Regulation of the Klebsiella pneumoniae Kpc
fimbriae by the site-specific recombinase KpcI
Chien-Chen Wu,
1Ying-Jung Huang,
1Chang-Phone Fung
2,3and Hwei-Ling Peng
1Correspondence Hwei-Ling Peng hlpeng@cc.nctu.edu.tw
1Department of Biological Science and Technology, National Chiao Tung University, Hsin Chu,
Taiwan, ROC
2Division of Infectious Diseases, Department of Internal Medicine, Taipei Veterans General Hospital,
Taipei, Taiwan, ROC
3School of Medicine, National Yang-Ming University, Taipei, Taiwan, ROC
Received 13 January 2010 Revised 1 April 2010 Accepted 6 April 2010
In the genome of Klebsiella pneumoniae NTUH-K2044, nine fimbrial gene clusters were identified. Besides type 1 and type 3 fimbriae, the others are novel and were named Kpa, Kpb, Kpc, Kpd, Kpe, Kpf and Kpg fimbriae. Prevalence analysis among 105 K. pneumoniae clinical isolates revealed that the kpc genes were highly associated with the K1 serotype isolates. Induced expression of the recombinant kpcABCD genes in Escherichia coli resulted in Kpc fimbriation and increased biofilm formation. A putative site-specific recombinase encoding gene kpcI and a 302 bp intergenic DNA flanked by 11 bp inverted repeats, namely kpcS, were identified in the upstream region of the kpcABCD genes. Using LacZ as the reporter, a dramatic difference in promoter activity of kpcS in two different orientations was observed and accordingly assigned as ON and OFF phase. kpcI expression was found to be able to invert kpcS in trans from phase ON to OFF and vice versa. Using the two-plasmid system, expression of kpcA, encoding the major component of the Kpc fimbriae, could be observed upon the induced expression of kpcI. These results indicate that KpcI is involved in the regulation of Kpc fimbriation in a phase-variable manner.
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative pathogen which causes suppurative lesions, bacteraemia, and urinary as well as respiratory tract infections mostly in patients with underlying diseases (Podschun & Ullmann, 1998). In Taiwan, K. pneumoniae has surpassed Escherichia coli as the predominant isolate from patients with hepatic abscesses. Although more than 77 capsular serotypes have been reported for this micro-organism (Podschun & Ullmann, 1998), isolates of the K1 serotype have been shown to be significantly associated with liver abscesses in diabetic patients (Fung et al., 2002; Lee et al., 2005). Reports of Klebsiella liver abscess (KLA) in Western countries have also been accumulating in recent years (Lederman & Crum, 2005). Besides the K1 serotype, other serotypes (mainly K2) and other virulence factors, including the mucoid factor RmpA and the siderophore aerobactin, have been reported to be important determi-nants of KLA (Yu et al., 2008). However, the correlation
between KLA and the K1 serotype remains to be investigated.
Successful establishment of infection by bacterial patho-gens requires adhesion to host cells, colonization of tissues, and in certain cases cellular invasion, followed by intracellular multiplication, dissemination to other tissues, or persistence (Pizarro-Cerda & Cossart, 2006). Fimbriae, also called pili, are hair-like appendages that extend out of the bacterial cell surface as an adherence factor. Generally, the fimbrial rod is composed of repeating subunits of pilin, the major component, and of adhesin, the minor subunit, located at the tip to mediate its specific binding activity. There are at least four mechanisms that account for the assembly of fimbriae: the chaperone–usher-dependent pathway, the general secretion pathway, the extracellular nucleation-precipitation pathway, and the alternate cha-perone pathway (Soto & Hultgren, 1999).
Several different types of fimbriae have been identified in K. pneumoniae. Type 1 fimbriae, found in virtually all members of the family Enterobacteriaceae (Klemm & Schembri, 2000), are well known for their ability to bind to mannose-containing trisaccharides of host glycoproteins. Expression of type 1 fimbriae has been closely associated Abbreviation: KLA, Klebsiella liver abscess.
A supplementary table and five supplementary figures are available with the online version of this paper.
with urinary tract infections (Kil et al., 1997; Struve et al., 2008). Type 3 fimbriae, encoded by the mrk gene cluster, are produced by the majority of K. pneumoniae strains (Allen et al., 1991) and are involved in biofilm-associated infections (Struve et al., 2009). The afimbrial adhesin CF29K, KPF-28 fimbriae, and a capsule-like extracellular afimbrial adhesin have also been reported for some K. pneumoniae strains (Di Martino et al., 1995; Favre-Bonte et al., 1995).
Many bacteria differentially express multiple types of fimbriae in order to adhere to different receptors during infection (Humphries et al., 2001; van der Velden et al., 1998; Weening et al., 2005). In Salmonella enterica serovar Typhi (S. Typhi) CT18, 12 fimbrial gene clusters of the chaperone–usher family have been found. This repertoire of fimbrial sequences has been found to be unique to the serotype Typhi, suggesting a role in the determination of specific host adaptation (Townsend et al., 2001). Several of the fimbrial gene clusters identified in the genome of enterohaemorrhagic E. coli have been shown to be more prevalent in the O157 serotype (Low et al., 2006). In order to investigate whether fimbrial type is associated with KLA pathogenesis, we initiated the study using bioinformatic tools to identify the fimbrial genes in K. pneumoniae NTUH-K2044, a liver abscess isolate of serotype K1 (Wu et al., 2009). The prevalence of the fimbrial genes was then employed in the analysis of 105 K. pneumoniae clinical isolates from different infection sites. Subsequently, the genes encoding Kpc fimbriae which appeared to be most prevalent in the K1 serotype isolates were selected for further study.
