克雷白氏肺炎桿菌CG43中尿嘧啶雙磷酸葡萄醣去氫酶之磷酸酪胺酸殘基鑑定
全文
(2) 中文摘要 細菌尿嘧啶雙磷酸葡萄醣去氫酶(UDP-glucose dehydrogenase, Ugd)的酵素活性 受可逆的酪胺酸磷酸化的調控。經磷酸化的Ugd活性顯著提升,而經牛小腸鹼性 去磷酸酶處理後活性下降。已知,大腸桿菌和克雷白氏肺炎桿菌CG43的酪胺酸 激酶Wzc可藉由磷酸化Ugd來調控莢膜多醣體的生成。然而,受磷酸化調控的酪 胺酸殘基尚未被確認。本研究藉由質譜儀的技術和序列比對,在克雷白氏肺炎桿 菌Ugd所含的17個酪胺酸選了9個酪胺酸殘基,分別在10、91、150、210、217、 242、249、265和335的位置,經點突變技術換成苯丙胺酸。再將這幾個定點突變 後的蛋白質表現純化後,以磷酸化和去磷酸化的反應,分析其酵素活性。實驗結 果發現Y91F和Y210F突變的酵素活性明顯高於野生型;尤其是Y91F突變株最為 明顯。相反的,Y10F、Y242F和Y249F突變株測得的酵素活性遠比野生型的酵素 低。我們也利用西方墨點法去確認了這些突變株是否有被磷酸化。但是,所有的 突變Ugd被KpWzc磷酸化的程度都和野生型Ugd一樣好。由我們的結果可以得知 Tyr10、Tyr242和Tyr249殘基對於Ugd酵素的活性影響非常大,而將酪胺酸置換成苯 丙氨酸後的Ugd仍可被磷酸化。雖然,本研究結果並未確認被磷酸化調控的酪胺 酸殘基,但是經由酵素動力學的實驗結果證實Tyr10、Tyr242和Tyr249殘基對於Ugd 酵素活性的重要。. i.
(3) Abstract The enzymatic activity of bacterial UDP-glucose dehydrogenase (Ugd) has been shown to be under regulation by a reversible phosphorylation on the tyrosine residues. In Escherichia coli and Klebsiella pneumoniae CG43, the kinase Wzc is responsible for Ugd phosphorylation thereby modulates the synthesis of the polysaccharidic capsule. The phosphorylation of E. coli Ugd resulted in a significant increase of its activity whereas the treatment of Ugd with calf intestine alkaline phosphatase reduced its activity. However, the specific tyrosine residue subjected to the phosphorylation has not been identified. This study attempts to identify the critical tyrosine residues by site-directed mutagenesis of nine of the seventeen tyrosine residues contained in KpUgd, that were selected on the basis of their conserved nature and mass spectrum analysis. These single Tyr-to-Phe mutations of KpUgd were respectively at Tyr10, Tyr 91. , Tyr150, Tyr210, Tyr217, Tyr242, Tyr249, Tyr265, and Tyr335. The mutant proteins were. purified and their activities, either with or without incubation with KpWzc (Arg451-Lys722) were determined. The activities of UgdY91F and UgdY210F were higher than that of the wild-type Ugd. While UgdY10F, UgdY242F and UgdY249F exhibited a lower activity than that of the wild-type Ugd. We also used western blot hybridization to analyze if the mutations affected the tyrosine phosphorylation. The results indicated that all the Ugd mutants as well as the wild-type protein were able to. ii.
(4) be phosphorylated by the kinase KpWzc. Taken together, we have shown that the residue Tyr10, Tyr242 and Tyr249 were critical for the Ugd activity. In addition, the replacement of Tyr10, Tyr. 91. , Tyr150, Tyr210, Tyr217, Tyr242, Tyr249, Tyr265, and Tyr335. with Phe did not abolish the phosphorylation of Ugd. Although the specific tyrosine phosphorylation residue has not been identified, the enzymatic kinetic analysis of the mutant proteins demonstrated the critical role of the residue Tyr10, Tyr242 and Tyr249for Ugd activity.. iii.
(5) 致謝 很快地,碩士班生涯就這樣過了。還記得剛進入交大研究所的我挫折不斷, 但感謝我的指導教授-彭慧玲老師,給我這個機會進入這個實驗室,並且耐心教 導,讓我可以成長,能夠去挑戰自己並且超越自己,我才得以完成這個論文的內 容。感謝張晃猷老師,讓我有機會去清大學習酵素動力學和細菌訊息傳遞這個領 域,一路上給予我指導和幫助,並且提供了一個良好的研究環境,以及相當自由 的研究空間,才能夠讓我有這樣的研究成果。感謝邱顯泰老師,撥出寶貴的時間 來擔任我的口試委員,並且提供了我論文及研究內容上許多相當寶貴的建議和指 導。感謝林志生老師,雖然跟您相處只有短短的一段時間,但是我知道您一直很 關心我,真的很謝謝您! 感謝已經畢業的智凱學長,常常在當兵之餘抽空來關心我的實驗進度,且常 常耐著性子不厭其煩地回答我提出的一個又一個實驗問題,並且常常當我的垃圾 桶聽我一堆心事,在這裡我真的要大聲的說一聲謝謝你!感謝交大實驗室的靖 婷、盈蓉學姊和新耀、健誠學長,在實驗上帶我操作許多實驗、提供許多想法、 給予很多幫助與建議,讓我得以順利完成實驗。感謝清大實驗室的婕琳學姊、欣 瑜、幸瑜和璿琛,在我剛到清大實驗室人生地不熟的階段給予實驗及精神上的鼓 勵,我不會忘記在多少個夜晚我們幾個女生在實驗室熬夜、晚上結伴同行這樣溫 暖的情誼。 在這段期間認識了很多人-源德學長、韻如學姊、莉芳學姊、敏潔學姊、蕙. iv.
(6) 如學姊、漢聲、方瑜、婉禎、芊瑜、Manish、嘉怡、雅淑、靜柔學姊、志桓、秉 熹、雅雯、純珊、哲充和顗峰,真的非常謝謝你們,有你們才會有現在的我和這 篇論文。 最後感謝我的家人,在多少個日子裡無怨無悔地在背後默默支持我守護我, 幫我度過難關,並且讓我順利地完成碩士班學業。. 李媺如 謹致於 交通大學生化工程研究所 中華民國九十七年一月. v.
(7) Page Abstract in Chinese--------------------------------------. i. Abstract in English--------------------------------------. ii. Acknowledgement-------------------------------------. iv. Content---------------------------------------------------. vi. Abbreviations--------------------------------------------. vii. Introduction---------------------------------------------. 1. Materials and Methods---------------------------------. 7. Results---------------------------------------------------. 14. Discussion-----------------------------------------------. 20. References-----------------------------------------------. 24. Tables------------------------------------------------------. 29. Figures----------------------------------------------------. 34. vi.
(8) Abbreviations ATP bp CFU CIAP CPS DNA DTT EDTA EPS h IPTG kb kDa LB mM NAD+ NADH nm ORF PAGE PCR PTK PTP PVDF rpm SDS UDP UDP-Glc UDP-GlcUA Ugd µM. adenosine triphosphate base pairs colony forming unit(s) calf intestine alkaline phosphatase capsular polysaccharide deoxyribonucleic acid dithiothreitol ethylenediamine-tetraacetic acid exopolysaccharide hours isopropyl-1-thio-β-D-galactopyranoside kilobase(s) kilodalton(s) Luria-Bertani millimolar nicotinamide adenine dinucleotide nicotinamide adenine dinucleotide (reduced form) nanometer open reading frame polyacrylamide gel electrophoresis polymerase chain reaction protein-tyrosine kinase protein-tyrosine phosphatase polyvinylidene difluoride revolutions per minute sodium dodecyl sulfate uridine 5'-diphosphate UDP-glucose UDP-glucuronic acid UDP-glucose dehydrogenase micromolar. vii.
