an important role in the immune response ... 46 Appendix 7. MS analysis of (a) GE16567; (b) CG4105; (c) CG31779; (d) GM05777p; (e)
CG9062; (f) GA20134; (g) GA10647; (h) CG6058; (i) CG11793 ... 47
x
xi
Abbreviations
APPL Amyloid protein precursor-like
CCK Cholecystokinin
CCR5 Chemokine (C-C motif) receptor 5 CD4 Cluster of differentiation 4
D. melanogaster Drosophila melanogaster
gp120 Glycoprotein 120 HIV Human immunodeficiency virus
MALDI-TOF Matrix-assisted laser desorption ionization time of flight PAGE Polyacrylamide gel electrophoresis
PAPS 3’-phosphoadenosine 5’-phosphosulfated
polyQ Polyglutamine
PSGL-1 P-selectin glycoprotein ligand-1
PTM Post-translational modification RNAi Ribonucleic acid interference
RT-PCR Reverse transcription-polymerase chain reaction SDS Sodium dodecyl sulfate
SOD1 Superoxide dismutase [Cu-Zn]
STs Sulfotransferases
TPST Tyrosylprotein sulfotransferase UAS Upstream Activation Sequence
Chapter 1 Introduction
1.1 Post-translational modifications
Post-translational modifications (PTMs) are one of the most important biological mechanisms in both prokaryote and eukaryote proteins, including acetylation, acylation, glycosylation, methylation, phosphorylation, ubiquitination, and sulfation (Appendix 1). These modifications can have both structural and regulatory functions, which modulate the properties of proteins by proteolytic cleavage or by the addition of a modifying group to amino acid, which may involve proteins’ activity state, localization, turnover, and interaction with other proteins (Mann et al., 2003).
1.2 Sulfotransferase
Sulfation reactions can be classified by the sulfoconjugation of the acceptor group; for instance, O-sulfation (ester), N-sulfation (amide), S-sulfation (thioester). O-sulfation is dominant in cellular sulfation reaction, which includes a hydroxyl group of small endogenous compounds such as catecholamines, steroids, thyroid hormones, vitamins, and larger molecules such as glycosaminoglycans, proteoglycans, galactoglycerolipids, and proteins (Strott, 2002).
Sulfate-containing bimolecules were discovered in 1876, but the mechanism of sulfation remains unknown until the active 3’-phosphoadenosine 5’-phosphosulfated (PAPS) was isolated. The PAPS-binding region is conversed at the amino acid level for all sulfotransferase (STs), and all STs use PAPS as the universal sulfate group donor (SO3), to
catalyze the sulfuryl group into a variety of amine and hydroxyl substrates (Appendix 2). STs can be basically divided into two classes: cytosolic STs or membrane-associated STs. Cytosolic STs are soluble proteins located in cytoplasm, and generally sulfonate small compounds including hormones, bioamines, as well as drugs and xenobiotics. They are
involved in detoxification, hormone regulation, and drug metabolism. Membrane-associated STs are membrane anchored proteins located in the trans-Golgi network (TGN), which implied that they are involved in the post-translational modification of larger biomolecules including carbohydrates and protein. They are mainly involved in molecular-recognition events and biochemical signaling pathways (Chapman et al., 2004).
1.3 Tyrosylprotein sulfotransferase
Post-translational tyrosine O-sulfation of protein was first discovered by Bettelheim in bovine fibrinopeptide B in 1954 (Bettelheim, 1954). However, limited information was known about tyrosylprotein sulfation until 1982, when Huttner directly identified that this PTM was mediated by tyrosylprotein sulfotransferase (TPST), an enzyme that catalyzes the transfer of a sulfuryl group from PAPS to the hydroxyl group of tyrosine residue in the protein/peptide (Figure 1) (Moore, 2003). Furthermore, Huttner proved that TPST was membrane-bound and located in the trans-Golgi network (Appendix 3) (Baeuerle and Huttner, 1987), and also characterized and purified TPST from bovine adrenal medulla (Niehrs and Huttner, 1990). It is now known that TPST is a widespread enzyme in multicellular eukaryotic organisms throughout the plant and animal kingdoms, and can be detected in most tissues and cell types from humans (Appendix 4) and rats (Mishiro et al., 2006) (Nishimura and Naito, 2007). There are two TPST isoenzymes in most species: TPST1 and TPST2, both encoding type II transmembrane domain (17-residues) and are 65-67% identical. However, no evidence has supported the distribution and relative abundance of the two TPSTs at the protein level due to the lack of detecting tools such as isoenzyme-specific antibodies and substrates.
Although not many tyrosine-sulfated proteins had been indentified, based on the amino acid sequences flanking known tyrosine O-sulfation sites, it is clear that the dominant characteristics of sulfation sites are generally between 3 and 4 acidic amino acids within ±5
residues of the sulfotyrosine. The SwissPort Group developed a software, called Sulfinator (http://ca.expasy.org/tools/sulfinator) (Monigatti et al., 2002), which predicts possible proteins that can process tyrosine sulfation and also its tyrosine sulfation site. It has been estimated that up to approximately 1% of all tyrosine residues in eukaryotic cells are predicted to undergo tyrosine sulfation, but only a few hundred proteins have been identified presently (Seibert and Sakmar, 2008).
1.4 Biological roles of tyrosine sulfation
TPSTs catalyze the sulfation of tyrosine residues within specific peptide sequences, which have also been implicated in several crucial physiological events. The localization of TPST indicated that the tyrosine sulfation occurs only on proteins that transit the trans-Golgi network; these proteins usually belong to secreted or membrane proteins (Monigatti et al., 2006). Tyrosine sulfation has been implicated in intracellular trafficking and proteolytic processing of secreted proteins, and identified as a key modulator of extracellular protein-protein interaction by recognition of the sulfate group of proteins, which includes hormonal regulation, hemostasis, inflammation and infectious diseases (Seibert and Sakmar, 2008).
1.4.1 Chemokine receptor
Chemokine are small, secreted proteins that exert many biological functions through G-protein-coupled receptors, including leukocyte trafficking, angiogenesis, angiostasis, viral infections, and host immune response to cancer (Zlotnik et al., 1999). Several chemokine receptors (CXCR3, CXCR4, CCR2b, CCR5, and CX3CR1) have undergone tyrosine sulfation. A comparison in the sequences of the known chemokine receptors shows that their N-terminal domains are highly acidic and contain one or more tyrosine residues, which suggests that many, perhaps all, of the chemokine receptors may be tyrosine sulfated. Currently, the most
popular topic on the study of tyrosine sulfation focuses on CCR5 due to its involvement of HIV-1 entry. CCR5 is post-translationally modified by sulfation of its N-terminal tyrosines, and the sulfated tyrosines contribute to the binding of MIP-1α, MIP-1β, and HIV-1 gp120/CD4 complexes (Appendix 5). The four tyrosine sulfated residues are stepwise modified; tyrosines at positions 14 or 15 are sulfated first, followed by position 10 and finally the tyrosine residue at position 3 (Sasaki et al., 2007). Mutation of the four sulfotyrosine residues to phenylalanine and chlorate inhibition inhibits HIV infection by 50-75% (Farzan et
al., 1999). This information suggests that inhibiting tyrosine sulfation of CCR5 may provide
a basis for the design of therapeutic agents aimed at blocking HIV-1 cellular entry.