METHODS
Bacterial strains, growth conditions and serotyping.K. pneu-moniae NTUH-K2044, a highly invasive and hypermucous strain of K1 serotype (Fang et al., 2004), was provided by Dr Jin-Town Wang, National Taiwan University Hospital. The 105 K. pneumoniae clinical isolates were recovered from different tissue specimens of patients with a variety of infections at the Veterans General Hospital, Taipei, Taiwan, from 1991 to 1998. These strains have been identified and their serotypes determined, as previously described (Fung et al., 2002). Fourteen of the 18 isolates from liver abscess patients were capsular serotype K1. Bacteria were generally propagated overnight in Luria–Bertani (LB) broth with agitation at 37uC. M9 minimal medium and tryptic soy broth (TSB) were also used. The antibiotics used included ampicillin (100 mg ml21), chloramphenicol (35 mg ml21) and kanamycin (25 mg ml21). All the tissue culture media used were purchased from Gibco.
Bioinformatics.The 5.5 Mb K. pneumoniae NTUH-K2044 genome sequence (GenBank accession no. AP006725.1) has been determined and annotated (Wu et al., 2009). The fimbriae proteins were identified using the Pfam database (accession no. PF00419) and HMMER on the basis of a hidden Markov model (Bateman et al., 2004; Eddy, 1998). The fimbrial gene clusters and the neighbouring genes were analysed by homology search using the BLAST program provided online by the National Center for Biotechnology Information.
PCR detection of fimbrial genes.All the primer pairs used in this study are listed in Supplementary Table S1. The PCR mixture
contained 20 mM Tris/HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2,
200 mM of each deoxynucleoside triphosphate (dATP, dCTP, dGTP and dTTP) and 1 U recombinant Taq DNA polymerase (Violet Bioscience Inc.), along with K. pneumoniae genomic DNA and specific primers. The amplification cycle consisted of an initial 5 min hold at 95uC, followed by 35 cycles of denaturation at 95 uC for 1 min, annealing at 50uC for 1 min, and extension at 72 uC for 1.5 min, and finally an elongation step for 10 min at 72uC. The amplified PCR product was then analysed by electrophoresis on a 1 % agarose gel.
Construction of the expression plasmid pETQ. The DNA fragment containing the T5lac promoter, multiple cloning sites and the rrnB T1 transcription terminator from pQE30 (Qiagen) was inserted into pET30a (Novagen) using restriction enzymes XbaI and XhoI. The 6His-tag-coding sequence was subsequently removed by inverse PCR to yield plasmid pETQ.
Construction of fimbriae expression plasmids.The kpcABCD genes were PCR-amplified using primers pcc053 and pcc056, and the PCR product was cloned into the expression plasmids pET30a and pETQ to yield pKPC-7 and pKPC-36, respectively. The kpcABC genes were PCR-amplified using primers pcc053 and pcc203 and cloned into pETQ to yield pAW67. Genes encoding type 1 fimbriae (fimAICDEFGH) were PCR-amplified using primers pcc167 and pcc169 and cloned into pETQ to yield pAW69. The DNA containing kpcSOFF-kpcABCD was PCR-amplified by primers pcc081 and pcc056
and then cloned into yT&A (Yeastern Biotech Co.) to yield pAW73.
Transmission electron microscopy (TEM).After IPTG (0.05 mM) induction of exponential-phase E. coli Novablue (DE3) harbouring the expression plasmids pKPC-7 or pET30a for 3 h, the bacteria were collected by centrifugation, washed once with PBS, and subjected to TEM observation performed as described by Huang et al. (2006).
KpcA antiserum preparation.The coding region of kpcA was PCR-amplified using primers pcc023 and pcc024 and cloned into pET30 to yield pKPCA. Plasmid pKPCA was then introduced into E. coli Novablue (DE3) for overexpression of the His6: : KpcA recombinant
protein. The recombinant protein was expressed and purified according to the protocol in the pET manual (Novagen). KpcA antiserum was prepared by immunizing a New Zealand white rabbit with 0.5 mg of the purified His6: : KpcA recombinant protein and the
immunized rabbit was exsanguinated on day 45. The specificity of the obtained antibody was confirmed using Western blotting (Supplementary Fig. S1).
Anti-KpcA Western blot hybridization and immunofluores-cence microscopy.Induction of the kpc genes was carried out by adding 0.5 mM IPTG to an exponential-phase E. coli HB101 [pKPC-36] culture and incubating for an additional 3 h. Bacterial lysates were resolved by SDS-PAGE; the Western blot analysis and immuno-fluorescence microscopy using anti-KpcA antiserum were performed as previously described (Huang et al., 2009).
Biofilm formation assay. Bacteria diluted 1 : 100 in LB broth supplemented with 25 mg kanamycin ml21and 0.5 mM IPTG were inoculated into each well of a 96-well microtitre dish (Orange Scientific) and statically incubated at 37uC for 48 h. The biofilm-forming ability of bacteria was analysed as described by Huang et al. (2009).
Construction of the KpcI expression plasmid.The DNA fragment encoding KpcI196 was PCR-amplified from K. pneumoniae
NTUH-K2044 by primers pcc183 and pcc150, and cloned into pBAD33 (Guzman et al., 1995) to generate pKPCI196. pKPCI196was transformed
into K. pneumoniae NTUH-K2044, and 0.4 %L-arabinose was added to the culture of the transformant for the induction of KpcI196expression. Switch orientation assay. The kpcS (switch region) was PCR-amplified from the tested strain using the primers pcc081 and pcc082. The amplified product was then purified and digested with AflII. The restricted fragments were separated on 2 % agarose gels and the pattern was visualized for the determination of orientation by staining with ethidium bromide. The DNA fragments containing kpcSONand
kpcSOFFwere PCR-amplified from L-arabinose-induced K.
pneumo-niae NTUH-K2044[pKPCI196] using primers pcc081 and pcc082, and
subsequently cloned into yT&A to yield the ON and pKPC-OFF plasmids, respectively.
b-Galactosidase assay. kpcS DNA was PCR-amplified from K. pneumoniae NTUH-K2044 by primers YCY001 and YCY002, and cloned into the promoter reporter plasmid placZ15 (Lin et al., 2006) to yield plasmids pSY003 and pSY004, containing kpcS without the two inverted repeats in opposite orientations. pSY003 and pSY004 were introduced into the K. pneumoniae NTUH-K2044 DlacZ strain CCW01, which was constructed by using the gene replacement vector pKOV (Link et al., 1997). One-tenth of an overnight culture of the bacteria carrying either plasmid was subcultured in LB broth to OD600~0.6–0.7. The b-galactosidase activity assay was carried out as
described by Lin et al. (2006).