(9) Introduction Klebsiella pneumoniae is a member of the Enterobacteriaceae family, recognized over 100 years ago as a cause of community acquired pneumonia (Keynan et al., 2007). Most Klebsiella strains are encapsulated by a polysaccharidic capsule of considerable thickness responsible for the glistening and mucoid colonies on agar plates. The highly virulent clinical isolates are often carrying heavy capsules as an important virulence factor to protect the bacteria from phagocytosis and killing by serum factors (Simoons-Smit et al., 1986). K. pneumoniae could be classified into 77 serological K antigen types according to the diverse structures of the capsular polysaccharides. The emergence of virulent K. pneumoniae, with predominant K1/K2 capsular serotypes has been observed over the last 20 years, caused a distinct clinical syndrome consisting of pyogenic liver abscesses, sometimes accompanied by meningitis and abscesses elsewhere (Keynan et al., 2007). K. pneumoniae CG43, showing a strong virulence to Balb/c mice with 50% lethal dose of 10 CFU, is a highly encapsulated clinical isolate of K2 serotype (Chang et al., 1996). The structure of K. pneumoniae K2 capsular polysaccharides (CPS) has been determined as →)4-Glc-(1→3)-α-Glc-(1→4)-β-Man-(3←1)-α-GlcA)-(1→]n (Arakawa et al., 1991). The biosynthetic pathway of K. pneumoniae K2 capsular polysaccharides (CPS) is similar to that of Escherichia coli group 1 CPS (Whitfield et. 1.
(10) al., 1999). In E.coli, capsules are classified into four groups on the basis of genetic and biochemical criteria. The biosynthesis and assembly mechanisms for groups 1 and 4 are closely related and follow Wzy-dependent processes (Whitfield, 2006). Capsule assembly appears to involve a multiprotein complex spanning the cell envelope. The complex facilitates spatial and temporal coupling of polymer biosynthesis, export, and translocation. In the Wzy-dependent mechanism, undecaprenol-linked repeat units are used in a polymerization reaction that requires Wzx and Wzy proteins. Newly synthesized lipid-linked repeat units are exported across the inner membrane in a process requiring Wzx. The growing glycan is transferred from its lipid carrier to the newly exported lipid-linked repeat unit, elongating the polymer in a “block-wise” manner by addition of new repeat units at the reducing terminus. The polymerization reaction is dependent on the Wzy protein. In group 1 capsule assembly, Wza is linked to the inner-membrane biosynthesis process by Wzc. Wzc forms tetramers and undergoes transphosphorylation of several C-terminal tyrosine residues. Phosphorylation of Wzc and its dephosphorylation by the phosphatase Wzb is essential for group 1 and 4 capsule assembly, suggesting that Wzc must cycle its phosphorylated state (Whitfield, 2006). The cps (capsular polysaccharide synthesis) gene cluster that is responsible for K2 CPS synthesis of K. pneumoniae Chedid has been identified, which contains a. 2.
(11) total of 19 open reading frames (ORFs) organized into 3 transcriptional units (Arakawa et al., 1995). Among these genes, orf3 to orf6, a highly conserved four-gene-block, are counterparts of E. coli wzi-wza-wzb-wzc (Rahn et al., 1999). orf3 is homologous to the gene product required for surface expression of E. coli group 1 K antigen. Wzi, an orf3 encoding protein, is an outer membrane protein and wzi mutant showed a significant reduction in cell-bound CPS polymer with a corresponding increase in cell-free material. This proposed that Wzi plays a late role in capsule assembly, perhaps in the process that links high-molecular-weight capsule to the cell surface (Alvarez et al., 2000; Rahn et al., 2003). orf4 is a Wza homolog, which is a periplasmic protein involved in polysaccharide export. orf5, or KpWzb, is a low molecular weight protein tyrosine phosphatase (PTP). orf6, or KpWzc, is a protein tyrosine kinase (PTK). Enzymatic activities of the two proteins, KpWzb and KpWzc, which are respectively Wzb and Wzc homolog, have been demonstrated (Preneta et al., 2002). Tyrosine phosphorylation is a key device in numerous cellular functions in eukaryotes, but in bacteria this protein modification was largely ignored until mid-1990s (Grangeasse et al., 2007). The first conclusive evidence of bacterial tyrosine phosphorylation came only a decade ago. In E. coli, a PTK activity has been reported (Manai et al., 1983). However, it was not known whether the source of. 3.
(12) phosphotyrosine might be the nucleotidylation of proteins rather than phosphorylation. The genetic and biochemical characterization of a tyrosine kinase in Acinetobacter johnsonii in 1997 greatly advanced the understanding of tyrosine phosphorylation in bacteria (Grangeasse et al., 2007). This covalent modification is catalyzed by autophosphorylating ATP-dependent protein-tyrosine kinases that exhibit structural and functional features similar, but not identical, to those of their eukaryotic counterparts. Since then, several tyrosine kinases exhibiting unexpected features have been identified in a variety of bacteria. These enzymes use homologues of Walker motifs of nucleotide-binding proteins for their catalytic mechanism, thus defining an idiosyncratic type of bacterial tyrosine kinases (Cozzone et al., 2004). Recent data provide evidence that there exists a direct relationship between the reversible phosphorylation of proteins on tyrosine and the production of these polysaccharidic polymers, which are known to be important virulence factors (Cozzone et al., 2004). An increasing list of endogenous proteins have been found to be the substrate of bacterial tyrosine kinases. The first known substrates of these kinases are the kinases themselves because they are autophosphorylating enzymes. The later identified proteins were UDP-sugar dehydrogenases (Grangeasse et al., 2003; Mijakovic et al., 2003). Phosphorylation of the enzyme increases their activity thereby stimulates formation of the precursors for polysaccharide production. UDP-. 4.
(13) glucose dehydrogenases (Ugd) are also involved in other metabolic pathways, including resistance to cationic peptides and polymyxin-type antibiotics or phosphate metabolism in E. coli and B. subtilis, respectively (Breazeale et al., 2003; Soldo et al., 1999). The E. coli Etk was recently found to have a key role in polymyxin resistance, supposedly mediated by phosphorylation of Ugd (Lacour et al., 2006). Ugd catalyzes the NAD+-dependent two fold oxidation of UDP-glucose (UDP-glc) to give UDP-glucuronic acid (UDP-glcUA). A sequestered aldehyde intermediate is produced in the first oxidation step and a covalently bound thioester is produced in the second oxidation step. UDP-glcUA is a common substrate for the biosynthesis of exopolysaccharide (Ge et al., 2004). In mammals, UDP-glcUA is used in the biosynthesis of hyaluronan and various glycosaminoglycans such as heparin sulfate and chondroitin sulfate. UDP-glcUA also serves as a precursor to UDP-xylose which provides a major component of the cell wall polysaccharides in plants (Dalessandro et al., 1977). In many strains of pathogenic bacteria, such as group A streptoccoci and Streptococcus pneumoniae type 3, UDP-glcUA is used in the construction of the antiphagocytic capsular polysaccharide (Dougherty et al., 1993; Arrecubieta et al., 1994). Although a lot of protein kinases and phosphatases have been predicted and identified in a variety of bacterial species, classical biochemical approaches have so. 5.