1.4.2 Leukocyte adhesion and inflammatory response
P-selectin glycoprotein ligand 1 (PSGL-1) is a protein specifically expressed on the cell membrane of leukocyte. In immune defense, the leukocytes need to reach the inflammation site through passage of the blood circulation, the then roll on, adhere to, and finally transmigrate between the endothelial cells and infective site (Appendix 6a). The binding between PSGL-1 of leukocyte and P-selectin of endothelial cells is essential for leukocyte adhesion in this inflammatory response (Kehoe and Bertozzi, 2000). Furthermore, the N-terminal of PSGL-1 contains three sulfated-tyrosine residues, which is the key point for the binding (Pouyani and Seed, 1995) (Appendix 6b). Treating tyrosine-sulfated PSGL-1 with bacterial arylsulfatase reduced the binding ability to P-selectin (Wilkins et al., 1995), and the results were also supported by point mutagenesis of tyrosine (Sako et al., 1995). As a result, TPST has become a therapeutic target for autoimmune diseases caused by chronic inflammation, such as rheumatoid arthritis and multiple sclerosis (Hsu et al., 2005).
1.4.3 Hemostasis and anticoagulation
The biological function of tyrosine sulfation is also involved in hemostasis. Platelet
aggregate (thrombus) is formed when blood vessels are injured, and coagulation factor VIII and von Willebrand factor (vWF) play important roles in the regulation of hemostasis. The binding between the two can form a stable complex in circulating blood. The sulfation of tyrosine residue (Tyr1680) in Factor VIII is essential for this binding, thereby tyrosine sulfation increases the circulating half-life of factor VIII (Leyte et al., 1991). Moreover, the complete mechanism of platelet attachment is accomplished by vWF that bridges subendothelial collagen and platelet membrane protein GP Ibα. The binding between vWF and n GP Ibα is dependent upon the sulfation of three tyrosine residues (Tyr276, 278, 279) (Marchese et al., 1995).
In anticoagulation, hirudin, a potent anticoagulant protein secreted in the saliva of the leech, is sulfated at Tyr63. The tyrosine sulfation of hirudin has a 10-fold higher affinity for thrombin than unsulfated form, which prevents coagulation by inhibit thrombin (Stone and Hofsteenge, 1986).
1.4.4 TPST knockout animal model
TPST knock-out mice was generated by the Moore group in 2008. TPST 1 -/- mice appear healthy and with normal fertility, but have effects on body weight (5% lower), fecundity (smaller litters), and postnatal viability (Ouyang et al., 2002). The characteristics of these mice suggest that specific proteins are involved in the regulation of body weight and reproductive physiology, which require tyrosine sulfation for optimal function. TPST2 -/- female mice had normal reproductive performance, but males revealed a severe defect of fertility. Furthermore, the Moore group identified two proteins from mice epididymis, Rnase9 and Mfge8, which are tyrosine sulfated in wild type mice but not in TPST2 -/- mice. This result suggests that lack of tyrosine sulfation may contribute to the infertility of TPST2 -/- mice (Hoffhines et al., 2008). TPST double knock-out mice were born in normal size, but a majority dies in the early postnatal period with signs of cardiopulmonary insufficiency,
due to the failure of lungs expansion at birth resulting in acute pulmonary hypertension, and death by asphyxia. Some knockouts survive the postnatal period, but fail to thrive and may display delayed growth due to hypothyroidism (Westmuckett et al., 2007). In addition to hypothyroidism, the growth-retard (grt) mouse model has a severe thyroid hypoplasia, and shows a missense mutation of a highly conserved region of TPST2 gene. Since thyroid-stimulating hormone receptor (TSHR) is one of the substrate for TPST2 and grt leads to a loss of TPST2 activity, tyrosine sulfation of TSHR by TPST2 might be crucial for TSH signaling and resultant gland function (Sasaki et al., 2007).
1.5 Bottlenecks of protein sulfation researches
Recently, more and more evidence indicates how important protein sulfation is in eukaryotes and the possible physiological pathways that protein sulfation may be involved. However, in the last five decades of studies on this topic, many questions remain unknown about TPSTs and protein sulfation. The bottlenecks of studying TPSTs include the difficulty of characterizing TPST due to the lack of source of homogenous protein samples. It is not easy to develop a fast and accurate assay for quantitative kinetics analysis without a homogenous source of protein. Moreover, tyrosine 0-sulfate may not be stable on the tyrosine residue of TPST substrate, which makes it difficult to detect or isolate sulfated proteins and peptides. Previous researches on protein sulfation had always focus on few TPST substrates as described above, therefore the understandings of TPST’s roles are restricted by the biological regulations and pathways of those few substrates. In order to systematically analyze the roles of protein sulfation in physiological regulation, a large-scale screening tool must be developed. Proteomics is a powerful and well established technique for the studying of PTMs, especially for many successful researches on glycosylation and phosphorylation. Therefore, application of this technique seems to be the optimal choice in studying the physiological regulation and pathways of tyrosylprotein sulfation by identifying
the upstream and downstream of proteins that are related to TPST.
1.6 Tyrosylprotein sulfotransferase in Drosophila melanogaster
Most vertebrates (such as rat, cow, chicken, zebrafish, African clawed frog) and invertebrates (such as Anopheles gambiae (mosquito), and Caenorhabditis elegans) have two TPSTs. It is interesting to note that Drosophila melanogaster is the only specie that was discovered to contain a single TPST gene (Moore, 2003). Therefore, D. melanogaster is a good model to study TPST, which a complete elimination of protein sulfation modification can be reached by a simple knockout of a single gene. Many advantages of using D.
melanogaster as a study model include the short generation time and easy growth. The
completion of genomic database is helpful for protein identification and its function, any kind of transgenic fly including TPST RNAi lines can be easily purchased from commercial company, and finally mRNA of TPST is highly expressed in several tissues of D.
melanogaster (Table 1). The mRNA expression of TPST is abundant in reproductive organs,
salivary gland, and nervous system (brain, head, and thoracicoabdominal ganglion). The Moore group conducted a series of successful researches on the relationship between tyrosine sulfation and reproductive system, and also identified a number of tyrosine-sulfated proteins from mice epididymis. As for salivary gland, a peptide secreted from submandibular gland, called histatin, had confirmed the presence of sulfotyrosine residues (Cabras et al., 2007). Nevertheless, it is interesting to notice that TPST is highly expressed in the neuron, but its function of TPST in the nervous system is yet to be examined.
1.7 Tyrosylprotein sulfotransferase in nervous system
So far, cholecystokinin (CCK) is the only TPST substrate found in the nervous system, and it functions as hormonal regulators of various digestive processes and feeding behaviors. However, no neuronal function has been elucidated for CCK (Nichols et al., 1988), except it
has been suggested that CCK administration causes nausea and anxiety, and induces a satiating effect (Greenough et al., 1998). More importantly, the question remains in what the function of TPST is in the nervous system due to its highly expressed mRNA. In the present study, neuron-specific TPST knockout of transgenic D. melanogaster is used as an animal model to systematically identify the possible TPST-regulated protein based on proteomic system, and to further investigate the physiological roles of these proteins. The results may help to obtain a greater understanding about the biological roles of TPST in tyrosine sulfation post-translational modification.
Chapter 2 Experimental Procedures
2.1 Materials 2.1.1 Fly Strains
UAS-CG32632RNAi lines (transformant ID: 22915, 23777, 41596, 41597, and 41925) were purchased from Vienna Drosophila RNAi Center (VDRC), and a neuron-specific Gal4 driver line, Appl-Gal4 line, was kindly provided by Prof. Horng-Dar Wang’s lab (National Tsing Hua University, Taiwan). Appl-Gal4 was used to cross with UAS-CG32632RNAi to express double-stranded RNA interference (Figure 2), or each of them crossed with wild type fly w1118 as controls. All flies were incubated in 12 hr day/night cycle incubator at 25℃and 60-70% humidity, raised and maintained with standard fly food.