Statistical methods. An unpaired t test was used to determine statistical significance, and values of P,0.001 were considered significant. The results of the biofilm-forming activity and b-galactosidase activity assays were derived from a single experiment that was representative of three independent experiments. Each sample was assayed in triplicate and the mean activity and standard deviation are presented.
RESULTS
Identification of the fimbrial gene clusters in K. pneumoniae NTUH-K2044
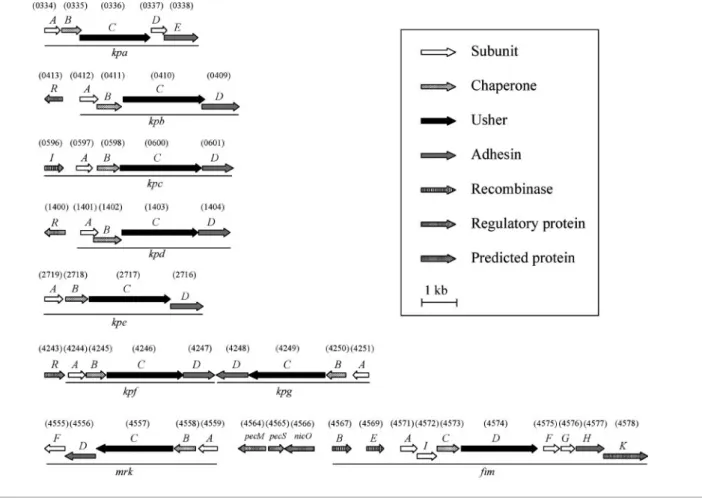
Nine fimbrial gene clusters were identified using the HMMER search of the genome of K. pneumoniae NTUH-K2044. Each contained at least four genes, encoding a putative major pilin, a chaperone, an usher and an adhesin for the biosynthesis of fimbriae belonging to the chaperone–usher-dependent assembly. As shown in Fig. 1, these included the type 1 and type 3 fimbrial gene clusters fim and mrk, and seven novel ones, namely kpa, kpb, kpc, kpd, kpe, kpf and kpg. Multiple sequence alignment byCLUSTAL Wshowed that the amino acid sequences of these pilins and adhesins shared 26.5–36.4 % similarity; chaperones and ushers shared a higher similarity ranging from 49.3 to 55.4 %. The mrk–fim fimbrial genes are clustered and transcribed divergently (Fig. 1). This gene organization including the pecM, pecS and nicO homologues has been found to be conserved in the genomes of K. pneumoniae C3091, MGH78578 and 342 (Fouts et al., 2008; Ogawa et al., 2005; Struve et al., 2009). Interestingly, the gene clusters kpf and kpg are also linked physically but transcribed convergently.
BLASTanalysis, using the sequences of the nine fimbrial gene clusters identified in K. pneumoniae NTUH-K2044 as templates, of the genome of K. pneumoniae strains
MGH78578 and 342 (Fouts et al., 2008; Ogawa et al., 2005) showed that, except for kpc and kpf, the genes were conserved in the three genomes. No homologue of the kpc genes was found in strains MGH78578 and 342, while homologues of the kpf genes were found in the genome of MGH78578 but not that of 342.
PCR screening for the presence of the fimbrial genes
To investigate the prevalence of the nine fimbrial gene clusters among K. pneumoniae strains, a total of 105 K. pneumoniae clinical isolates from different infection sites were collected. Two specific primer pairs corresponding to the pilin- and adhesin-encoding genes were designed for PCR detection. Prevalence was determined on the basis of the presence of the PCR amplicons (Supplementary Fig. S2). The analysis revealed the presence of the kpa, kpd, kpe, kpg, fim and mrk genes in most of the isolates, and the prevalence percentages were 99, 82, 93, 97, 84 and 100 %, respectively. The prevalence percentages for the kpb, kpc and kpf genes were lower at 52, 33 and 70 % of the isolates, respectively. No obvious correlation between fimbrial type and disease could be identified. However, the kpb and kpc genes were shown to be more prevalent in K1 isolates (P,0.0001). As shown in Table 1, most of the clinical isolates of the K1 serotype harboured kpb and kpc genes, while non-K1 isolates carrying the kpb and kpc genes were much less frequent (32 and 1 %, respectively). The close association of kpc fimbriae with the K1 serotype prompted the selection of the kpc fimbriae for further study of the KLA pathogenic mechanism.
