0021-9193/11/$12.00 doi:10.1128/JB.05359-11
Copyright © 2011, American Society for Microbiology. All Rights Reserved.
Negative Effect of Glucose on ompA mRNA Stability: a Potential Role
of Cyclic AMP in the Repression of hfq in Escherichia coli
䌤
†
Hsiao-Hsien Lin, Chi-Cheng Hsu, Chi-Dung Yang, Yih-Wei Ju, Yi-Pei Chen, and Ching-Ping Tseng*
Department of Biological Science and Technology, College of Biological Science and Technology,National Chiao Tung University, Hsinchu, Taiwan, Republic of China
Received 20 May 2011/Accepted 25 July 2011
Glucose is a carbon source that is capable of modulating the level of cyclic AMP (cAMP)-regulated genes. In the present study, we found that the stability of ompA mRNA was reduced in Escherichia coli when glucose (40 mM) was present in Luria-Bertani (LB) medium. This effect was associated with a low level of cAMP induced by the glucose. The results were confirmed with an adenylyl cyclase mutant with low levels of cAMP that are not modulated by glucose. Northern blot and Western blot analyses revealed that the host factor I (Hfq) (both mRNA and protein) levels were downregulated in the presence of cAMP. Furthermore, we showed that a complex of cAMP receptor protein (CRP) and cAMP binds to a specific P3hfqpromoter region of hfq and
regulates hfq expression. The regulation of the hfq gene was confirmed in vivo using an hfq-deficient mutant transformed with an exogenous hfq gene containing the promoter. These results demonstrated that expression of hfq was repressed by the CRP-cAMP complex. The presence of glucose resulted in increased Hfq protein levels, which decreased ompA mRNA stability. An additional experiment showed that cAMP also increased the stability of fur mRNA. Taken together, these results suggested that the repression of Hfq by cAMP may contribute to the stability of other mRNA in E. coli.
The degradation of mRNA appears to be a major player in controlling the level of gene expression, in addition to the regulation of transcription and translation. The stability of mRNA from a single gene can be modulated in response to growth conditions, environmental signals, and translational ef-ficiency. Therefore, the regulation of mRNA stability can pro-vide a means to actively adapt the cellular translation to the changes in growth conditions.
The degradation of the outer membrane protein A (OmpA) mRNA is used as a model to study the regulation of mRNA decay in Escherichia coli. OmpA is abundant in the outer membrane, with a copy number of⬃100,000 per cell, and plays important roles in a variety of functions (30). Previous publi-cations have shown that the stability of ompA mRNA increased as the cell growth rate increased due to the medium composi-tion (25). In other words, the stability of mRNA in bacteria can be adjusted in response to growth conditions. The 5⬘ untrans-lated region (5⬘ UTR) of the ompA transcript is important for regulating mRNA stability in response to changing growth rates (8, 11). For example, the binding of host factor I (Hfq), an RNA chaperon protein, to the 5⬘ UTR of the ompA tran-script negatively regulates growth rate-dependent degradation of ompA mRNA (36). Hfq stimulates ompA mRNA decay by binding to an A/U-rich sequence of the ompA 5⬘ UTR region which overlaps an endoribonuclease RNase E (rne) cleavage site and thus appears to interfere with ribosome binding, by which Hfq stimulates ompA mRNA decay (1, 22, 37).
Glucose is a major source of carbon for the growth of E. coli. For glucose metabolism, E. coli has developed pleiotropic reg-ulators for the transcriptional regulation, such as cyclic AMP (cAMP) and its receptor protein (CRP) complex. It is known that cAMP forms a complex with CRP (cAMP-CRP complex) that binds to a specific promoter region of genes to either activate or inhibit transcription depending upon the position bound by the complex relative to the promoter (3). As a result, the cAMP-CRP complex regulates the expression of hundreds of genes (10, 38). However, little is known about the relation-ship between glucose regulation and hfq expression in post-transcriptional regulation. In this study, we found that glucose significantly enhanced the decay of ompA mRNA with a half-life of⬃11.5 to ⬃8.1 min. This change in the decay rate was associated with a decrease in cAMP levels. Given the fact that Hfq is involved in the destabilization of ompA mRNA, we tested whether there is a relationship between the cAMP levels and the expression of hfq. Interestingly, we found that cAMP-CRP was capable of binding to a specific region of the P3hfq promoter and subsequently repressed hfq expression. The find-ing was further substantiated by expressfind-ing an exogenous hfq gene using hfq and lacZ promoters in an hfq-deficient mutant. Overall, we provide the first evidence and mechanism(s) for the regulation of mRNA stability by glucose. In the absence of glucose, cAMP can repress hfq expression and thereby stabi-lizes ompA mRNA. In contrast, enhanced hfq expression in the presence of glucose destabilizes ompA mRNA.