(14) far revealed only a few substrate proteins and even fewer phosphorylation sites. Via in vitro phosphorylation assay, we have previously demonstrated that the Wzc of E. coli (EcWzc) and K. pneumoniae CG43 (KpWzc) was able to phosphorylate UDP-glucose. dehydrogenase (Ugd) which is required for the synthesis of sugar nucleotide precursor (Zhi-Kai Li , 2006) and the phosphorylation appeared to enhance the Ugd activity. But the phosphotyrosine residues in Ugd required for regulation of the enzymatic activity have not been determined. In this study we performed site directed mutagenesis to identify the phosphotyrosine residues of Ugd in K. pneumoniae. We produced single Tyr-to-Phe mutations at Tyr10, 91, 150, 210, 217, 242, 249, 265, and 335 of KpUgd, and compared the activities of the unphosphorylated with the phosphorylated proteins.. 6.
(15) Materials and methods Bacterial strains, plasmids, and growth conditions. The bacterial strains and plasmids used in this study are listed in Table 1. K. pneumoniae CG43 is a clinical isolates recovered from Chang Gung Memorial Hospital, Linkou. E. coli and K. pneumoniae CG43 were routinely cultured at 37°C in Luria-Bertani (LB; 10 g/l tryptone, 5 g/l yeast extract, 10 g/l sodium chloride) broth or on LB agar. When indicated, the following antibiotic concentrations were used for selection of E. coli: kanamycin 25 μg/ml, ampicillin 100 μg/ml and tetracycline 12.5 μg/ml. The concentration of streptomycin for the selection of K. pneumoniae CG43-S3 was 50 μg/ml.. DNA manipulation Plasmids were purified by using the High-Speed Plasmid Mini kit (GeneMark, Taiwan). All recombinant DNA experiments were carried out by standard procedures as described (Sambrook et al., 2001). Restriction endonucleases and DNA modifying enzymes were purchased from either New England Biolab (Beverly, MA) or MBI Fermentas (Hanover, MD), and were used according to the recommendation of the suppliers. PCR amplifications were performed with Taq DNA polymerase (Geneaid, Taiwan). PCR products and DNA fragments were purified 7.
(16) using the Gel/PCR DNA Fragments Extraction kit (Geneaid, Taiwan). The primers used in this study were synthesized by MDBio, Inc, Taiwan.. Construction of His6 tag kpwzc domain expression plasmids The 774-bp kpwzc (nucleotides 1342–2115) gene fragment, with appropriate restriction sites at both ends, encoding the C-terminal domain of KpWzc without the tyrosine cluster, were synthesized by PCR amplification using genomic DNA from K. pneumoniae CG43 strain as a template. For kpwzc cloning, the sequences of the two primers were 5’-GCCGAGCTCATTCTATTACGTAAAGGA ATTGAAACTC-3’ at the N-terminus (the SacI site is italicized) and 5’AAAAACTGCAGATTAGTTGCTTTTTTTACCACACC-3’ at the C-terminus (the PstI site is italicized; the stop codon of kpwzc is underlined). The DNA fragment synthesized was restricted by SacI and PstI and ligated into pQE30 vector opened with the same enzymes. The resulting plasmids were termed pQE30-42KpWzc (Table II).. Site-directed mutagenesis Ugd mutants were produced with the QuikChange site-directed mutagenesis method (Stratagene) according to the manufacturer’s protocols. The. 8.
(17) primers used are listed in Table II. The resulting plasmids were respectively termed pETUgdY10F~Y335F (Table II) and the mutant Ugd proteins were expressed in E. coli NovaBlue(DE3).. Overproduction and purification of His6 tag fusion Ugd and the derived mutants The bacterial cells were incubated in 100 ml of LB medium supplemented with kanamycin and tetracycline at 37℃ with shaking until OD600 reached 0.5-0.6. Isopropyl-1-thio-β-D-galactopyranoside (IPTG) was then added to a final concentration of 1 mM and the growth was continued for 4 h at 37℃. Subsequently, the cells were harvested by centrifugation at 5000 rpm for 10 min, resuspended in binding buffer (20 mM Tris-HCl, 500 mM NaCl, 5 mM imidazole, pH 7.9), and the cell suspension disrupted by sonication and then the cell debris removed by centrifugation at 14000 rpm for 20 min. Finally, the His6-tagged proteins were purified from the supernatant via affinity chromatography using His-Bind resin (pharmacia), and the elution was carried out with elute buffer (20 mM Tris-HCl, 500 mM NaCl, 1 M imidazole, pH 7.9). Aliquots of the collected fractions were analyzed by SDS-PAGE and the fractions containing His6-Ugd were dialyzed against the buffer containing 50 mM Tris-HCl (pH7.5), 100 mM NaCl, 1 mM EDTA, and 10% glycerol.. 9.
(18) Overproduction and purification of His6 tag fusion KpWzc cytoplasmic domain E. coli M15 cells were transformed with pQE30 vector derivatives expressing mutated KpWzc cytoplasmic domain, His6–KpWzc (Arg451-Lys722) (see Table II). The overproduction procedure was the same for Ugd but the overnight cell culture supplemented with ampicillin and kanamycin. IPTG was then added at a final concentration of 0.5 mM. Cells were harvested by centrifugation at 5000 g for 10 min, resuspended in 10 ml binding buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol), and the cell suspension disrupted by sonication and then the cell debris removed by centrifugation at 14,000 rpm for 20 min. The supernatant was added to Ni+2-NTA-agarose matrix and was first washed with binding buffer, then protein elution was carried out with elute buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 100 mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol), and eluted fractions were analyzed by SDS-PAGE. Fractions containing purified His6-tagged proteins were pooled and dialyzed at 4°C against a large volume of dialysis buffer (50 mM sodium phosphate, pH 7.4, 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 5 mM dithiothreitol).. SDS-polyacrylamide gel electrophoresis Protein preparations were treated for 5 min at 95℃ in loading buffer. 10.
(19) (0.0626 M Tris-HCl buffer pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, and 100 mM dithiothreitol). Twenty microliters of sample was applied in a 12.5% SDS polyacrylamide slab gel. Electrophoresis was carried out at room temperature until the tracking dye ran off the bottom of the slab gel. The gel was stained for 5 min using solution containing 2.5% Coomassie Blue R250, 45% methanol, and 10% acetic acid, and destained briefly in boiling water.. Western blot analysis of the phosphotyrosine proteins The whole cell protein lysate was analyzed by SDS-PAGE. The resolved proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane. The transfer buffer was 25 mM Tris, 192 mM glycine, 0.37% SDS, and 20 % methanol. The membrane was detected by anti-phosphotyrosine, clone 4G10 antibody (Upstate, catalog number 05-321) and HRP conjugated secondary antibody.. In vitro phosphorylation assay The phosphorylation reaction was carried out as described (Grangeasse et al., 2003) by incubating the mixture (20 μl) containing about 2 μg of the purified kinase and/or Ugd with 10 μM ATP in 25 mM Tris-HCl (pH 7.0), 1 mM DTT, 5 mM MgCl2, 1 mM EDTA at 37℃for 1 h. The reaction was stopped by addition of the sample buffer and heating at 95℃ for 5 min. After electrophoresis (12.5% 11.