2.1.2 Oligonucleotide primers
Oligonucleotide primers were synthesized by Mission Biotech Co., Ltd. (Taiwan). The primers for CG32632 are 5’-AATGGCAGCTGCTTTATCGT-3’ (forward) and 5’-CATGCTGTCCGTGCTCG-3’ (reverse); the primers for Superoxide dismutase [Cu-Zn] (SOD1) are 5’-TTGACTTGCTCAGCTCGTGT-3’ (forward) and 5’-CACGGTTTTCTTCGAACAGG-3’ (reverse).
2.1.3 Proteomics
The 13-cm Immobiline DryStrip (pH 4-7), IPG strip buffer (pH4-7), 87% glycerol, and DryStrip cover fluid were purchased from GE Healthcare Bio-Science (NJ, USA). The protein maker kit was purchased from Fermentas (Harrinton, Canada). The RC DC protein assay kit and 40% acrylamide/bis solution 29:1 were obtained from BioRad (Richmond, CA, USA). Sequencing-grade modified trypsin was purchased from Promega (Mannheim, Germany). Other chemicals: agarose was purchased from Amresco; acetic acid was
purchased from Fluka; 37% formaldehyde, glycine, trifluoracetic acid (TFA), and urea were purchased from J. T. Baker; methanol was purchased from Mallinckrodt; isopropanol was purchased from Merck; potassium hexaxyano ferrate (III) and Zinc sulfate were purchased from Panreac; sodium carbonate was purchased from Riedel-de Haën; ammonium bicarbonate, bromophenol blue, ethylenediaminetetraacetic acid (EDTA), imidazole, iodoacetamide (IAA), silver nitrate, sodium acetate, sodium thiosulphate, tetramethylenediamine (TEMED), triton X-100, and trypsin were purchased from Sigma-Aldrich; ammonium persulfate (APS), 3-[(3-cholamidopropyl) dimethylammonio]
-1-propanesulfonate (CHAPS), dithiotreitol (DTT), sodium dodecyl sulfate (SDS), and tris were purchased from USB.
2.2 Methods 2.2.1 RT-PCR
1- to 3-days old male flies were homogenized by pestle, total RNA was extracted with REzole, and then 5 μg of the total RNA was reverse transcribed with random hexamer primer. For each cDNA preparation, a control synthesis reaction was treated with DNase on 37°C for 25 mins to ensure that there was no contaminating genomic DNA. The resulting cDNA library was subjected to PCR with primers. The products of PCR were analyzed by electrophoresis on 1% agarose gels, and then visualized using ethidium bromide (EtBr). rp49 gene was used as an internal standard among different lines.
2.2.2 Sample Preparation
Whole flies were washed 3 times with PBS, and then the total protein of the whole fly were extracted using ultrasonication with lysis buffer containing 8 M urea, 2% CHAPS, 1% v/v Triton X-100, 0.1% w/v SDS, and 5 mM DTT. The extract was boiled at 95℃ for 5 minutes following centrifugation for 10 mins at 15000¯g for several times until the
supernatant was clear.
2.2.3 Protein Quantitation
Using BSA as a standard, protein quantitation of the homogeneous from D. melanogaster was estimated by a colorimetric assay (RC DC protein assay, Bio-Rad). Reagent A’ (2.5μl DC Reagent S and 125μl DC Reagent A) was prepared for each sample. Mix 25μl of sample (25X dilution with extraction buffer) with 125μl RC Reagent I. After gently vortex, 125μl of RC Reagent II was added, and centrifuge at 15000Xg for 10minutes. The protein pellet was mixed with 127μl Reagent A’, and then after the incubation for 5 minutes until the pellet was dissolved, added 1ml of DC Reagent B into the solution. Incubate the solution at room temperature for 15 minutes, and finally absorbance can be read at 750nm with UV/VIS spectrophotometer.
2.2.4 Two Dimensional Electrophoresis
IEF: 200μg or 500μg of the total proteins were loaded depending on the detection methods (silver/SYPRO Ruby stain or negative stain). The proteins were mixed with the rehydration buffer (8 M urea, 2% w/v CHAPS, 0.5% v/v IPG buffer pH 4-7, 0.002% bromophenol blue, and 18.2 mM DTT), and then eletrofocused in DryStrip using the following protocol with 50μA per strip at 20℃: (1) rehydration for 12 hours; (2) 500V for 500VHr (step and hold); (3) 1000V for 1000VHr (step and hold); (4) 3000V for 3000VHr (gradient); (5) 5000V for 5000VHr (gradient); (6) 8000V for 8000VHr (gradient); (7) 8000V for 40000VHr (step and hold). For silver stain and SYPRO Ruby stain, the strips were shaken for 15 minutes with first equilibration buffer (6 M urea, 29.3% v/v glycerol, 2% w/v SDS, and 75 mM Tris-HCl at pH 8.8) which contain 1% DTT, and then were shaken for 15 minutes with second equilibration buffer which contain 2.5% IAA. For reverse stain, the strips were shaken for 2X15 minutes with equilibration buffer.
SDS-PAGE: the strips were transferred onto 12.5% second-dimensional SDS-PAGE (1.5mm for silver stain or 1.0mm for negative stain) with the following protocol: (1) 15mA/strip for 30minutes at 4℃; (2) 30mA/strip for 5.5 hours at 4℃.
2.2.5 Protein Staining and analysis of 2-DE gels
Silver stain: fixation (40% methanol, 10% acetic acid) for overnight; sensitization (30% methanol, 0.2% w/v sodium thiosulphate, 0.5M sodium acetate) for 30 minutes; washing (ddH2O) for 3X5 minutes; silver reaction (0.25% w/v silver nitrate) for 20 minutes; washing
(ddH2O) for 2X1 minute; developing (2.5% w/v sodium carbonate, 0.02% formaldehyde) for
3-5 minutes; stopping (0.05M EDTA) for 10 minutes; washing (ddH2O) for 3X5 minutes;
preserving (8.7% glycerol, 30% methanol) for 1 hour.
Reverse stain: fixation (40% methanol, 10% acetic acid) for overnight; neutralization (tris, glycine, SDS) for 2X30 minutes; solution I (200mM imidazole, 0.1% SDS) for 1 hour; washing (ddH2O) for 2X 1 minute; solution II (300mM zinc sulfate) for 1-2 minutes; stopping
(ddH2O).
SYPRO Ruby staining: fixation (40% methanol, 10% acetic acid) for overnight; the gel was staining with SYPRO Ruby protein gel stain (Bio-Rad) for 16-18 hours; Rinse the gel in 10% methanol and 7% acetic acid for 1 hour, which to decrease the background fluorescence; finally was the gel before imaging. In SYPRO Ruby staining gel, it is readily visualized using a UV or blue light source box.
Digital images of the gels were scanned by ImageScanner, and analyzed using ImageMaster 2D Platinum software V5.0 (GE Healthcare Bio-Science). The spots were detected and the background was subtracted (mode: average on boundary), and the gels were aligned and matched. A quantitative determination of the spots volumes was performed (mode: total spot volume normalization). Specific spots, either upregulated or downregulated, were excised for further identifying by MS analysis.