MATERIALS AND METHODS
Bacterial strains.The strains and plasmids used in this work are listed in Table 1. All primers are shown in Table S1 in the supplemental material. E. coli strain BW25113 (wild type) and strains JW3778 (cAMP-deficient or cya mutant) and JW4130 (hfq-deficient mutant), knocked out by the Keio collection system (2), were kindly provided by the National Institute of Genetics of Japan. An Hfq and cya double mutant (CPT021) was produced in our laboratory by transforming
* Corresponding author. Mailing address: Department of Biological Science and Technology, National Chiao Tung University, 75 Po-Ai Street, Hsinchu, Taiwan, 30050 Republic of China. Phone: 886-3-571-2121, ext. 56908. Fax: 886-3-572-9288. E-mail: cpts@cc.nctu.edu.tw.
† Supplemental material for this article may be found at http://jb .asm.org/.
䌤Published ahead of print on 12 August 2011.
plasmid pCP20 (6) with the kanamycin marker removed into JW4130. A P1 phage containing a⌬cya::kan fusion from JW3778 was used to construct a CPT021 double deletion using a standard protocol of OpenWetWare from Rob-ert Sauer’s Laboratory at the Massachusetts Institute of Technology (http: //openwetware.org/wiki/Sauer:P1vir_phage_transduction).
Plasmid construction.To construct the P3hfq-lacZ fusion, the P3hfqpromoter region (containing the 275 bp upstream of the hfq start codon) was inserted into pRW50 (kindly provided by S. J. Busby, University of Birmingham [19, 28]) to get plasmid pRW50-P3hfq-lacZ. The Hfq expression plasmid (pSP73-Phfq-hfq) containing the same P3hfqpromoter region, and a putative CRP binding se-quence, was constructed by insertion of the hfq gene (including a 309-bp coding region and 25-bp terminator from the genomic DNA of E. coli wild-type BW25113) into a pSP73 vector, in which the T7 promoter was deleted by HpaI and BamHI. A pSP73-PNCH-hfq plasmid was similarly constructed, except that
the putative CRP binding region at TGGGA AGGGGT TCACT was replaced by a non-CRP binding region or ACGCT AGGGGT AGAGT (namely, NCH). This replacement was done with a complementary sequence described by Hollands et al. (13) using a KOD-Plus mutagenesis kit (Toyobo, Osaka, Japan). To construct the pSP73-PlacZ-hfq plasmid, the promoter region (total of 275 bp) of pSP73-Phfq-hfq was replaced with a lacZ promoter (containing the 264 bp upstream of the start codon) using PCR-driven overlap extension (12). For expression and purification of CRP protein, the 633-bp crp gene was amplified in our laboratory using primers crpF and crpR by PCR. The PCR products were inserted into the BamHI and KpnI sites of the pQE30 vector to obtain pQE30-crp.
Media and growth conditions.Cells were grown in Luria-Bertani (LB) me-dium with or without 40 mM glucose at 37°C until reaching the exponential phase with an optical density at 600 nm (OD600) of 0.3 to 0.5. Tetracycline, ampicillin,
or kanamycin was added as needed to the medium at a concentration of 50 g/ml. Initiation of mRNA transcription was inhibited by the addition of rifam-pin to a final concentration of 500g/ml before harvesting the total cellular RNA. After adding rifampin, cultured cells were harvested at different time points and chilled in a dry ice/ethanol bath for 1 min prior to RNA extraction.