(20) SDS-PAGE), the gel was stained with Coomassie Blue and the result visualized by Western Blot with 4G10 anti-phosphotyrosine antibody (Upstate).. Phosphatase activity assay Alkaline phosphatase activity was monitored at 37°C by using a continuous detection method based on the detection of p-nitrophenol formed from p-nitrophenyl phosphate (pNPP). Rates of dephosphorylation were determined at 405 nm and pNPP at a concentration 0.1 mM (Cirri et al., 1993).. Enzymatic activity measurement and kinetic characterizations Enzymatic activities of the Ugd were determined by monitoring the change in absorbance at 340 nm that accompanies the reduction of NAD+ to NADH. The enzyme assay was performed at room temperature in 100 mM Tris-HCl (pH 9.0), 100 mM NaCl, 2 mM DTT, 2 mM NAD+ and 5 mM UDP-glucose (Pagni et al., 1999). The Km and Vmax for UDP-glc and NAD+ were determined independently using standard assay conditions. Constants for UDP-glc as substrate were measured by holding NAD+ constant and varying UDP-glc from 0.01 to 5 mM. Similarly, NAD+ kinetic measurements were made by holding UDP-glc constant and varying NAD+ from 0.005 to 2 mM. Data were plotted with GraphPad Prism 4 software. Km and Vmax were calculated by fitting the data to the Michaelis-Menten equation and assuming a single binding site each for substrate and cofactor.. 12.
(21) Software Structural prediction were performed at Swiss-Model (http://swissmodel.expasy.org//SWISS-MODEL.html), using Swiss-PdbViewer 3.7 and RasMol version 2.6 respectively. Multiple alignments were performed by Vector NTI 6.0.. In-Gel digestion The His6-tagged fusion protein containing the Ugd and mutants were in vitro phosphorylated by KpWzc. Thereafter, the protein were boiled in SDS-PAGE sample buffer, separated on a 12.5% SDS-PAGE, and stained with Coomassie Blue. The piece of gel containing the Ugd or mutants fusion protein was excised and in-gel-digested with trypsin according to standard procedures. Tryptic peptides were extracted with 5% formic acid/50% acetronitrile, dried, and stored at -20°C until analysis by mass spectrometry.. 13.
(22) Results Localization and predicted structure of point mutations in KpUgd. As shown in Fig. 1, 17 tyrosine residues could be identified in KpUgd. The nine tyrosine residues including Tyr10, Tyr. 91. , Tyr150, Tyr210, Tyr217, Tyr242, Tyr249,. Tyr265, and Tyr335, which are likely important in the enzyme function, have been selected mainly on the consensus property. In comparing with the crystal structure of Streptococcus pyogenes Ugd (Fig. 2A) (Campbell et al., 2000), similar structure was predicted by the Swiss-Model (http://swissmodel.expasy.org//SWISS-MODEL.html) for KpUgd with 55% sequence identity (Fig. 2B). By superimposing the context of the Ugd crystal structure of S. pyogenes, Cys 253 in KpUgd was found at the position equivalent to catalytic nucleophile Cys 260 in S. pyogenes Ugd. The Tyr10 was found to be strictly conserved among UDP-sugar dehydrogenases, accessible to solvent, and situated very close to the interface of NAD+ and UDP-glc bound to the active site of the enzyme. The tyrosine residues Tyr150 and Tyr242 in this region were probably important in substrate and cofactor binding (Fig. 2C). Information regarding the effect of specific mutations on enzyme function is limited mainly to those affecting the active site. Nevertheless, Fig. 2C also revealed that central α-helix (α 9) serving as the core of the dimer interface could be important. There are a total of 24 hydrogen bonds to stabilize the dimer interface, though none of. 14.
(23) the amino acids involved are strictly conserved. The aromatic residues including Phe199, Tyr203, Tyr210, Tyr217, and Tyr265 were found to be located within the dimer interface. Among them, Tyr210 is noteworthy as it exhibits strong conservation of aromatic or hydrophobic character in all bacteria, though higher order species have a serine at this position (Campbell et al., 2000). Beside, the Tyr210 and Tyr217 residing in α 9 and the Tyr242 and Tyr249 residues that forms the binding pocket of Ugd could also be important in determining the enzymatic activity.. Mass spectrographic analysis of the phosphorylation state of Ugd. We have also carried out a MALDI-TOF MS analysis of the resulting peptides from the trypsin-digested Ugd and phosphorylated-Ugd. The peptides containing 14 of the 17 tyrosine residues in Ugd could be identified. Among them, two peptides with the molecular mass of the phosphorylated form were noted. One was TNYFNTSTVEAVIR (residues 89–102 of Ugd) with the molecular mass corresponding to that of a phosphor-tyrosine containing peptide. The second peptide has the mass of the phosphorylated peptide ALYDNLHPSR (residues 148–157). The deduced analysis indicated that the phosphorylation sites were probably at Tyr91 and Tyr150, which have been selected for site-directed mutagenesis.. 15.
(24) Construction, expression and purification of the KpUgd mutants. To identify the tyrosine residues subjected to phosphorylation in KpUgd, we constructed nine single tyrosine to phenylalanine mutants. The wild-type and mutant Ugd cloned in pET30b were expressed in E. coli NovaBlue (DE3) and the proteins purified through Ni+2-NTA-agarose matrix (Fig.3). In general, about 3.8 mg of Ugd and the derived mutant proteins of high purity (> 95%) could be obtained from a 100 ml culture. The purified recombinant Ugd mutant proteins were then subjected to SDS-PAGE and stained with Coomassie Blue, or western blot hybridization probed with an anti-His Tag mouse monoclonal antibody (Novegen catalog 70796-4). These enzymes appeared to be stable in the buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 10% glycerol.. The cytoplasmic PTK domain of KpWzc could catalyze Tyrosine phosphorylation of Ugd. It has been shown that the protein-tyrosine kinase Wzc of E. coli is able to phosphorylate Ugd (Grangeasse et al., 2003). Sequence analysis revealed that the KpUgd and KpWzc respectively exhibit 83% sequence identity with that of the E. coli enzymes. It is hence expected that KpUgd could also be phosphorylated by KpWzc, the counterpart of E.coli protein PTK Wzc.. 16.
(25) To verify this hypothesis, phosphorylation experiments were carried out with KpUgd and His6–KpWzc (Arg451-Lys722), the catalytic PTK domain of KpWzc with the C-terminal tyrosine cluster. When the two purified proteins were incubated together with 10 μM ATP, phosphorylation of Ugd was indeed detected using western blotting analysis (Fig. 4). The labeled band with an apparent molecular mass of 31 kDa corresponded to the phosphorylated His6–KpWzc (Arg451-Lys722), whereas the labeled-band with higher molecular weight (44 kDa) was the phosphorylated His6-Ugd. This indicated that the phosphorylation of Ugd was dependent on the KpWzc. All the Ugd mutants were phosphorylated by KpWzc as shown in Fig. 4, indicating that none of the mutated tyrosine residues impaired the phosphorylation.. Phosphorylation enhances the Ugd activity. As shown in Fig. 5, the enzymatic activity analysis indicated that none of the mutations dramatically affected the Ugd activity. To test whether the phosphorylation of Ugd affects its catalytic activity, the His6-Ugd mutants purified from E. coli were first treated with CIAP and then re-purified by Ni+2-NTA-agarose matrix. The efficiency of the removal of CIAP was tested with p-nitrophenyl phosphate (pNPP) as shown in Fig. 6. Finally, the dephosphorylated-Ugd was incubated with KpWzc for 1 h and the activity of the phosphorylated Ugd measured.. 17.