2.2.6 In-gel Digestion of Protein Spots in 2-DE gels
Specific spots, either upregulated or downregulated, were excised from the gels. The gel particles were washed with 50mM NH4HCO3/acetonitrile (1:1) for 15 minutes, and then
silver destain solution (15mM K3Fe(CN)6, 50mM Na2S2O3) was used to destain the gel
particles within 2X10 minutes. After removing the solution, we washed the gel particle in 20mM ammonium bicarbonate until it became colorless. The remaining liquid was removed, and then the gel particles were shrunken by adding just enough acetonitrile to cover the gel. Finally, the gel particles were dried down for 15-20 minutes at room temperature. (In reverse stain, the gel particles were swelled in 10mM DTT/25mM NH4HCO3 for 1 hour at
56℃, and then we replaced the solution by 55mM IAA/25mM NH4HCO3 for 30minutes at
room temperature. The gel particles were washed with 50mM NH4HCO3/acetonitrile (1:1) for
15 minutes, then shrunk and dried down following the steps described before.) The proteins in the gel particles were digested in the enzyme solution (25mM NH4HCO3 with 5ng/μl of
trypsin) at 4℃ for 1 hour, and then incubated at 37℃ overnight by adding 3μl of 25mM NH4HCO3. The digests were sonicated in a water bath for 10 minutes, and then 50%
acetonitrile with 1% TFA was added. Finally, the supernatants were collected for analysis of MALDI-TOF-MS.
2.2.7 Protein Identification by MALI-TOF Mass Spectrometry
The MS raw data were processed by searching the protein databases (Swiss-Prot, MSDB, NCBInr) using MASCOT (http://www.matrixscience.com). To denote a protein as unambiguously identified, the Mowse scoring algorithms were used. Only proteins whose score exceeded the significance threshold (P<0.05) were concerned.
2.2.8 Oxidation Stress Assay of Flies from APPL-GAL4>UAS-TPSTRNAi, APPL-GAL4/+, and UAS-TPSTRNAi /+
The young male flies about 1- to 3-days old were collected for oxidative stress test. The flies were fed with 10mM paraquat in 5% sucrose water once a day (16:30). The dead fly number was counted every 4 hours (00:30, 8:30, 12:30, 16:30, 20:30) till all flies were dead. Each tube contained about 20 flies, and at least 100 flies were included for each line. The statistical significance of the observed change in the stress test was evaluated by using Student’s t test.
Chapter 3 Results
3.1 TPST mRNA distribution of wild type D. melanogaster
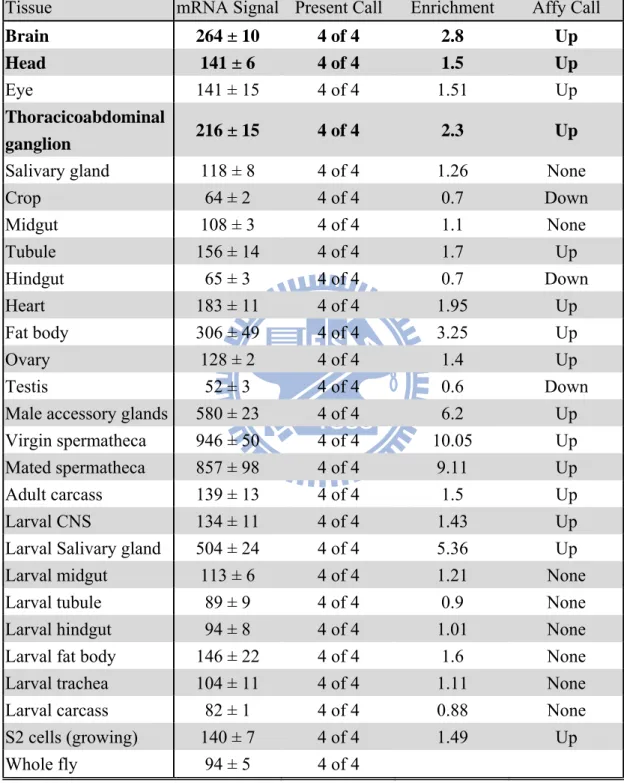
The expression of TPST mRNA of TPST varied in different tissues from the analysis of Affymatrix Dros2 expression arrays whose results deposited in Flyatlas (http://flyatlas.org/). Flyatlas showed that TPST gene was highly abundant in several tissues, such as spermatheca, male accessory glands, salivary gland, and in the nervous system, including head, brain, and thoracicoabdominal ganglion as shown in Table 1. The mRNA signal of over 100 indicated as being abundant and over 1000 as remarkable. The present call showed how many of the four arrays for each sample actually gave a detectable expression. The enrichment displayed how much higher the signal is in a particular tissue than in the whole fly, which indicated whether the gene is tissue-specific. The biological rules of the higher mRNA expression on these tissues will need to be further clarified.
3.2 TPST mRNA expression in neuron-specific knockdown D. melanogaster
To determine whether TPSTRNAi can sufficiently knockdown TPST in the nervous system of flies, five different RNAi lines were crossed with Appl-Gal4 line. By using rp49 as an internal control, total mRNA of APPL-GAL4>UAS-TPSTRNAi, UAS-TPSTRNAi alone, and APPL-GAL4 alone were extracted from male flies and proceeded with RT-PCR, respectively (Figure 3). APPL-GAL4>UAS-TPSTRNAi representing the TPST gene was knocked down in the nervous system of flies; UAS-TPSTRNAi/+ and APPL-GAL4/+ represents the control groups. The relative quantities of TPST mRNA were analyzed and showed that only one (41596) of the five transgenic flies had a statistically significant decrease in TPST mRNA content compared to Appl-Gal4 and UAS-TPSTRNAi lines alone (Figure 4). This decrease of TPST mRNA content might correspond to the relative quantity of TPST in the pan-neuron showed in Table 1. Therefore, this RNAi transgenic line (41596) was chosen for further
experiments.
3.3 Proteome analysis of neuron-specific TPST knockdown D. melanogaster
The two-dimensional electrophoreses was utilized to spread the protein spots of D.
melanogaster proteome, and 13-cm drystrip with pH 4-7 as well as 12.5% SDS-PAGE were
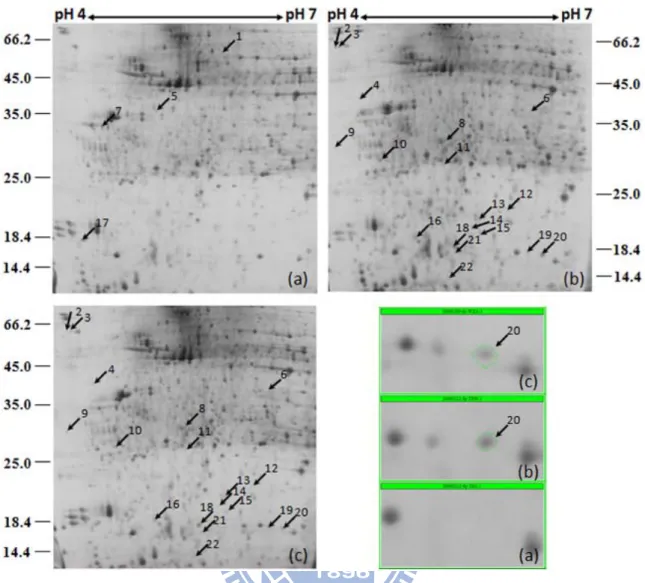
optimized for analytical condition. The gels were repeated at least three times in each line of flies, and combined all together for further spots analysis. Approximately 1200 protein spots were detected under silver stain for each gel. However, only the protein spots that up and down-expressed in APPL-GAL4>UAS-TPSTRNAi fly compared to both of the control groups were considered as shown in Table 2. The protein expression of 22 proteins spots were considered to be significantly changed, including 4 proteins were up-regulated (spot 1, 5, 7, 17) and 18 proteins were down-regulated (spot 2-4, 6, 8-16, 18-22). Arrows represented that the protein spots were up-expression (Figure 5a, b, c).