RNA preparation.RNA isolation was performed according to the procedures of the TRI reagent-RNA kit (Molecular Research Center, Cincinnati, OH). Cells collected by centrifugation (8,000⫻ g for 10 min) at 4°C were resuspended in 1.5 ml TRI reagent. The homogenate was incubated at 65°C for 10 min, 0.3 ml chloroform was added, and the mixture was vortexed vigorously for 30 s and incubated at room temperature for 5 min. The mixture was then centrifuged at 12,000⫻ g for 15 min. The supernatant was mixed with 0.75 ml isopropanol, and the RNA was precipitated at⫺20°C for 1 h before centrifugation at 12,000 ⫻ g for 10 min. The pellet was washed twice with cold 75% ethanol and dissolved in 50l of deionized water pretreated with 0.1% diethyl pyrocarbonate (DEPC).
Northern blot analyses.Equal amounts of total RNA (determined by mea-suring its absorbance at the OD260) were loaded and separated on a 1.2% or 2%
agarose-formaldehyde gel for the analysis of ompA, hfq, or fur. The integrity and quality of RNA samples were confirmed by the presence of 23S and 16S rRNA. Following electrophoresis, gels containing RNA were transferred to a positively charged nylon membrane (Millipore, Bedford, MA) using a 10⫻ saline sodium
citrate (SSC) buffer solution (l⫻ SSC is 0.15 M NaCl and 15 mM sodium citrate, pH 7.0), and the membrane was cross-linked with UV light. The membrane was equilibrated in a hybridization buffer containing 5⫻ Denhardt’s solution (1⫻ Denhardt’s solution contains 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% bovine serum albumin), 5⫻ SSC, 1% SDS, 5% dextran sulfate, and 50% formamide and hybridized with digoxigenin (DIG)-labeled DNA probes for at least 12 h at 42°C. DNA probes specific to E. coli ompA, hfq, and fur were amplified with dNTPs and a DIG-11-dUTP mixture (Roche Applied Science, Mannheim, Germany) as previously described (18). Following hybridization, membranes were washed five times (twice with 2⫻ SSC and 0.1% SDS at 25°C for 5 min and three times with 0.1⫻ SSC and 0.1% SDS at 37°C for 15 min). After washes, the membranes were incubated with anti-DIG antibody conjugated with alkaline phosphatase and detected by adding disodium 3-(4-methox-yspiro{1,2-dioxetane-3,2⬘-(5⬘-chloro)tricyclo[3.3.1.13,7
]decan}-4-yl) pheryl phos-phate (CSPD) chemiluminescent substrate (Roche Applied Science, Indianap-olis, IN). The blots were then developed by exposure to Kodak films. The half-life of mRNAs was represented as the mean⫾ standard deviation (SD) of the band intensity analyzed by Northern blotting in triplicates, while the band intensity was determined by the Zero-Dscan image analysis system (Scanalytics, Billerica, MA).
Real-time PCR analyses.Cells were grown to reach to an exponential phase. Total RNA was isolated as described above and treated with DNase I (Promega, Madison, WI). Briefly, cDNA was synthesized from 5g of total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA), with the cor-responding primers designed for hfq or rne by the use of Primer Express software (Applied Biosystems; Foster City, CA). Real-time PCR using SYBR Premix ExTag (Takara, Tokyo, Japan) was executed with an ABI PRISM 7000 (Applied Biosystems) according to the standard operation procedures. The relative level of 16S rRNA was used as an internal control to normalize hfq and rne expression. The relative expression of hfq and rne was calculated by the comparative thresh-old cycle (⌬⌬CT) method according to the manufacturer’s recommendation. SD was calculated using Microsoft Excel (Redmond, WA), and Student’s t tests were used for statistical analysis. Differences were considered statistically significant if P values were⬍0.05.
-Galactosidase assay. Plasmid pRW50-Phfq-lacZ was transformed into BW25113 and JW3778. Cells were cultured as indicated above. The activity of -galactosidase was measured as described previously (21). The -galactosidase values were represented as the average of results from at least three independent experiments, with a variation of no more than 5% from the mean.