(26) As shown in Fig. 7, the results showed that the Ugd activity that has been subjected to phosphorylation by Kp-Wzc was enhanced about 6- fold higher than the untreated Ugd (Fig. 7). By contrast, the incubation of His6-Ugd with CIAP led to a drastic loss of its activity. This indicated that the phosphorylated Ugd has higher level of enzyme activity and the purified His6-Ugd from E. coli probably contained a small fraction of phosphorylated forms which were responsible for the observed weak Ugd activity.. Phosphorylation of the single-site-mutants. To compare the phosphorylation activity between the wild-type Ugd and the mutant Ugd, a continuous spectrophotometric assay was used. KpUgd with or without KpWzc was incubated with ATP and then Ugd activity measured. The phosphorylation activation measured in the Y91 and Y210F mutants were respectively 2.23 mM/min and 1.71 mM/min, in comparing with 0.05 mM/min for the wild-type Ugd. The Ugd activity of phosphorylated-Y91F or phosphorylated-Y210F was significant higher than that of wild-type Ugd, suggesting that the hydrogen bond to stabilize the dimer interface by Tyr210 could be broken thus affecting the enzyme activity of KpUgd.. Determination of kinetic parameters of Ugd and the derived mutants. To obtain kinetic constants for wild-type and mutant forms of Ugd, the 18.
(27) initial velocity of the reaction by measuring the unique NADH absorbance at 340 nm was determined. The kinetic constants of His6-tagged wild-type and mutant forms of Ugd for UDP-glc were calculated by fixing the concentration of NAD+ with various concentrations of UDP-glc (Fig. 8). On the other hand, varying the concentration of NAD+ with a constant concentration of UDP-glc was also used to measure the activity (Fig. 9). The saturation kinetics of both wild-type and mutant forms of Ugd were observed for both substrates. For UDP-glc, the data were fitted to Lineweave-Burk equation and the results shown in Table 4. The Kcat/Km showed that the catalytic efficiency of Y91F or Y210F for UDP-glc was much higher than that of the wild type. While the Kcat/Km of Y91F for NAD+ was much higher than that of the wild type.. 19.
(28) Discussion The majority of PTK are encoded by genes located in operons for the biosynthesis of polysaccharides. Tyrosine autophosphorylation of PTK has been reported to control exopolysaccharide production both in Firmicutes and Proteobacteria. The findings that the Ugd of the model organisms for Gram-positive and Gram-negative bacteria, B. subtilis and E. coli, were phosphorylated by PTK, suggests that Ugd is a general substrate for PTK in bacteria. The orthologous Ugd and PTK could be found in many other bacteria including Salmonella typhimurium (Ugd and Wzc), Staphylococcus aureus (CapL and CapB), and Vibrio vulnificus (genes vv10781 and vv10774). However, Ugd is believed not the only substrate for bacterial PTK. For example, RpoH was reported to be phosphorylated at amino acid position 260 in E. coli, which is located in the conserved region 4.2, and that this phosphorylation event attenuates RpoH activity as a sigma factor. Additionally, another antisigma factor RseA was shown to be phosphorylated at the N-terminally located Tyr-38 (Klein et al., 2003). In Streptomyces griseus, SSBs was identified as a novel target of the bacterial tyrosine kinase (Mijakovic et al., 2006). Herein, we have demonstrated that the protein-tyrosine kinase Wzc of K. pneumoniae was able to phosphorylate Ugd, and the phosphorylation of Ugd resulted in a significant increase of its dehydrogenase activity. The dephosphorylation by calf. 20.
(29) intestine alkaline phosphatase reduced its enzyme activity. The activation of Ugd activity leads to synthesis of a large amount of UDP sugar, the precursor for the bacterial polysaccharide production. It is hence a role directly linked to virulence has be suggested. The use of mass spectrometry in protein phosphorylation site determination has increased significantly in the past few years. Recently, the in-depth phosphorylation site-resolved dataset from B. subtilis and E. coli have been presented. A recently developed proteomics approach based on phosphopeptide enrichment and high accuracy of mass spectrometric analysis was used. There are 81 phosphorylation sites on 79 E. coli proteins, with 68/23/9% distribution on the Ser/Thr/Tyr phosphorylation sites. In B. subtilis, 103 unique phosphopeptides from 78 proteins and 78 phosphorylation sites including 54 on serine, 16 on threonine, and 8 on tyrosine, were determined (Macek et al., 2007). No phosphorylation sites on Ugd was reported in the studies, although the in vitro analysis has demonstrated a quite efficient phosphorylation on E. coli Ugd (Grangeasse et al., 2003). It is likely that the peptides containing phosphor-tyrosine residues were too hydrophobic to be eluted from the gel under the conditions employed, and different strategy has to be used to recover the tyrosine-phosphorylated peptides. In this study, an in vitro system to explore the phosphorylation-induced. 21.
(30) activation of the activity of Ugd was used. On the basis of the structure modeling of KpUgd, the relative position on the space and the UDP-glc binding pocket could help to explain the critical site for the Ugd activity. The UDP-glc binding pocket which can be divided into two regions: the UMP binding pocket composed solely of the residues from the C-terminal domain, and the glucose 1-phosphate binding pocket consisting primarily the residues from the N-terminal domain. The UMP binding pocket was lined with a stretch of coil (Tyr 242~Gly 250) that makes three main chain hydrogen bonds, two side chain hydrogen bonds, and a π-edge stacking interaction of Tyr 242 with the UMP moiety. The glucose 1-phosphate binding pocket was found at the dimer interface, limited to a small region (Phe 142-Glu 145) between β 7 and α 7 of the N-terminal domain that forms three main hydrogen bonds to the glucose 1-phosphate moiety (Campbell et al., 2000). Two of the selected nine tyrosine residues, Y242F and Y249F, are responsible for UDP-glc binding. If the hydroxyl group of the tyrosine residue was retained, KpWzc may introduce a fixed negative charge through phosphorylation to stabilize the Ugd. The formation of a salt bridge between the negative charged-residue 249 and a nearby positively charged residue. Rotation of the loop could allow the side chain of residue 249 to reach the surface where a salt bridge might form with the several positively charged residues located in this surface vicinity. More detailed studies of Kp-UgdY242F or Y249F might be of. 22.
(31) some interests as a model for structural changes occurring upon phosphorylation.. 23.
(32) References 1. Arakawa Y, Ohta M, Wacharotayankun R, Mori M, Kido N, Ito H, Komatsu T, Sugiyama T, Kato N. 1991. Biosynthesis of Klebsiella K2 capsular polysaccharide in Escherichia coli HB101 requires the functions of rmpA and the chromosomal cps gene cluster of the virulent strain Klebsiella pneumoniae Chedid (O1:K2). Infect Immun. 59(6):2043-50. 2. Alvarez D, Merino S, Tomás JM, Benedí VJ, Albertí S. 2000. Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect Immun. 68(2): 953-955. 3. Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. 1995. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 177(7): 1788-96. 4. Arrecubieta C, López R, García E. 1994. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: Sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J. Bacteriol. 176(20): 6375–6383. 5. Breazeale SD, Ribeiro AA, Raetz CR. 2002. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-L-arabinose. J Biol Chem. 277(4):2886-96. 6. Campbell RE, Mosimann SC, van De Rijn I, Tanner ME, Strynadka NC. 2000. The first structure of UDP-glucose dehydrogenase reveals the catalytic 24.