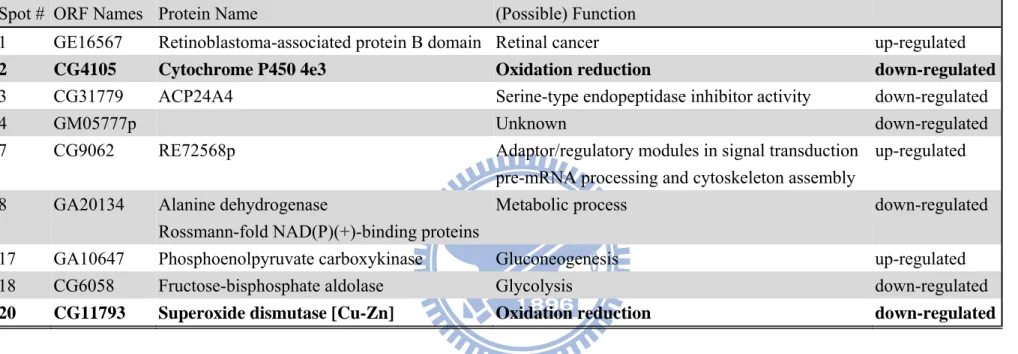
3.4 Identification of proteins changes in neuron-specific TPST knockdown of D. melanogaster
According to the analysis of two-dimensional SDS PAGE, 22 protein spots were excised from the gel and in-gel digested by trypsin after SYPRO Ruby staining, only 9 protein spots were identified by MALDI-TOF mass spectrometer as shown in Table 3. The up-regulated proteins were retinoblastoma-associated protein B (GE16567, Appendix 7a) which may involve in retinal cancer; phosphoenolpyruvate carboxykinase (GA10647, Appendix 7g) which may involve in gluconeogenesis; and CG9062 which may involve in pre-mRNA processing and cytoskeleton assembly (Appendix 7e). The down-regulated identified proteins were cytochrome P450 4e3 (CG4105, Appendix 7b) and superoxide dismutase [Cu-Zn] (CG11793, Appendix 7i) that were responsible for oxidative stress; fructose-bisphosphate aldolase (CG6058, Appendix 7h) that is involved in glycolysis; alanine
dehydrogenase (GA20134, Appendix 7f) that is involved in metabolic process; CG31779, which has serine-type endopeptidase inhibitor activity (Appendix 7c); and GM05777p with an unknown function (Appendix 7d). From these identified proteins, the TPST somehow regulated the stress tolerance by down-regulated cytochrome P450 4e3 and superoxide dismutase [Cu-Zn].
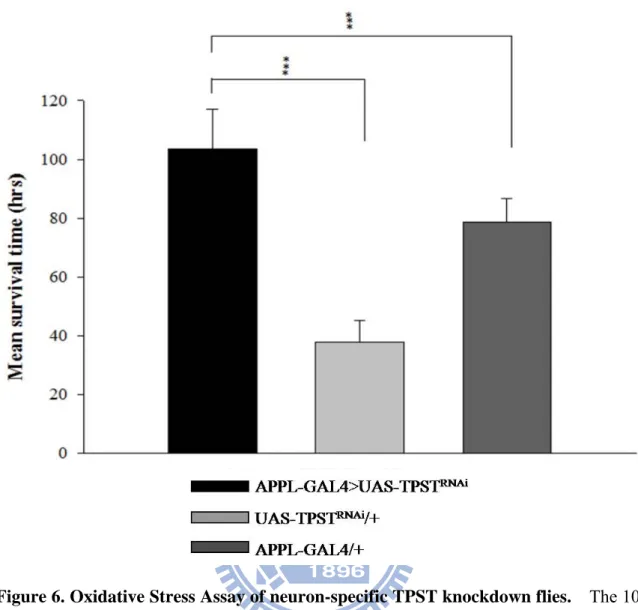
3.5 Oxidative stress assay of neuron-specific TPST knockdown in D. melanogaster
In order to identify the possible physiological pathway of TPST in the nervous system of
D. melanogaster, stress tolerance was chosen since proteins had been identified to involve in
oxidative stress. The oxidative stress induced by 10mM paraquat revealed that the neuron-specific TPST knockdowned flies whose mean survival time (103.6 ± 13.3 hrs) were significant long compared to the UAS-TPSTRNAi alone (37.8 ± 7.3 hrs) and APPL-GAL4 alone (78.6 ± 8.3 hrs) in Figure 6. The neuron-specific TPST knockdowned flies displayed an increasing in survival time of 2.74-fold compared to UAS-TPSTRNAi flies and of 1.32-fold compared to APPL-GAL4 flies.
3.6 mRNA expression of superoxide dismutase [Cu-Zn] in neuron-specific knockdown D. melanogaster
Although the protein spots of superoxide dismutase [Cu-Zu] (SOD1) disappeared on the two-dimensional electrophoresis gel, however Figure 7 shown that the mRNA expression of SOD1 in neuron-specific TPST knockdowned flies remained to have no difference compared to the two controls. Therefore, the down-regulation of SOD1 did not result from the mRNA expression in gene level. Moreover, Flyatlas performed that SOD1 was highly abundant in a majority of D. melanogaster tissues as shown in Table 4, which also indicates that the SOD1 was expressed ubiquitously in D. melanogaster.
Chapter 4 Discussions
Tyrosine sulfation was discovered in 1950s by the first sulfated protein, bovine fibrinogen (Bettelheim, 1954), and then tyrosylprotein sulfotransferase was denoted as the enzyme that catalyzed this reaction in 1983 (Moore, 2003). Protein sulfation has been largely researched over the past 50 years, however, a number of bottlenecks serve as challenges in previous studies, such as the difficulty of sourcing the homogeneous enzyme, limited information of enzyme characteristics (kinetics), unstable sulfated groups on the substrate, and lack of sensitive detecting methods for the sulfate group. Tyrosine-sulfated proteins have been indentified in physiological processes, including coagulation, leukocyte adhesion, chemokine signaling, and HIV entry (Seibert and Sakmar, 2008). At present, the understandings of TPST function are confined on the specific substrate involved in biological functions as described above. In order to systematically analyze the roles of protein sulfation in physiological regulation, we designed a proteome-wide screening tool that basically relied on proteomic techniques. Drosophila melanogaster was chosen as the source of study animal, due to it can grow easily, short generation span, well-established genomic database, commercial transgenic lines, and more importantly, D. melanogaster only has a single TPST gene (Moore, 2003). The amino acid sequence of TPST in D.
melanogaster shares 58% and 56% with human TPST1, and TPST2, respectively (Figure 8).
Approximately 75% of known human disease genes have a recognizable match in the genetic code of D. melanogaster, and 50% of D. melanogaster protein sequences have mammalian analogues (Reiter et al., 2001) which makes D. melanogaster an appropriate animal model for pathological studies on TPST.
To study the biological function of protein sulfation, knockdown endogenous TPST by RNAi technique in D. melanogaster is the most direct way. At present, TPST knockout mice was developed by Moore group, and the dysfunction of TPST has been indicated to
cause a severe defection of sperm motility which reveals that protein sulfation may be involved in the reproductive system (Hoffhines et al., 2008). Moreover, there is no information available about the relationship between protein sulfation and neurons, we are interested in focusing on neuron-specific knockdown due to the highly mRNA expression of TPST in the D. melanogaster nervous system (Table 1). The neuron-specific driver, APPL-GAL4, was used to cross with five different UAS-TPSTRNAi transgenic flies as our animal sources; only one had accomplished the neuron-specific knockdown in statistical significance compared to the controls, APPL-GAL4 alone and UAS-TPSTRNAi alone (Figure 3, 4). The mRNA was extracted from whole flies, therefore, the result of RT-PCR does not directly indicate the knockdown in the nervous system, and instead, represents the knockdown of TPST in the whole fly. Theoretically, the two controls, APPL-GAL4 alone and UAS-TPSTRNAi alone, should have similar quantity of TPST mRNA expression. Figure 3 shows the two controls have different relative quantity of TPST, which may contribute to the distinct genetic background by different maintenance and growing conditions between the two controls.