EMSA.Electrophoretic mobility shift assays (EMSA) were carried out with modifications as described previously (26). In addition to hfq and NCH frag-ments which were commercially synthesized, the rest of the fragfrag-ments were amplified by PCR. All DNA fragments used in the assays were labeled with [␥-32
P]dATP and purified over G25 columns (GE-Healthcare, Uppsala, Swe-den). The pQE30-crp plasmid was transformed into E. coli BL21(DE3), and the CRP recombinant protein was isolated using a His tag column system purchased from Qiagene (Hilden, Germany) (26). The binding reactions (20l) were performed by incubating 10 ng of labeled DNA fragments with various amounts
TABLE 1. Strains and plasmids used in this study
Strain or plasmid Characteristics Source or
referencea
Strains
BW25113 lacIqrrnB3⌬lacZ4787 hsdR514 ⌬(araBAD)567 ⌬(rhaBAD)568 rph-1 NIG
JW3778 BW25113⌬cya::kan NIG
JW4130 BW25113⌬hfq::kan NIG
CPT021 JW4130⌬hfq ⌽(⌬cya::kan) This study
Plasmids
pRW50 Vector for creating hfq promoter-lacZ reporter fusions; Tetr 28
pRW50-P3hfq-lacZ pRW50 cut with EcoRI and HindIII containing 275-bp P3hfqpromoter region; Tet
r This study
pCP20 Plasmid for removal of the kan marker from JW4130; Ampr NIG
pSP73 Vector for hfq gene expression; Ampr Promega
pSP73-Phfq-hfq pSP73 cut with HpaI and BamHI containing 275-bp hfq promoter region and 334-bp hfq gene; Ampr This study
pSP73-PNCH-hfq Non-CRP binding site was corrected with mutagenic primers from pSP73-Phfq-hfq; Amp
r This study
pSP73-PlacZ-hfq pSP73 cut with HpaI and BamHI containing 264-bp lacZ promoter region and 334-bp hfq gene; Ampr This study
pQE30 Vector for CRP protein expression; Ampr Laboratory stock
pQE30-crp pQE30 cut with BamHI and KpnI containing 633-bp crp gene; Ampr This study a
of purified CRP (0 to 200 nM) and cAMP (0 to 200M) in a binding buffer [2 g/ml of sonicated herring sperm DNA, 4 mM Tris-HCl (pH 7.0), 5 mM sodium chloride, 2 mM magnesium chloride, 2 mM dithiothreitol, 50 mg/ml bovine serum albumin (BSA), and 15g/ml poly(deoxyinosinic-deoxycytidylic) acid]. After incubation at 37°C for 30 min, samples were electrophoresed in a native polyacrylamide gel (6%) in 0.5⫻ Tris-borate (TBE) buffer (1⫻ TBE contains 0.089 M Tris-borate, 0.089 M boric acid, and 0.002 M EDTA, pH 7.0). After electrophoresis, the gel was dried on a 3M paper for 80 min and autoradio-graphed on Kodak film.
DNase I footprinting.A DNase I footprinting assay was performed as de-scribed previously (5, 16, 39). The DNA fragment from nucleotide position⫺251 to⫺32 (as indicated in Fig. 3) was generated by PCR using the 5⬘ 6-carboxy-fluorescein (FAM)-labeled primers. Binding of the cAMP-CRP complex to the labeled DNA fragment was carried out as described for EMSA. Then, the binding mixtures were partially digested with 0.5 units DNase I containing 40 mM Tris-HCl (pH 8.0), 10 mM MgSO4, and 1 mM CaCl2. After incubation for
1 min at 37°C, the reaction was stopped by heating at 65°C for 10 min in the presence of 20 mM EGTA, pH 8.0. The digested DNA fragments were purified using the MinElute PCR purification kit from Qiagen. The purified DNA frag-ments were detected with a 3730 DNA analyzer (Applied Biosystems). The protected regions were then analyzed with GeneMarker (SoftGenetics, State College, PA) and then aligned to the sequences generated using the same primers.
Western blot analysis.The Western blot analysis used for Hfq protein expres-sion was essentially similar to that described previously (34). In brief, cells were harvested at an OD600of 0.3 to 0.5 and resuspended in 100 mM potassium
phosphate buffer (pH 7.6), and sonication for 5 min and centrifugation at 12,000⫻ g for 15 min at 4°C followed. A total of 8 g of extract protein per lane was electrophoresed on 12% SDS-polyacrylamide gel for 2 h at 100 V. After transferring to nitrocellulose paper and washing, the protein was detected using an antibody specific against Hfq (kindly provided by U. Bla¨si, University of Vienna) and developed by an ECL detection system (Amersham, Arlington Heights, IL).