(33) residues necessary for the two-fold oxidation. Biochemistry. 39 (23):7012-23. 7. Chang HY, Lee JH, Deng WL, Fu TF, Peng HL. 1996. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb Pathog. 20(5):255-61. 8. Cirri P, Chiarugi P, Camici G, Manao G, Raugei G, Cappugi G, Ramponi G. 1993. The role of Cys12, Cys17 and Arg18 in the catalytic mechanism of low-Mr cytosolic phosphotyrosine protein phosphatase. Eur J Biochem. 214(3):647-57. 9. Cozzone AJ, Grangeasse C, Doublet P, Duclos B. 2004. Protein phosphorylation on tyrosine in bacteria. Arch Microbiol. 181(3):171-81. 10. Dalessandro G, Northcote DH. 1977. Changes in enzymic activities of nucleoside diphosphate sugar interconversions during differentiation of cambium to xylem in sycamore and poplar. Biochem. J. 162 (2):267-79. 11. Dougherty BA, van de Rijn I. 1993. Molecular characterization of hasB from an operon required for hyaluronic acid synthesis in group A streptococci. J. Biol. Chem. 268(10):7118-24. 12. Ge X, Penney LC, van de Rijn I, Tanner ME. 2004. Active site residues and mechanism of UDP-glucose dehydrogenase. Eur J Biochem. 271(1):14-22. 13. Grangeasse C, Obadia B, Mijakovic I, Deutscher J, Cozzone AJ, Doublet P. 2003. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J Biol Chem. 10; 278 (41):39323-9. 14. Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. 2007. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci. 32(2):86-94.. 25.
(34) 15. Keynan Y, Rubinstein E. 2007. The changing face of Klebsiella pneumoniae infections in the community. Int J Antimicrob Agents. 30(5):385-9. 16. Kirstein J, Zühlke D, Gerth U, Turgay K, Hecker M. 2005. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 24(19):3435-45. 17. Klein G, Dartigalongue C, Raina S. 2003. Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol Microbiol. 48(1):269-85. 18. Lacour S, Doublet P, Obadia B, Cozzone AJ, Grangeasse C. 2006. A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res Microbiol. 157(7):637-41. 19. Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 6(4):697-707. 20. Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. 2007. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 15. 21. Mijakovic I, Poncet S, Boël G, Mazé A, Gillet S, Jamet E, Decottignies P, Grangeasse C, Doublet P, Le Maréchal P, Deutscher J. 2003. Transmembrane modulator-dependent. bacterial. tyrosine. kinase. activates. UDP-glucose. dehydrogenases. EMBO J. 22 (18):4709-18. 22. Mijakovic I, Petranovic D, Macek B, Cepo T, Mann M, Davies J, Jensen PR, Vujaklija D. 2006. Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res. 34(5):1588-96. 23. Mouslim C, Groisman EA. 2003. Control of the Salmonella ugd gene by three. 26.
(35) two-component regulatory systems. Mol Microbiol. 47(2):335-44. 24. Pagni M, Lazarevic V, Soldo B, Karamata D. 1999 Assay for UDPglucose 6-dehydrogenase in phosphate-starved cells: gene tuaD of Bacillus subtilis 168 encodes the UDPglucose 6-dehydrogenase involved in teichuronic Acid synthesis. Microbiology. 145 (5): 1049-1053. 25. Preneta R, Jarraud S, Vincent C, Doublet P, Duclos B, Etienne J, Cozzone AJ. 2002. Isolation and characterization of a protein-tyrosine kinase and a phosphotyrosine- protein phosphatase from Klebsiella pneumoniae. Comp Biochem Physiol B Biochem Mol Biol. 131(1):103-12. 26. Rahn A, Drummelsmith J, Whitfield C. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J Bacteriol. 181(7):2307-13. 27. Rahn A, Beis K, Naismith JH, Whitfield C. 2003. A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J Bacteriol. 185(19):5882-90. 28. Sambrook J, D W Russell. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor laboratory Press, Cold Spring Harbor, New York. 29. Simoons-Smit AM, Verweij-van Vught AM, MacLaren DM. 1986. The role of K antigens as virulence factors in Klebsiella. J Med Microbiol. 21(2):133-7. 30. Soldo B, Lazarevic V, Pagni M, Karamata D. 1999. Teichuronic acid operon of Bacillus subtilis 168. Mol. Microbiol. 31(3):795-805. 31. Whitfield C, Roberts IS. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 31(5):1307-19. 27.
(36) 32. Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 75:39-68. 33. Wu J, Ohta N, Zhao JL, Newton A. 1998. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci U S A. 96 (23):13068-73. 34. Vincent C, Doublet P, Grangeasse C, Vaganay E, Cozzone AJ, Duclos B. 1999. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol. 181(11):3472-7. 35. Vincent C, Duclos B, Grangeasse C, Vaganay E, Riberty M, Cozzone A J, Doublet P. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J Mol Biol. 304 (3):311-21. 36. 白平輝 (2004) 克雷白氏肺炎桿菌CG43莢膜多醣體的產生和酪胺酸磷酸化 作用的研究. 國立交通大學 生物科技研究所 碩士論文 37. 李智凱 (2005) 克雷白氏肺炎桿菌CG43中參與莢膜多醣體生合成之核心蛋 白Wza、Yor5、Yco6 和Wzx 的功能性研究. 國立交通大學 生物科技研究所. 碩士論文. 28.
(37) Table 1 Bacterial strains used and constructed in this study Strain E. coli: NovaBlue(DE3). Genotype or relevant characteristic. Reference or source. endA1 hsdR17(rk12-mk12+) supE44 thi-1 recA1 gyrA96 Novagen relA1 lac[F,pro AB lacqZ△M15:: Tn10](DE3);Tetr RecA1 supE44 endA1 hsdR17 gyrA96 rolA1 thi △ (lac-proAB) F- ompT hsdSB(rB-mB-)gal dcm(DE3). Laboratory stock. K. pneumoniae: CG43. Clinical isolate of K2 serotype. Laboratory stock. CG43-S3. rspl mutant, Strepr. Laboratory stock. JM109 BL21-RIL M15(pREP4). Laboratory stock S S S – – + + – – + NaI , Str , Rif , Thi , Lac , Ara , Gal , Mtl , F , RecA , Qiagen Uvr+, Lon+.. 29.
(38) Table 2 Plasmids used and constructed in this study Plasmids pET-30a-c pQE30-32 pET-KpWzcE23 pET-Ugd pET-UgdY10F pET-UgdY91F pET-UgdY150F pET-UgdY210F pET-UgdY217F pET-UgdY242F pET-UgdY249F pET-UgdY265F pET-UgdY335F pQE30-42KpWzc. Description Overexpression of His6 fusion proteins, Kmr Overexpression of His6 fusion proteins, Ampr Encoding KpWzc from Arg451 to Lys722, His6–KpWzc(Arg451-Lys722), cloned in EcoRΙ/SalΙ sites, Kmr Encoding KpUgd from Met1 to Asp388, His6–KpUgd, cloned in EcoRΙ/SalΙ sites, Kmr Same as pET-Ugd, but mutated on Y10F Same as pET-Ugd, but mutated on Y91F Same as pET-Ugd, but mutated on Y150F Same as pET-Ugd, but mutated on Y210F Same as pET-Ugd, but mutated on Y217F Same as pET-Ugd, but mutated on Y242F Same as pET-Ugd, but mutated on Y249F Same as pET-Ugd, but mutated on Y265F Same as pET-Ugd, but mutated on Y335F Overexpression of His6–KpWzc(Ser448-Ala705), Ampr. 30. Reference or source Novagen Qiagen 李智凱, 2005 白平輝,2004 This study This study This study This study This study This study This study This study This study This study.