Total proteins were extracted from 1-7 day-old male flies with ultrasonication, and 200μg protein were loading onto each drystrip. pH4-7 Drystrips were chosen because the protein spots mostly tended to be distribute in the acidic region. Approximately 1200 proteins spots were visualized for each SDS-PAGE after silver staining, and different patterns of protein spots varied by different extracting methods. It has been estimated that only about 8% of the protein encoded by the genome could be analyzed in a two-dimensional SDS-PAGE of a total protein extract of D. melanogaster (Ericsson, 1999). Twenty-two protein spots showed a significant difference when compared with the controls, with 4 proteins up-regulated and 18 down-regulated. Among those protein spots, a number of spots seemed to have a pI shifting on the gel. Only 9 proteins were identified from MALDI-TOF mass spectrometry after preceding an in-gel trypsin digestion with trypsin. The protein loss
during the process of in-gel digestion could be a reason to decrease the protein identification of mass spectrometry. More importantly, however, the visualizing method, silver staining in this study, was the major cause for the low efficiency of protein identification by mass. Although silver staining methods have raised the detection limit to the nanogram range, the protein identification of excised spots was often an obstacle that cannot be overcome easily (Poland et al., 2005). In order to solve the problem, the silver staining gels are usually treated as an analytical gel; other staining methods are then used as preparative gel for mass identification, such as coomassie staining, reverse staining, and SYPRO Ruby staining (Sasse and Gallagher, 2004). We chose SYPRO Ruby and reverse staining for the preparative gel, and the mass analysis results were shown in Table 3 and Appendix 7a-i.
The neuron-specific knockdown flies down-regulated two oxidative stress proteins: cytochrome P450 4e3 (Cyp4e3) and superoxide dismutase [Cu-Zn] (SOD1). Superoxide dismutases is an ubiquitous enzymes that functions to efficiently catalyze the dimutation of superoxide anions (Zelko et al., 2002), which is known to protect organisms from reactive oxygen metabolites (Goulielmos et al., 2003). SOD1 is widely distributed and comprised 90% of the total SOD (Noor et al., 2002), the mRNA expression of D. melanogaster is shown in Table 4. It is obvious to notice, however, that the protein spot of SOD1 (spot 20) in the gel was completely disappeared in TPST knockdown flies (Figure 5). Interestingly, TPST is only knocked down in nervous system, but it caused SOD1, expressed ubiquitously, to completely vanish in silver staining vision. Besides the actual down-regulation of the protein, the disappearance of protein spot could be contributed to the change of either isoelectric focusing point or the molecular weight of the protein, which caused a spot shift. Further confirmation is needed for the clarification.
Based on the down-regulated Cyp4e3 and SOD1, implications can be made that the neuron-specific knockdown flies tended to suffer stress more easily, especially oxidative stress. In the oxidative stress assay, we used paraquat to increase the quantity of free radical
in D. melanogaster. An unexpected result revealed that the mean survival time of the TPST knockdown flies was much longer than UAS-TPSTRNAi alone for 66 hours and APPL-GAL4 alone for 25 hours, (Figure 6). The longevity of the APPL-GAL4 alone might need to be clarified in advance. The incredible longevity of neuron-specific TPST knockdown flies seems to be an opposite result as we expected. Oxidative stress has been reported to be a common underlying mechanism in the pathogenesis of many neurodegenerative disorders such as Alzheimer, Huntington, and Parkinson disease (Gruenewald C et al., 2009). Previous researches on SOD1 and neuron have indicated that overexpression of SOD1 in D.
melanogaster can reduce oxidative damage (Landis and Tower, 2005), extend lifespan (Parkes et al., 1998) (Sampayo et al., 2003), and neuron protection (Botella et al., 2008). The
overall evidence reveals that down-regulated SOD1 should decrease the survival rate of TPST knockdown flies in oxidative stress assay. Nevertheless, our result is in conflict with previous findings. The disappearance of the SOD1 (spot 20) on the two-dimensional SDS-PAGE did not result from the mRNA depletion on the gene level, which was proved by RT-PCR (Figure 7).
Moreover, there is only a single TPST gene in D. melanogaster by the analysis from BLAST. The TPST gene, however, might express two isoforms, Tango-PB and Tango-PC, with different length of amino acids. The difference between these two isoforms is that Tango-PB possessed extended C-terminal 150 amino acid residues with polyglutamine (polyQ) and polyasparagine (polyN) (Figure 9). A number of neurodegenerative diseases are characterized by the formation of intracellular protein aggregates and neurodegeneration. The polyQ sequence can easily cause protein misfolding and the formation of inclusion body (Nagai and Popiel, 2008). In the neurons, polyQ protein inclusion are aggregated which probably induce neurotoxicity (Li et al., 2008). Therefore, the neurons are protected by knockdowning TPST in nervous system, which decreases the inclusion proteins conducted from the aggregation of Tango-PB. Finally, it increased the survival rate of D. melanogaster.
Further confirmation is certainly needed for this inference.
Chapter 5 Conclusion
Protein sulfation had been extensively researched for more than 50 years, but there remains a lot of questions and basic knowledge that still needs to be investigated. The present study serves as the first to examine the tissue-specific sulfoproteomics by using
Drosophila melanogaster as the animal model. TPST in nervous system is specifically
knockdown due to the highly TPST mRNA expression in the head and brain of the D.
melanogaster. By the manipulation of RNA interference technique and GAL4-UAS system,
the TPST in the nervous system was successfully knockdowned with statistical significance. Proteomic analysis following protein identification from mass spectrometry revealed that protein tyrosine sulfation might be involved in metabolism and oxidative stress. Moreover, the oxidative stress assay showed the surprisingly result that the neuron-specific TPST knockdown flies had remarkably longer survival rates. These results are important to comprehend the biological roles and regulatory actions of protein tyrosine sulfation in the nervous system.
References
Baeuerle PA, and Huttner WB. (1987) Tyrosine sulfation is a trans-Golgi-specific protein modification. J. Cell. Biol. 105:2655-64.
Bettelheim FR. (1954) Tyrosine-O-sulfate in a peptide from fibrinogen. J. Am. Chem. Soc. 76:2838-39.
Botella JA, Bayersdorfer F, and Schneuwly S. (2008) Superoxide dismutase overexpression protects dopaminergic neurons in a Drosophila model of Parkinson's disease. Neurobiol. Dis. 30(1):65-73.
Cabras T, Fanali C, Monteiro JA, Amado F, Inzitari R, Desiderio C, Scarano E, Giardina B, Castagnola M, and Messana I. (2007) Tyrosine polysulfation of human salivary histatin 1. A post-translational modification specific of the submandibular gland. J. Proteome Res. 6(7):2472-80.
Chapman E, Best MD, Hanson SR, and Wong CH. (2004) Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed. Engl. 43(27):3526-48.
Ericsson, C. (1999) 2-D protein extracts from Drosophila melanogaster. Methods Mol. Biol. 112:35-41.
Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, and Choe H. (1999) Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96(5):667-76.