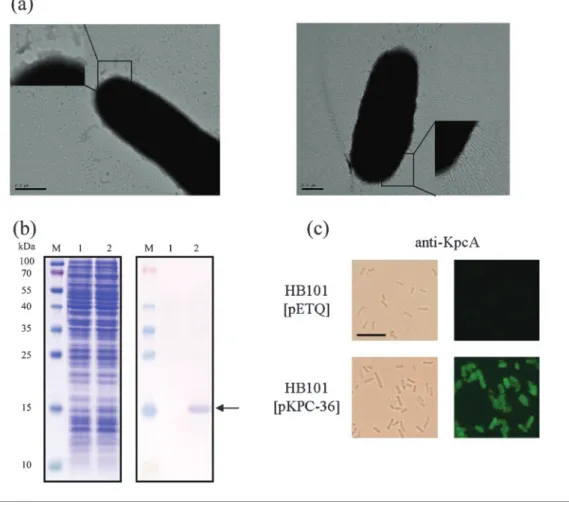
Display of the Kpc fimbriae on the E. coli surface In order to determine whether the kpcABCD genes encode a fimbrial apparatus, an E. coli fimbriae display system was used. The kpcABCD genes were PCR-amplified and cloned into pET30a, designated pKPC-7, to allow controlled expression by IPTG induction. Numerous thin and rigid fimbriae on the surface of E. coli Novablue (DE3) harbouring pKPC-7 could be observed, while E. coli harbouring pET30a was afimbriate (Fig. 2). Expression of KpcA, the putative major pilin, could also be detected by Western blot analysis (Supplementary Fig. S1). These results indicated that the kpcABCD genes are sufficient to produce a fimbrial apparatus. However, the growth rate of E. coli harbouring pKPC-7, in comparison with that of E. coli Novablue (DE3) [pET30a], was obviously decreased even without IPTG induction. This could be a result of the overexpression of kpc genes, especially kpcA (Supplementary Fig. S1, lane 3). In case the biased growth rate impeded the characterization of the Kpc fimbriae, another expression plasmid, pETQ, which can be used for protein expression under T5lac promoter control in E. coli and K. pneumoniae, was used. The pETQ plasmid carrying the kpcABCD genes, pKPC-36, was then transformed into E. coli HB101, which is an afimbriate bacterium. Expression of the kpcABCD genes
was tightly controlled under the T5lac promoter, and no obvious difference in growth rate between E. coli HB101 harbouring pKPC-36 and E. coli HB101 harbouring pETQ
was noted. Using polyclonal anti-KpcA antibodies, Western blot analysis revealed that KpcA was expressed (Fig. 2b), and fimbriation on the surface of the recombinant E. coli HB101
Table 1. Prevalence of kpb and kpc genes in K. pneumoniae isolates
K. pneumoniae isolate No. of strains containing kpb/total no. of strains isolated (%)
No. of strains containing kpc/total no. of strains isolated (%)
K1 serotype Other serotype K1 serotype Other serotype
Liver abscess 14/14 3/4 14/14 1/4 Bile 2/7 1/7 Urine 1/1 3/13 1/1 0/13 Sputum 3/3 3/11 2/3 1/11 Wound 6/6 5/15 6/6 1/15 Blood 5/5 4/17 5/5 2/17 Ascites 4/9 1/9 Total 29/29 (100)* 24/76 (32)* 28/29 (97)* 7/76 (1)* *P,0.0001.
Fig. 1. Fimbrial gene clusters of the chaperone–usher-dependent assembly class in K. pneumoniae NTUH-K2044. The designation of putative fimbrial genes and the locus tag (KP1_number) of ORFs annotated in the K. pneumoniae NTUH-K2044 genome are indicated. A total of nine fimbrial gene clusters and genes encoding putative regulators are shown. Each of the putative fimbrial operons is underlined. The putative functions of the ORFs are also shown.
was also observed by fluorescence microscopy (Fig. 2c). E. coli HB101[pKPC-36] was thus used for the characterization of the recombinant Kpc fimbriae.
Expression of Kpc fimbriae increased biofilm-forming activity
Activity assessment of the recombinant Kpc fimbriae, including haemagglutination, cell adherence and biofilm formation, was subsequently carried out. However, no haemagglutination activity against red blood cells from guinea pig, rabbit or human (type A and type B) could be observed for the recombinant bacteria. Besides human epithelial cell lines Int407 (intestine), HCT-8 (intestine), Hep-2 (larynx) and T24 (bladder), two human hepatocel-lular liver carcinoma cell lines, HepG2 and SK-HEP-1, were used to assess the cell adherence activity of the Kpc fimbriae. The cell adherence assay was performed as described by Huang et al. (2006). Although the recombin-ant E. coli HB101 exhibited differential adherence to these cell lines, Kpc fimbriation did not increase the adherence
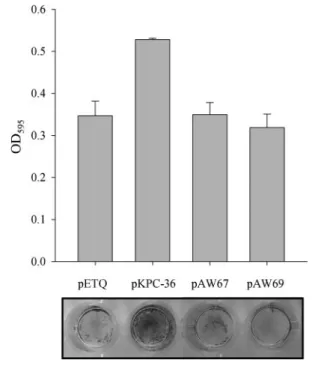
activity of the bacteria to any of the cells. On the other hand, as shown in Fig. 3, E. coli HB101 harbouring pKPC-36 exhibited a higher level of biofilm-forming activity on the abiotic surface than that observed for E. coli HB101 [pETQ], as assessed by direct observation after crystal violet staining or by quantitative measurement (P,0.001). However, E. coli HB101 harbouring the type 1 fimbriae expression plasmids pAW69 or pAW67 (pETQ carrying the kpcABC genes) exhibited levels of biofilm-forming activity similar to those of the bacteria carrying pETQ. This suggested a specific binding of the Kpc fimbriae to polystyrene wells and a positive role of Kpc fimbriae in biofilm formation. The expression of type 1 fimbriae encoded on pAW69 was also confirmed by yeast agglu-tination analysis (Supplementary Fig. S3b).
The KpcI recombinase is probably the regulator for the Kpc fimbriae
A gene encoding a putative DNA recombinase of the lambda integrase family (Esposito & Scocca, 1997;
Nunes-Fig. 2. Expression of Kpc fimbriae on recombinant E. coli. (a) Transmission electron micrographs of recombinant Kpc fimbriae. Left panel, E. coli Novablue (DE3) [pET30a]; right panel, E. coli Novablue (DE3) [pKPC-7]. Bars, 0.5 mm. (b) Anti-KpcA Western blot analysis of E. coli HB101 harbouring pETQ (lane 1) or pKPC-36 (lane 2). The expressed KpcA is indicated by an arrow. (c) Bright-field (left panel) and anti-KpcA immunofluorescence (right panel) microscopic analysis of E. coli HB101 harbouring pETQ or pKPC-36 (magnification ¾630). Bar, 10 mm.