(39) Table 3 Primers used in this study Primer KpWzcE2 KpWzcE3 KpWzc-2 KpWzc-4 UGDE1 UGDE2 Y10FF Y10FR Y91FF Y91FR Y150FF Y150FR Y210FF Y210FR Y217FF Y217FR Y242FF Y242FR Y249FF Y249FR Y265FF Y265FR Y335FF Y335FR. Sequence (5’ to 3’) CGTAAAGGAATTGAAACTCCAGA AAGGGGATTCTTCGTCCCCT GCCGAGCTCATTCTATTACGTAAAGGAATTGAAACTC AAAAACTGCAGATTAGTTGCTTTTTTTACCACACC CGAATGAAAATTACTATTTCCGG CCAGTGTCAGACAGGCAGAA CTATTTCCGGCACAGGTTTTGTTGGTTTATCGAACGG CCGTTCGATAAACCAACAAAACCTGTGCCGGAAATAG CGATCCCAAAACTAACTTCTTCAACACCTCTACGG CCGTAGAGGTGTTGAAGAAGTTAGTTTTGGGATCG GGACGTGCGCTGTTCGACAACCTGCACC GGTGCAGGTTGTCGAACAGCGCACGTCC GGCGCTGCGCGTTGCCTTCTTCAACGAGCT AGCTCGTTGAAGAAGGCAACGCGCAGCGCC TTCAACGAGCTAGACAGCTTTGCTGAAAGCCAG CTGGCTTTCAGCAAAGCTGTCTAGCTCGTTGAA GTATCGGCAACCACTTCAACAACCCGTCCTTTG CAAAGGACGGGTTGTTGAAGTGGTTGCCGATAC CCGTCCTTTGGCTTTGGCGGCTACTGC GCAGTAGCCGCCAAAGCCAAAGGACGG GCTGCTGGCGAACTTTGAATCGGTCCCG CGGGACCGATTCAAAGTTCGCCAGCAGC GGTATTCCGGTTATTATCTTTGAACCGGTGATGCAGG CCTGCATCACCGGTTCAAAGATAATAACCGGAATACC. 31.
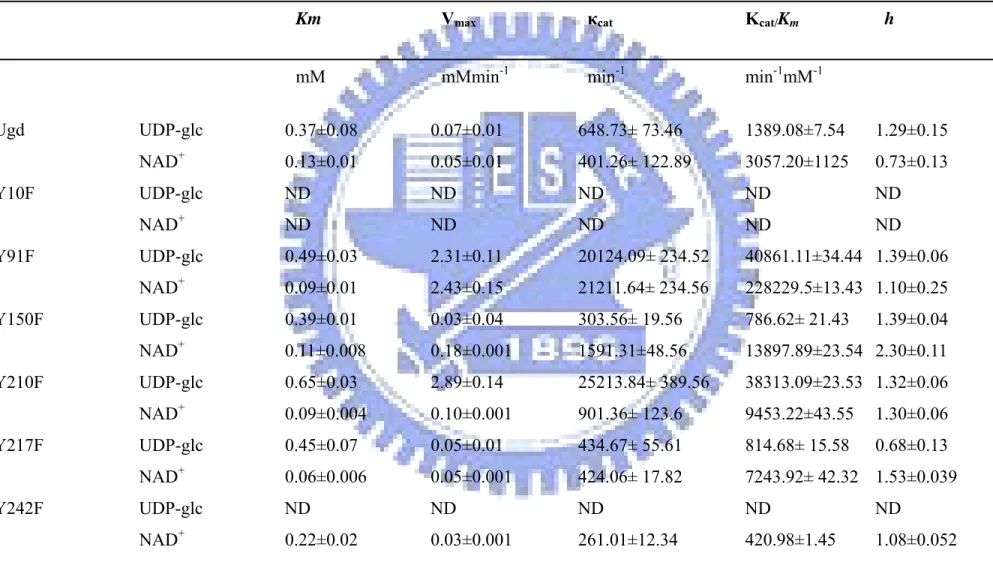
(40) Table 4 Kinetic properties of Ugd and mutants. Ugd Y10F Y91F Y150F Y210F Y217F Y242F. Km. Vmax. κcat. Κcat/Km. mM. mMmin-1. min-1. min-1mM-1. h. UDP-glc. 0.37±0.08. 0.07±0.01. 648.73± 73.46. 1389.08±7.54. 1.29±0.15. NAD+. 0.13±0.01. 0.05±0.01. 401.26± 122.89. 3057.20±1125. 0.73±0.13. UDP-glc. ND. ND. ND. ND. ND. NAD+. ND. ND. ND. ND. ND. UDP-glc. 0.49±0.03. 2.31±0.11. 20124.09± 234.52. 40861.11±34.44 1.39±0.06. NAD+. 0.09±0.01. 2.43±0.15. 21211.64± 234.56. 228229.5±13.43 1.10±0.25. UDP-glc. 0.39±0.01. 0.03±0.04. 303.56± 19.56. 786.62± 21.43. NAD+. 0.11±0.008. 0.18±0.001. 1591.31±48.56. 13897.89±23.54 2.30±0.11. UDP-glc. 0.65±0.03. 2.89±0.14. 25213.84± 389.56. 38313.09±23.53 1.32±0.06. NAD+. 0.09±0.004. 0.10±0.001. 901.36± 123.6. 9453.22±43.55. 1.30±0.06. UDP-glc. 0.45±0.07. 0.05±0.01. 434.67± 55.61. 814.68± 15.58. 0.68±0.13. NAD+. 0.06±0.006. 0.05±0.001. 424.06± 17.82. 7243.92± 42.32 1.53±0.039. UDP-glc. ND. ND. ND. ND. ND. NAD+. 0.22±0.02. 0.03±0.001. 261.01±12.34. 420.98±1.45. 1.08±0.052. 32. 1.39±0.04.
(41) Y249F. UDP-glc. ND. ND. ND. ND. ND. NAD. 0.12±0.002. 0.02±0.001. 174.00±11.45. 1450.07±12.34. 1.4±0.10. UDP-glc. 0.46±0.03. 0.07±0.12. 609.03±2.45. 1323.97±45.88. 1.29±0.03. NAD+. 0.14±0.01. 0.06±0.13. 522.02±3.24. 3728.7±64.67. 1.40±0.26. UDP-glc. 0.36±0.01. 0.33±0.001. 2871.14±11.11. 7975.39±36.78. 1.19±0.05. 0.31±0.04. 0.48±0.01. 4176.20±23.45. 13471.63±46.54 2.20±0.14. +. Y265F Y335F. +. NAD. The values of wild-type Ugd and mutants were obtained from the initial velocity data and nonlinear regression analysis of from V =Vmax [S]h /([S]h +Kmh) as described under “Material and Methods.” ND: Not detectable under standard reaction conditions and found less than 0.05% activity relative to wild-type enzyme after measuring activity for 200 s.. 33.
(42) Fig.1. Sequence comparison of Ugds. Sequence alignment of Ugd from K. pneumoniae, E. coli K-12, S. pyogenes, B. subtilis, E. amylovora, and S. pneumoniae is shown. The star signs indicate the position of the nine tyrosine residues mutated in this study. The conserved residues are highlighted. Secondary structural elements are shown schematically with cylinders representing α-helices and arrows representing β-strands.. 34.
(43) A.. B.. C.. 35.
(44) Fig.2. Ribbon diagram of the Ugd structure. (A) Structure of S. pyogenes Ugd. (B)The predicted structure for K . pneumoniae Ugd. (C)The nine tyrosine residues (Y10, Y91, Y150, Y210, Y217, Y242, Y249, Y265 and Y335) are shown in stick format. The Ugd catalytic site residues are shown in magenta and displayed in ball-and-stick format. N means the N terminus and C means the C terminus. The NAD+ binding site is colored yellow, and UDP-glc-binding site is colored red.. 36.