Goulielmos GN, Arhontaki K, Eliopoulos E, Tserpistali K, Tsakas S, and Loukas M. (2003) Drosophila Cu,Zn superoxide dismutase gene confers resistance to paraquat in
Escherichia coli. Biochem. Biophys. Res. Commun. 308(3):433-8.
Greenough A, Cole G, Lewis J, Lockton A, and Blundell J. (1998) Untangling the effects of hunger, anxiety, and nausea on energy intake during intravenous cholecystokinin octapeptide (CCK-8) infusion. Physiol. Behav. 65(2):303-10.
Gruenewald C, Botella JA, Bayersdorfer F, Navarro JA, and Schneuwly S. (2009) Hyperoxia-induced neurodegeneration as a tool to identify neuroprotective genes in Drosophila melanogaster. Free Radic. Biol. Med. 46(12):1668-76.
Hoffhines AJ, Jen CH, Leary JA, and Moore KL. (2009) Tyrosylprotein sulfotransferase-2 expression is required for sulfation of RNase 9 and Mfge8 in vivo. J. Biol. Chem.
284(5):3096-105.
Hsu W, Rosenquist GL, Ansari AA, and Gershwin ME. (2005) Autoimmunity and tyrosine sulfation. Autoimmun. Rev. 4(7):429-35.
Kehoe JW, and Bertozzi CR. (2000) Tyrosine sulfation: a modulator of extracellular protein-protein interactions. Chem. Biol. 7(3):R57-61.
Landis GN, and Tower J. (2005) Superoxide dismutase evolution and life span regulation. Mech. Ageing. Dev. 126(3):365-79.
Leyte A, van Schijndel HB, Niehrs C, Huttner WB, Verbeet MP, Mertens K, and van Mourik JA. (1991) Sulfation of Tyr1680 of human blood coagulation factor VIII is essential for the interaction of factor VIII with von Willebrand factor. J. Biol. Chem. 266(2):740-6. Li X, Li H, and Li XJ. (2008) Intracellular degradation of misfolded proteins in
polyglutamine neurodegenerative diseases. Brain Res. Rev. 59(1):245-52.
Mann M, and Jensen ON. (2003) Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21(3):255-61.
Marchese P, Murata M, Mazzucato M, Pradella P, De Marco L, Ware J, and Ruggeri ZM. (1995) Identification of three tyrosine residues of glycoprotein Ib alpha with distinct roles in von Willebrand factor and alpha-thrombin binding. J. Biol. Chem. 270(16):9571-8. Mishiro E, Sakakibara Y, Liu MC, and Suiko M. (2006) Differential enzymatic characteristics
and tissue-specific expression of human TPST-1 and TPST-2. J. Biochem. 140(5):731-7. Monigatti F, Gasteiger E, Bairoch A, and Jung E. (2002) The Sulfinator: predicting tyrosine
sulfation sites in protein sequences. Bioinformatics. 18(5):769-70.
Monigatti F, Hekking B, and Steen H. (2006) Protein sulfation analysis--A primer. Biochim. Biophys. Acta. 1764(12):1904-13.
Moore KL. (2003) The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 278(27):24243-6.
Nagai Y and Popiel HA. (2008) Conformational changes and aggregation of expanded polyglutamine proteins as therapeutic targets of the polyglutamine diseases: exposed beta-sheet hypothesis. Curr. Pharm. Des. 14(30):3267-79.
Nichols R, Schneuwly SA, and Dixon JE. (1988) Identification and characterization of a Drosophila homologue to the vertebrate neuropeptide cholecystokinin. J. Biol. Chem.
263(25):12167-70.
Niehrs C, and Huttner WB. (1990) Purification and characterization of tyrosylprotein sulfotransferase. EMBO J. 9(1):35-42.
Nishimura M, and Naito S. (2007) Tissue-specific mRNA expression profiles of human carbohydrate sulfotransferase and tyrosylprotein sulfotransferase. Biol. Pharm. Bull. 30(4):821-5.
Noor R, Mittal S, and Iqbal J. (2002) Superoxide dismutase--applications and relevance to human diseases. Med. Sci. Monit. 8(9):RA210-5.
Ouyang YB, Crawley JT, Aston CE, and Moore KL. (2002) Reduced body weight and
increased postimplantation fetal death in tyrosylprotein sulfotransferase-1-deficient mice. J. Biol. Chem. 277(26):23781-7.
Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, and Boulianne GL. (1998) Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat. Genet. 19(2):171-4.
Poland J, RabilloudT, and Sinha P. (2005) The Proteomics Protocols Handbook. Humana Press. 177-184.
Pouyani T, and Seed B. (1995) PSGL-1 recognition of P-selectin is controlled by a tyrosine sulfation consensus at the PSGL-1 amino terminus. Cell 83(2):333-43.
Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. (2001) A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res.
11(6):1114-25.
Sako D, Comess KM, Barone KM, Camphausen RT, Cumming DA, and Shaw GD. (1995) A sulfated peptide segment at the amino terminus of PSGL-1 is critical for P-selectin
binding. Cell 83(2):323-31.
Sampayo JN, Gill MS, and Lithgow GJ. (2003) Oxidative stress and aging--the use of superoxide dismutase/catalase mimetics to extend lifespan. Biochem. Soc. Trans.
31:1305-7.
Sasaki N, Hosoda Y, Nagata A, Ding M, Cheng JM, Miyamoto T, Okano S, Asano A, Miyoshi I, and Agui T. (2007) A mutation in Tpst2 encoding tyrosylprotein
sulfotransferase causes dwarfism associated with hypothyroidism. Mol. Endocrinol. 21(7):1713-21.
Sasse J and Gallagher SR. (2004) Staining proteins in gels. Curr. Protoc. Immunol. Chapter 8: Unit 8.9.
Seibert C, Cadene M, Sanfiz A, Chait BT, and Sakmar TP. (2002) Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence. Proc. Natl. Acad. Sci. USA 99(17):11031-6.
Seibert C, and Sakmar TP. (2008) Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers. 90(3):459-77. Stone SR, and Hofsteenge J. (1986) Kinetics of the inhibition of thrombin by hirudin.
Biochemistry 25(16):4622-8.
Strott CA. (2002) Sulfonation and molecular action. Endocr. Rev. 23(5):703-32.
Westmuckett AD, Hoffhines AJ, Borghei A, and Moore KL. (2008) Early postnatal pulmonary failure and primary hypothyroidism in mice with combined TPST-1 and TPST-2
deficiency. Gen. Comp. Endocrinol. 156(1):145-53.
Wilkins PP, Moore KL, McEver RP, and Cummings RD. (1995) Tyrosine sulfation of
P-selectin glycoprotein ligand-1 is required for high affinity binding to P-selectin. J. Biol. Chem. 270(39):22677-80.
Zelko IN, Mariani TJ, and Folz RJ. (2002) Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33(3):337-49.
Zlotnik A, Morales J, and Hedrick JA. (1999) Recent advances in chemokines and chemokine receptors. Crit. Rev. Immunol. 19(1):1-47.
Tables
Table 1. The tyrosylprotein sulfotransferase gene expression analysis of adult D. melanogaster (FlyAtlas).