Duby et al., 1998) was identified in the upstream region of the kpcABCD genes (Fig. 1) and was named kpcI. Multiple sequence alignment of the family of site-specific recombi-nases, including FimB/E of the type 1 fimbriae, MrpI of the MR/P fimbriae, FotS/T of the CS18 fimbriae and KpcI, revealed four conserved residues, R42, H136, R139 and Y171, of the tyrosine recombinase family (Supplementary Fig. S4) (Abremski & Hoess, 1992; Han et al., 1994). Between kpcI and kpcA, a 302 bp region flanked by a pair of 11 bp inverted repeats (IRs) was identified (Fig. 4). These findings suggested a recombinase-mediated phase variation
control of Kpc fimbriae, as reported for type 1 fimbriae, MR/P fimbriae and CS18 fimbriae (Honarvar et al., 2003; Klemm, 1986; Li et al., 2002). KpcI is thus predicted to be able to invert the DNA segment between the two inverted repeats to regulate the expression of Kpc fimbriae. The putative invertible DNA segment was designated kpcS (switch region).
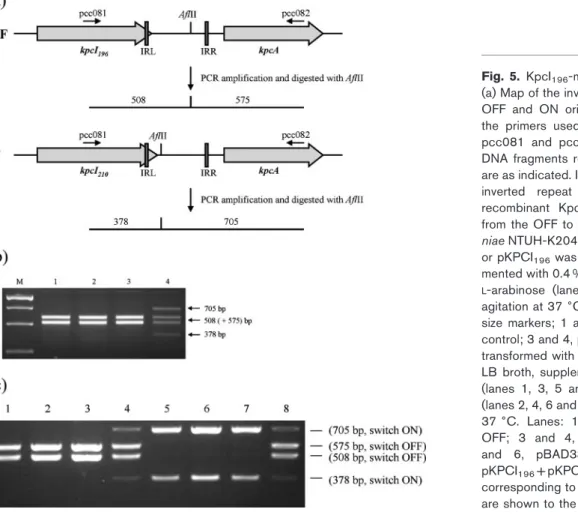
Expression of kpcI leads to inversion of kpcS A PCR-based switch orientation assay (Fig. 5a) was subsequently employed to detect the kpcS inversions in K. pneumoniae NTUH-K2044 grown in various growth condi-tions, including LB broth, TSB and M9 minimal medium with or without agitation at 25, 30 and 37uC. Since enteropathogenic E. coli bundle-forming pili are expressed when the bacteria are grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Vuopio-Varkila & Schoolnik, 1991), several tissue-culture media, including DMEM, Roswell Park Memorial Institute (RPMI) medium, minimum essential medium (MEM), Basal Medium Eagle (BME) and McCoy’s 5a medium, were also used to grow K. pneumoniae NTUH-K2044 (statically incubated at 37uC in 5 % CO2). However, the kpcS inversion was not observed under all the above growth conditions. The expression of KpcA was also not detected by Western blot analysis, suggesting an ‘OFF’ phase of kpcS (kpcSOFF) in K. pneumoniae NTUH-K2044. In the kpcSOFF phase, a polypeptide of 196 amino acid residues, KpcI196, could be deduced (Supplementary Fig. S4). To determine whether expression of the KpcI recombinase could invert the DNA, pKPCI196,an expression plasmid encoding KpcI196 driven by an araBAD promoter, was transformed into K. pneumoniae NTUH-K2044. As shown in Fig. 5(b), the addition ofL-arabinose to induce expression of KpcI196 was able to invert the DNA. After the inversion, the DNA was isolated, sequenced and named kpcSON (Fig. 4). To assess the promoter activity of kpcSON or kpcSOFF, each DNA was cloned in front of the promoterless lacZ gene in pLacZ15 by transcriptional fusion, and the resulting plasmids were named pSY003 and pSY004, respectively. After the transformation of pSY003 or pSY004 into K. pneumoniae
Fig. 3. Biofilm-forming ability of E. coli expressing the Kpc fimbriae. The development of biofilms of E. coli HB101 harbouring pETQ, pKPC-36, pAW67 and pAW69 was observed (lower panel) and quantified (upper panel) as described in Methods. A higher biofilm-forming activity could be observed for E. coli HB101 [pKPC-36].
Fig. 4. Sequence analysis of the putative promoter region of the kpc gene cluster. The 500 bp upstream and 50 bp downstream regions of the kpcA translation start codon ATG (underlined) in the kpcSONphase are shown. The 11 bp inverted
repeats are outlined by square boxes. The predicted ”10 and ”35 promoter regions are shaded. The kpcI210translation stop
NTUH-K2044 DlacZ strain CCW01, the LacZ activity of the transformants was measured. The kpcSON phase activity (21 359.5±233.4 Miller units) was much higher than that of kpcSOFF (109.2±3.2 Miller units) and pLacZ15 (111.6±1 Miller units), implying that the orientation of kpcS determines the transcription efficiency of the kpc genes. Effect of the recombinant KpcI196on the
switching of kpcS
The fimbrial recombinases have been shown to have different activities towards the orientation control of the invertible element. The FimB recombinase inverts the fim switch in the ON-to-OFF and OFF-to-ON directions, whereas FimE inverts it predominantly in the ON-to-OFF direction (Blomfield et al., 1991; Gally et al., 1993; McClain et al., 1991); MrpI is able to invert the mrp switch in both directions (Zhao et al., 1997). As shown in Fig. 5(b), the induced expression of KpcI196inverted kpcS in the OFF-to-ON direction. Since the growth condition for K. pneumo-niae to switch ON kpcS is yet to be identified, a two-plasmid system in E. coli was used to analyse the activity of KpcI196towards kpcS inversion in both directions. pKpcI196 was introduced into E. coli JM109 harbouring either pKPC-ON or pKPC-OFF. As shown in Fig. 5(c), the induced
expression of KpcI196in the recombinant E. coli was able to invert kpcS in both the ON-to-OFF and OFF-to-ON directions.