(45) A.. B.. Fig.3. Purification and western blot analysis of the recombinant Ugd proteins. The purified recombinant Ugd mutant proteins were subjected to SDS-PAGE and stained with Coomassie blue (A), and Western blotting probed with anti-(His)6 antibodies (B). Lane M: protein markers with molecular weights as indicated. Lanes 1, Ugd; 2, Y10F; 3, Y91F; 4, Y150F; 5, Y210F; 6, Y217F; 7, Y242F; 8, Y249F; 9, Y265F; 10, Y335F Ugd mutant proteins. The His-tagged Ugd proteins are indicated by an arrow.. 37.
(46) A.. B.. Fig. 4. All the Ugd mutants could be phosphorylated by KpWzc(Arg451-Lys722). In vitro phosphorylation was performed in the absence (upper) and presence (lower panel) of exogenously added KpWzc. The molecular weight markers are indicated on left. The gels were stained with Commassie Blue (left panel) and analyzed by Western blot with 4G10 antibody.. 38.
(47) A.. B.. C.. 39.
(48) D.. E.. F.. 40.
(49) G.. H.. I.. 41.
(50) J.. Fig.5. Measurement of the reaction rates of different Ugd variants. The reaction mixture contains 100 mM Tris-HCl (pH 9.0), 100 mM NaCl, 2 mM DTT, 2 mM NAD+ and 5 mM UDP-glc and Ugd or the Ugd mutant. UDP-glc was added after 20 s, and the presence of NADH detected at 340 nm. (A) Ugd WT, (B). Y10F,. (C) Y91F, (D) Y150F, (E) Y210F, (F) Y217F, (G) Y242F, (H)Y249F, (I) Y265F, (J) Y335F.. 42.
(51) Fig.6. Phosphatase activity assessment in the Ugd preparation Alkaline phosphatase activity was monitored at 37°C by using a continuous method based on the detection of p-nitrophenol formed from p-nitrophenyl phosphate (pNPP). Rates of dephosphorylation were determined at 405 nm (Sigma) and pNPP at a concentration of 0.1 mM.. 43.
(52) Time [sec]. Fig.7. Activation of Ugd activity by tyrosine phosphorylation. Ugd activity was monitored at 340 nm for 200 s by measuring NADH formation of the purified His6-Ugd (. ), CIAP treated His6-Ugd (. phosphorylated with His6–KpWzc (Arg451-Lys722) (. 44. ).. ), CIAP treated His6-Ugd.
(53) A. 1/V (mM NADH/min)-1. 200. 100. 0 0.0. 2.5. B.. 5.0. 7.5. 10.0. 12.5. 1/[UDP-glc] (mM-1). 1/V (mM NADH/min)-1. 5 4 3 2 1 0 0.0. 2.5. 5.0. 7.5. 10.0. 12.5. -1. 1/[UDP-glc] (mM ). C. 1/V (mM NADH/min)-1. 250 200 150 100 50 0 0.0. 2.5. 5.0. 7.5. 1/[UDP-glc] (mM-1). 45. 10.0. 12.5.
(54) 1/V (mM NADH/min)-1. D.. 4.5 4.0 3.5 3.0 2.5 2.0 1.5 1.0 0.5 0.0 0.0. 2.5. 5.0. 7.5. 10.0. 12.5. 10.0. 12.5. 10.0. 12.5. -1. 1/[UDP-glc] (mM ). E. 1/V (mM NADH/min)-1. 250 200 150 100 50 0 0.0. F.. 2.5. 5.0. 7.5 -1. 1/[UDP-glc] (mM ). 1/V (mM NADH/min) -1. 200. 100. 0 0.0. 2.5. 5.0. 7.5. 1/[UDP-glc] (mM-1). 46.
(55) G. 1/V (mM NADH/min)-1. 200. 100. 0 0.0. 2.5. 5.0. 7.5. 10.0. 12.5. 1/[UDP-glc] (mM-1). Fig. 8. Kinetics studies of the wild type Ugd and the derived mutants Kinetics of Ugd using variable amounts of UDP-glucose with 2 mM of NAD+. Five micrograms of the purified Ugd were used for each determination. (A) Ugd WT, (B) Y91F, (C) Y150F (D) Y210F (E) Y217F (F) Y265F (G) Y335F.. 47.
(56) A. 1/V (mM NADH/min)-1. 125 100 75 50 25 0 0. 5. 10. 15. 20. 25. 20. 25. 20. 25. 1/[NAD+ ] (mM-1). B. 1/V (mM NADH/min)-1. 1.25 1.00 0.75 0.50 0.25 0.00 0. 5. 10. 15. 1/[NAD+ ] (mM-1). C. 1/V (mM NADH/min)-1. 100 75 50 25 0 0. 5. 10. 15. 1/[NAD+ ] (mM-1). 48.
(57) D. 1/V (mM NADH/min)-1. 35 30 25 20 15 10 5 0 0. 5. 10. 15. 20. 25. 20. 25. 20. 25. 1/[NAD+ ] (mM-1). E. 1/V (mM NADH/min)-1. 50 40 30 20 10 0 0. 5. 10. 15. 1/[NAD+ ] (mM-1). F. 1/V (mM NADH/min)-1. 350 300 250 200 150 100 50 0 0. 5. 10. 15 +. -1. 1/[NAD ] (mM ). 49.
(58) G. 1/V (mM NADH/min)-1. 300. 200. 100. 0 0. 5. 10. 15. 20. 25. 20. 25. 20. 25. 1/[NAD+ ] (mM-1). H. 1/V (mM NADH/min)-1. 125 100 75 50 25 0 0. 5. 10. 15 +. -1. 1/[NAD ] (mM ). I. 1/V (mM NADH/min)-1. 100 75 50 25 0 0. 5. 10. 15. 1/[NAD+ ] (mM-1). 50.
(59) Fig. 9. Kinetics studies of the wild type Ugd and the derived mutants Kinetics of Ugd using variable amounts of NAD+ with 5 mM of UDP-glc. Five micrograms of the purified Ugd were used for each determination. (A) Ugd WT, (B) Y91F, (C) Y150F (D) Y210F (E) Y217F (F) Y242F (G) Y249F (H) Y265F (I) Y335F.. 51.
(60)
數據




相關文件
We explicitly saw the dimensional reason for the occurrence of the magnetic catalysis on the basis of the scaling argument. However, the precise form of gap depends
For pedagogical purposes, let us start consideration from a simple one-dimensional (1D) system, where electrons are confined to a chain parallel to the x axis. As it is well known
The observed small neutrino masses strongly suggest the presence of super heavy Majorana neutrinos N. Out-of-thermal equilibrium processes may be easily realized around the
incapable to extract any quantities from QCD, nor to tackle the most interesting physics, namely, the spontaneously chiral symmetry breaking and the color confinement..
(1) Determine a hypersurface on which matching condition is given.. (2) Determine a
• Formation of massive primordial stars as origin of objects in the early universe. • Supernova explosions might be visible to the most
The difference resulted from the co- existence of two kinds of words in Buddhist scriptures a foreign words in which di- syllabic words are dominant, and most of them are the
(Another example of close harmony is the four-bar unaccompanied vocal introduction to “Paperback Writer”, a somewhat later Beatles song.) Overall, Lennon’s and McCartney’s