Tissue mRNA Signal Present Call Enrichment Affy Call
Brain 264 ± 10 4 of 4 2.8 Up
Head 141 ± 6 4 of 4 1.5 Up
Eye 141 ± 15 4 of 4 1.51 Up
Thoracicoabdominal
ganglion 216 ± 15 4 of 4 2.3 Up
Salivary gland 118 ± 8 4 of 4 1.26 None
Crop 64 ± 2 4 of 4 0.7 Down Midgut 108 ± 3 4 of 4 1.1 None Tubule 156 ± 14 4 of 4 1.7 Up Hindgut 65 ± 3 4 of 4 0.7 Down Heart 183 ± 11 4 of 4 1.95 Up Fat body 306 ± 49 4 of 4 3.25 Up Ovary 128 ± 2 4 of 4 1.4 Up Testis 52 ± 3 4 of 4 0.6 Down
Male accessory glands 580 ± 23 4 of 4 6.2 Up Virgin spermatheca 946 ± 50 4 of 4 10.05 Up Mated spermatheca 857 ± 98 4 of 4 9.11 Up
Adult carcass 139 ± 13 4 of 4 1.5 Up
Larval CNS 134 ± 11 4 of 4 1.43 Up
Larval Salivary gland 504 ± 24 4 of 4 5.36 Up Larval midgut 113 ± 6 4 of 4 1.21 None
Larval tubule 89 ± 9 4 of 4 0.9 None
Larval hindgut 94 ± 8 4 of 4 1.01 None Larval fat body 146 ± 22 4 of 4 1.6 None Larval trachea 104 ± 11 4 of 4 1.11 None Larval carcass 82 ± 1 4 of 4 0.88 None S2 cells (growing) 140 ± 7 4 of 4 1.49 Up
Whole fly 94 ± 5 4 of 4
Table 2. Criteria of selecting spots in 2-D gel for protein analysis. APPL-GAL4>
UAS-TPSTRNAi UAS-TPST
RNAi/+ APPL-GAL4/+ o o o o x o o o x
o
x
x
up-regulationx
o
o
down-regulationx x o
x o x x x x O indicated the up-expression of protein spotX indicated the down-expression of protein spot
indicated the up/down-regulation of protein spot that actually changing by TPST knockdown system.
30
Table 3. Protein changes in neuron-specific TPST knockdown Drosophila melanogaster.
Spot # ORF Names Protein Name (Possible) Function
1 GE16567 Retinoblastoma-associated protein B domain Retinal cancer up-regulated
2 CG4105 Cytochrome P450 4e3 Oxidation reduction down-regulated
3 CG31779 ACP24A4 Serine-type endopeptidase inhibitor activity down-regulated
4 GM05777p Unknown down-regulated
7 CG9062 RE72568p Adaptor/regulatory modules in signal transduction up-regulated
pre-mRNA processing and cytoskeleton assembly
8 GA20134 Alanine dehydrogenase Metabolic process down-regulated
Rossmann-fold NAD(P)(+)-binding proteins
17 GA10647 Phosphoenolpyruvate carboxykinase Gluconeogenesis up-regulated
18 CG6058 Fructose-bisphosphate aldolase Glycolysis down-regulated
31
Table 4. The superoxide dismutase [Cu-Zn] gene expression analysis of adult D. melanogaster (FlyAtlas).
Tissue mRNA Signal Present Call Enrichment Affy Call
Brain 1789 ± 20 4 of 4 1 None
Head 2451 ± 92 4 of 4 1.4 Up
Eye 3497 ± 77 4 of 4 1.93 Up
Thoracicoabdominal
ganglion 2042 ± 28 4 of 4 1.1 Up
Salivary gland 1642 ± 45 4 of 4 0.91 None
Crop 3783 ± 180 4 of 4 2.1 Up Midgut 3002 ± 31 4 of 4 1.7 Up Tubule 3580 ± 98 4 of 4 2 Up Hindgut 2844 ± 58 4 of 4 1.6 Up Heart 5032 ± 335 4 of 4 2.77 Up Fat body 3763 ± 432 4 of 4 2.07 Up Ovary 1413 ± 22 4 of 4 0.8 Down Testis 554 ± 10 4 of 4 0.3 Down
Male accessory glands 951 ± 45 4 of 4 0.5 Down Virgin spermatheca 2537 ± 41 4 of 4 1.4 Up Mated spermatheca 2402 ± 117 4 of 4 1.32 Up Adult carcass 4082 ± 123 4 of 4 2.2 Up
Larval CNS 1432 ± 69 4 of 4 0.79 Down
Larval Salivary gland 2308 ± 134 4 of 4 1.27 Up
Larval midgut 1816 ± 29 4 of 4 1 None
Larval tubule 2354 ± 84 4 of 4 1.3 Up Larval hindgut 1438 ± 36 4 of 4 0.79 Down Larval fat body 2970 ± 294 4 of 4 1.6 Up Larval trachea 1994 ± 52 4 of 4 1.1 Up Larval carcass 2034 ± 97 4 of 4 1.12 None S2 cells (growing) 1201 ± 10 4 of 4 0.66 Down Whole fly 1814 ± 55 4 of 4
Figures
Figure 1. The tyrosylprotein sulfotransferase reaction. TPSTs catalyze the transfer of sulfate from the universal sulfate donor PAPS to the hydroxyl group a luminally oriented peptidyltyrosine residue to form a tyrosine O4-sulfate ester and 3’, 5’-ADP.
Figure 2. Directed gene expression in D. melanogaster. To generate transgenic lines expressing GAL4 in neuron-specific patterns, the GAL4 gene is inserted randomly into the genome, driving GAL4 expression from genomic enhancer (APPL). A GAL4-dependent target gene can then be constructed by subcloning any sequence behind GAL4 binding sites. The target gene is silent in the absence of GAL4. To activate the target gene in neuron-specific pattern, flies carrying the target (UAS-TPSTRNAi) are crossed to flies expressing GAL4 (Enhancer Trap GAL4). In the progeny of this cross, it is possible to activate UAS- TPSTRNAi in cells where GAL4 is expressed and to observe the effect of this directed misexpression on development.
Figure 3. TPST mRNA expression of five neuron-specific TPST knockdown lines detected by RT-PCR. The neuron-specific TPST (black) was knockdowned compared to flies carrying UAS-TPSTRNAi alone (light gray) and APPL-GAL4 alone (dark gray).
Figure 4. RNAi knockdown of TPST in the fly detected by RT-PCR. The neuron-specific TPST knockdown by APPL-GAL4. The neuron-specific TPST (black) was knockdowned compared to flies carrying UAS-TPSTRNAi alone (light gray) and APPL-GAL4 alone (dark
gray), whose P<0.05. The p values were calculated by Student’s t test. All experiments were carried out with at least three independent replicates.
Figure 5. 2-D electrophoresis of neuron-specific TPST knockdown fly and two controls. (a)APPL-GAL4>UAS-TPSTRNAi; (b) UAS-TPSTRNAi/+; (c) APPL-GAL4/+. The arrow indicates the up-regulated protein expression. The flies ranging from 50 to 100 were homogenized by ultrasonication. The 2-D electrophoresis was performed under 13-cm Immobiline DryStrips with pH 4–7 for isoelectric focusing and then 12.5% SDS polyacrylamide gels with silver staining. Spot 20 is the protein spot of superoxide dismutase [Cu-Zn].
Figure 6. Oxidative Stress Assay of neuron-specific TPST knockdown flies. The 10 mM paraquat was treated to neuron-specific TPST knockdowned flies (black), UAS-TPSTRNAi alone (light gray), and APPL-GAL4 alone (dark gray). The knockdown flies have remarkably longer survival rate compared with the two controls, whose P<0.001. The p values were calculated by Student’s t test.
Figure 7. The mRNA of superoxide dismutase [Cu-Zn] (SOD1) remains intact in neuron-specific TPST knockdown flies as measured by RT-PCR.

![Table 4. The superoxide dismutase [Cu-Zn] gene expression analysis of adult D.](https://thumb-ap.123doks.com/thumbv2/9libinfo/8582718.189426/44.892.126.745.166.893/table-superoxide-dismutase-cu-gene-expression-analysis-adult.webp)