KpcI-mediated expression of KpcA
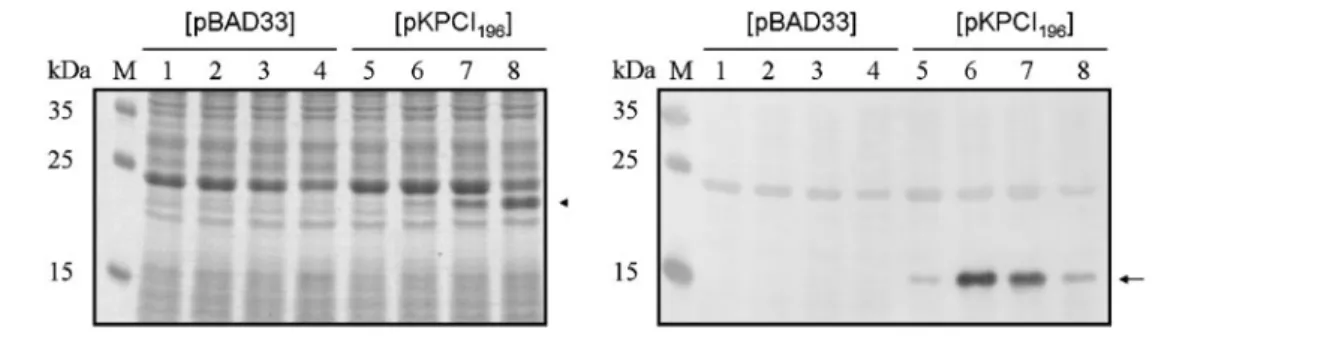
Although the recombinant KpcI196was able to switch ON kpcS, the induced expression of KpcI196 in K. pneumoniae did not lead to the expression of KpcA (data not shown). This result implies the involvement of further regulators in regulation of the expression of Kpc fimbriae. To avoid possible factors in K. pneumoniae that impede the expression of KpcA, another two-plasmid system was constructed to analyse the KpcI-mediated phase variation in E. coli. The DNA fragment containing the kpcSOFF– kpcABCD region was PCR-amplified and cloned into plasmid yT&A to yield pAW73. Subsequently, pKPCI196 was introduced into E. coli harbouring pAW73. Upon induction with L-arabinose, the induced expression of KpcI196 was able to switch ON the expression of KpcA as detected by anti-KpcA antiserum (Fig. 6). This indicated that the KpcI196-mediated phase variation was able to control the expression of KpcA.
To investigate whether the production of KpcA could only be detected in the E. coli system (Fig. 6) but not in the
Fig. 5. KpcI196-mediated inversion of kpcS.
(a) Map of the invertible region, kpcS, in both OFF and ON orientations. The positions of the primers used in the PCR amplification, pcc081 and pcc082, and the sizes of the DNA fragments resulting from AflII digestion are as indicated. IRL, inverted repeat left; IRR, inverted repeat right. (b) Expression of recombinant KpcI196 resulted in a switch
from the OFF to the ON phase. K. pneumo-niae NTUH-K2044 transformed with pBAD33 or pKPCI196was grown in M9 broth
supple-mented with 0.4 % glucose (lanes 1 and 3) or
L-arabinose (lanes 2 and 4) for 16 h with
agitation at 37 6C. Lanes: M, DNA molecular size markers; 1 and 2, pBAD33 vector as a control; 3 and 4, pKPCI196. (c) E. coli JM109
transformed with two plasmids was grown in LB broth, supplemented with 0.4 % glucose (lanes 1, 3, 5 and 7) or 0.4 % L-arabinose (lanes 2, 4, 6 and 8), for 16 h with agitation at 37 6C. Lanes: 1 and 2, pBAD33 +pKPC-OFF; 3 and 4, pKPCI196+pKPC-OFF; 5
and 6, pBAD33+pKPC-ON; 7 and 8, pKPCI196+pKPC-ON. The fragment sizes
corresponding to the position of the switches are shown to the right of the panel.
phase ON K. pneumoniae cells, which may be due to no transcription of mRNA, or the instability of kpcA mRNA or the KpcA protein, plasmid pKPC-36 was introduced into K. pneumoniae NTUH-K2044. KpcA production was readily observed upon IPTG induction (Supplementary Fig. S5), which implied that transcription of kpcA was low in the ON phase of K. pneumoniae cells. Further studies will be needed to identify regulators of Kpc fimbriae expres-sion.
DISCUSSION
A large number of fimbrial gene clusters are known to be present in a bacterial genome (Low et al., 2006; Townsend et al., 2001; Vallet et al., 2001). It has been demonstrated that multiple types of fimbriae are required for the full virulence and long-term persistence of Salmonella enterica serovar Typhimurium in mice (Humphries et al., 2005; van der Velden et al., 1998; Weening et al., 2005). Although these fimbrial gene clusters identified using bioinformatic tools should be considered as putative ones, we speculate that, in addition to the type 1 and type 3 fimbriae, the others may also be required for K. pneumoniae infection in different environments.
A prevalence study has indicated that S. Typhi possesses a unique repertoire of fimbrial gene sequences (Townsend et al., 2001). A specific repertoire of fimbrial operons was therefore proposed as a complex virulence factor involved in infections (Humphries et al., 2001). Although no obvious correlation between disease types and fimbrial types was noted in the K. pneumoniae clinical isolates, we found that out of 29 K. pneumoniae K1 isolates, 28 possessed an identical repertoire of fimbrial gene clusters. Since a correlation of the K1 serotype with KLA has been reported, the conserved fimbrial repertoire possessed by the K. pneumoniae strains of serotype K1 may also play a role in pathogenesis.
Like many of the fimbrial operons identified that are not expressed in vitro (Nuccio et al., 2007), the expression of Kpc fimbriae also could not be observed with various environmental stimuli, including temperature, starvation and aeration. Western blot analysis using the anti-KpcA antiserum was employed to investigate whether the 35 K. pneumoniae clinical isolates which possessed kpc genes (Table 1) expressed Kpc fimbriae. However, no expression of Kpc fimbriae could be identified for the bacteria grown statically overnight in LB broth or M9 medium at 25 or 37uC (data not shown). To further characterize the Kpc fimbriae, the kpcABCD genes were heterologously expressed in the afimbriate E. coli HB101. The displayed Kpc fimbriae were shown to confer upon the recombinant E. coli a higher biofilm-forming activity. Bacterial biofilm formation on indwelling devices, such as catheters and endotracheal tubes, is a significant medical problem. The Kpc fimbriae may play a role in the development of infections in catheterized patients. However, this possibility awaits further investigation.
As shown in Fig. 5(c), the recombinant KpcI196possessed activity to switch kpcS in both directions; therefore, the induced expression of KpcI196in K. pneumoniae may lead to both ON-to-OFF and OFF-to-ON inversions that occur simultaneously in different cells in the bacterial population (Fig. 5b, lane 4). However, whether KpcI196 could invert kpcS in the ON-to-OFF direction in K. pneumoniae remains to be investigated. Unlike all the characterized ‘fimbrial recombinases’, in which the inverted repeat left (IRL) is located in the non-coding region between the recombinase-encoding gene and the fimbrial operon, the IRL of kpcS is located in the coding region of kpcI (Fig. 4). The inversion of kpcSOFFcould result in a polypeptide of 210 residues, KpcI210 (Supplementary Fig. S4). The DNA fragment containing kpcI210 could be amplified by PCR from the genomic DNA of the L-arabinose-induced K. pneumoniae NTUH-K2044[pKPCI196]. However, when the plasmid containing the PCR amplicon of kpcI210 was
Fig. 6. KpcI196-mediated expression of KpcA in E. coli. Plasmid pBAD33 or pKPCI196, as marked above the panels, was
introduced into E. coli JM109[pAW73]. E. coli carrying the two plasmids was grown in LB broth, and when growth reached mid-exponential phase, the expression of KpcI196was induced by varying concentrations ofL-arabinose. Lanes: 1 and 5, no
induction; 2 and 6, 0.002 %; 3 and 7, 0.02 %; 4 and 8, 0.2 %L-arabinose induction. After 3 h induction, the bacteria were analysed by SDS-PAGE (left panel) and anti-KpcA Western blot hybridization (right panel). The expression of KpcI196
introduced into E. coli JM109, unexpected point mutations and recombination adjacent to the IRL were found in the kpcI210-coding region. In contrast, we found no evidence of mutations or recombination in pKPCI196. recA-independ-ent recombination has also been described in the study of FimE recombinase (McClain et al., 1991). Since the IRL of kpcS is located in the coding region of kpcI210, and the expression of kpcI210changes its own coding sequence, the activity of KpcI210 is difficult to assess and remains to be studied.
In this study, we report the identification of nine fimbrial gene clusters in the genome of K. pneumoniae NTUH-K2044. The Kpc fimbria was found to be most prevalent in the clinical isolates of serotype K1. The heterologous expression of the kpcABCD genes in E. coli resulted in Kpc fimbriation and an increased biofilm-forming activity. Moreover, the involvement of KpcI in the regulation of Kpc fimbriation in a phase-variable manner was elucidated.
ACKNOWLEDGEMENTS
We thank Dr Shih-Feng Tsai’s group in the Division of Molecular and Genomic Medicine, National Health Research Institutes, for provid-ing the genome sequence data of K. pneumoniae NTUH-K2044 prior to publication. We are also grateful to Dr Jin-Town Wang, National Taiwan University Hospital, for providing K. pneumoniae NTUH-K2044. The work is supported by grants from the National Research Program for Genomic Medicine (NPGRM) of the National Science Council (NSC 96-3112-B-009-004 and NSC 97-2320-B-009-001-MY3) and from the Veterans General Hospital University System of Taiwan Joint Research Program (VGHUST94-P5-27).
REFERENCES
Abremski, K. E. & Hoess, R. H. (1992). Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng 5, 87–91.
Allen, B. L., Gerlach, G. F. & Clegg, S. (1991).Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J Bacteriol 173, 916–920.
Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S. & other authors (2004). The Pfam protein families database. Nucleic Acids Res 32, D138–D141.
Blomfield, I. C., McClain, M. S., Princ, J. A., Calie, P. J. & Eisenstein, B. I. (1991).Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J Bacteriol 173, 5298–5307.
Di Martino, P., Bertin, Y., Girardeau, J. P., Livrelli, V., Joly, B. & Darfeuille-Michaud, A. (1995). Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involved in nosocomial infections. Infect Immun 63, 4336– 4344.
Eddy, S. R. (1998).Profile hidden Markov models. Bioinformatics 14, 755–763.
Esposito, D. & Scocca, J. J. (1997).The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res 25, 3605–3614.
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C. & Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains
causing primary liver abscess and septic metastatic complications. J Exp Med 199, 697–705.
Favre-Bonte, S., Darfeuille-Michaud, A. & Forestier, C. (1995).
Aggregative adherence of Klebsiella pneumoniae to human intestine-407 cells. Infect Immun 63, 1318–1328.
Fouts, D. E., Tyler, H. L., DeBoy, R. T., Daugherty, S., Ren, Q., Badger, J. H., Durkin, A. S., Huot, H., Shrivastava, S. & other authors (2008).
Complete genome sequence of the N2-fixing broad host range
endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet 4, e1000141.
Fung, C. P., Chang, F. Y., Lee, S. C., Hu, B. S., Kuo, B. I., Liu, C. Y., Ho, M. & Siu, L. K. (2002).A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50, 420–424.
Gally, D. L., Bogan, J. A., Eisenstein, B. I. & Blomfield, I. C. (1993).
Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol 175, 6186–6193.
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995).Tight regulation, modulation, and high-level expression by vectors contain-ing the arabinose PBADpromoter. J Bacteriol 177, 4121–4130. Han, Y. W., Gumport, R. I. & Gardner, J. F. (1994). Mapping the functional domains of bacteriophage lambda integrase protein. J Mol Biol 235, 908–925.
Honarvar, S., Choi, B. K. & Schifferli, D. M. (2003).Phase variation of the 987P-like CS18 fimbriae of human enterotoxigenic Escherichia coli is regulated by site-specific recombinases. Mol Microbiol 48, 157–171.
Huang, Y. J., Wu, C. C., Chen, M. C., Fung, C. P. & Peng, H. L. (2006).
Characterization of the type 3 fimbriae with different MrkD adhesins: possible role of the MrkD containing an RGD motif. Biochem Biophys Res Commun 350, 537–542.
Huang, Y. J., Liao, H. W., Wu, C. C. & Peng, H. L. (2009).MrkF is a component of type 3 fimbriae in Klebsiella pneumoniae. Res Microbiol 160, 71–79.
Humphries, A. D., Townsend, S. M., Kingsley, R. A., Nicholson, T. L., Tsolis, R. M. & Baumler, A. J. (2001).Role of fimbriae as antigens and intestinal colonization factors of Salmonella serovars. FEMS Microbiol Lett 201, 121–125.
Humphries, A., Deridder, S. & Baumler, A. J. (2005). Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect Immun 73, 5329–5338.
Kil, K. S., Darouiche, R. O., Hull, R. A., Mansouri, M. D. & Musher, D. M. (1997). Identification of a Klebsiella pneumoniae strain associated with nosocomial urinary tract infection. J Clin Microbiol 35, 2370–2374.
Klemm, P. (1986).Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J 5, 1389–1393.
Klemm, P. & Schembri, M. A. (2000).Bacterial adhesins: function and structure. Int J Med Microbiol 290, 27–35.
Lederman, E. R. & Crum, N. F. (2005).Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol 100, 322–331.
Lee, C. H., Leu, H. S., Wu, T. S., Su, L. H. & Liu, J. W. (2005).Risk factors for spontaneous rupture of liver abscess caused by Klebsiella pneumoniae. Diagn Microbiol Infect Dis 52, 79–84.
Li, X., Lockatell, C. V., Johnson, D. E. & Mobley, H. L. (2002).
Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol Microbiol 45, 865–874.
Lin, C. T., Huang, T. Y., Liang, W. C. & Peng, H. L. (2006).Homologous response regulators KvgA, KvhA and KvhR regulate the synthesis of capsular polysaccharide in Klebsiella pneumoniae CG43 in a coordinated manner. J Biochem 140, 429–438.
Link, A. J., Phillips, D. & Church, G. M. (1997).Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179, 6228–6237.
Low, A. S., Holden, N., Rosser, T., Roe, A. J., Constantinidou, C., Hobman, J. L., Smith, D. G., Low, J. C. & Gally, D. L. (2006).Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157 : H7. Environ Microbiol 8, 1033–1047.
McClain, M. S., Blomfield, I. C. & Eisenstein, B. I. (1991).Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol 173, 5308–5314.
Nuccio, S. P., Chessa, D., Weening, E. H., Raffatellu, M., Clegg, S. & Baumler, A. J. (2007). SIMPLE approach for isolating mutants expressing fimbriae. Appl Environ Microbiol 73, 4455–4462.
Nunes-Duby, S. E., Kwon, H. J., Tirumalai, R. S., Ellenberger, T. & Landy, A. (1998).Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res 26, 391–406.
Ogawa, W., Li, D. W., Yu, P., Begum, A., Mizushima, T., Kuroda, T. & Tsuchiya, T. (2005).Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol Pharm Bull 28, 1505–1508.
Pizarro-Cerda, J. & Cossart, P. (2006).Bacterial adhesion and entry into host cells. Cell 124, 715–727.
Podschun, R. & Ullmann, U. (1998).Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and patho-genicity factors. Clin Microbiol Rev 11, 589–603.
Soto, G. E. & Hultgren, S. J. (1999).Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol 181, 1059–1071.
Struve, C., Bojer, M. & Krogfelt, K. A. (2008).Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 76, 4055–4065.
Struve, C., Bojer, M. & Krogfelt, K. A. (2009). Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun 77, 5016–5024.
Townsend, S. M., Kramer, N. E., Edwards, R., Baker, S., Hamlin, N., Simmonds, M., Stevens, K., Maloy, S., Parkhill, J. & other authors (2001). Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun 69, 2894–2901.
Vallet, I., Olson, J. W., Lory, S., Lazdunski, A. & Filloux, A. (2001).The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci U S A 98, 6911–6916.
van der Velden, A. W., Baumler, A. J., Tsolis, R. M. & Heffron, F. (1998).Multiple fimbrial adhesins are required for full virulence of Salmonella Typhimurium in mice. Infect Immun 66, 2803–2808.
Vuopio-Varkila, J. & Schoolnik, G. K. (1991).Localized adherence by enteropathogenic Escherichia coli is an inducible phenotype associated with the expression of new outer membrane proteins. J Exp Med 174, 1167–1177.
Weening, E. H., Barker, J. D., Laarakker, M. C., Humphries, A. D., Tsolis, R. M. & Baumler, A. J. (2005).The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun 73, 3358– 3366.
Wu, K. M., Li, L. H., Yan, J. J., Tsao, N., Liao, T. L., Tsai, H. C., Fung, C. P., Chen, H. J., Liu, Y. M. & other authors (2009). Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191, 4492–4501.
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, C. C., Lai, C. C. & Chuang, Y. C. (2008).Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 62, 1–6.
Zhao, H., Li, X., Johnson, D. E., Blomfield, I. & Mobley, H. L. (1997).In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol Microbiol 23, 1009–1019.