RESULTS
Effects of glucose and cAMP on the stability of ompA mRNA.
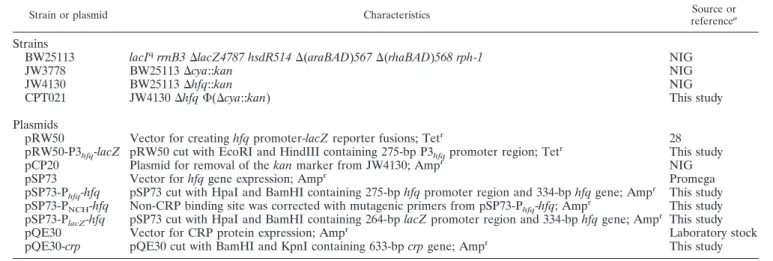
ompA is abundantly expressed in E. coli and is often used to investigate mRNA stability. In the present study, we found that glucose had a negative effect on the stability of ompA mRNA when comparing E. coli grown in LB medium supplemented with or without glucose. Figure 1A reveals that ompA mRNA was degraded faster when E. coli was grown in the presence of glucose. Since it is well established that cAMP levels are low when growing in the presence of glucose (23), we further tested whether the addition of exogenous cAMP could stabilize the ompA mRNA. Figure 1B shows that the addition of cAMP restored the stability of the ompA mRNA, suggesting that cAMP played a role in stabilizing the ompA mRNA. To sub-stantiate this hypothesis, we showed that an E. coli mutant lacking cAMP (JW3778 or the⌬cya mutant) caused even faster decay of ompA mRNA (Fig. 1C, top) compared to the wild type (BW25113) (Fig. 1A, top). The addition of glucose to this mutant did not further destabilize the mRNA (Fig. 1C, bot-tom), indicating that such a negative effect of glucose on ompA mRNA stability in wild-type E. coli is cAMP dependent. Taken together, our data suggest that glucose destabilization of the ompA mRNA is attributable to a decrease in cAMP levels.
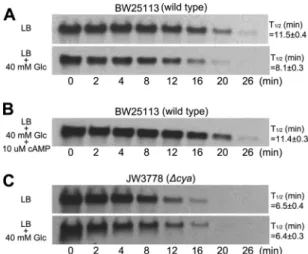
hfq expression in cAMP-deficient mutant. One study re-ported that Hfq facilitates the degradation of ompA mRNA (36). We attempted to determine whether there is a linkage between cAMP and the expression of hfq. We examined the effect of cAMP on hfq gene expression in a cAMP-deficient mutant. This mutant expressed significantly higher hfq mRNA levels (P ⬍ 0.05) than wild-type E. coli as determined by a real-time PCR assay (Fig. 2A). On the other hand, the
expres-sion of rne, known to participate in ompA mRNA degradation, was not affected in the cAMP-deficient mutant (Fig. 2A, right). Previous studies have shown that hfq can be transcribed from three different promoters, P1hfq, P2hfq, or P3hfq(32). In order to determine which promoter is regulated by cAMP, we exam-ined the hfq mRNA level transcribed from each of the pro-moters. The levels of hfq mRNA transcribed from the P1hfq and P2hfqpromoter did not markedly change (data not shown) in response to glucose. In contrast, significant changes were seen in the level of hfq transcribed from the P3hfqpromoter in this study. We showed that growing wild-type E. coli in glucose-supplemented medium, a culture condition that reduces cAMP levels, upregulated the hfq mRNA in the wild type. A similar increase in hfq mRNA was seen in the cAMP-deficient mutant (Fig. 2B). To determine whether the P3hfqpromoter is regu-lated by cAMP, we examined the expression of the P3hfq-lacZ construct consisting of 275 bp of the P3hfq promoter cloned into a lacZ expression vector, pRW50. The -galactosidase levels were about 2-fold higher in LB medium supplemented with glucose than in LB medium. There was a⬎2-fold increase in P3hfq-lacZ expression by the cAMP-deficient mutant (Fig. 2C). Increases in Hfq protein expression was confirmed using Western blotting. Our results show that a glucose-induced re-duction in cAMP levels in the wild type or cAMP-deficient mutants resulted in increased Hfq protein expression (Fig. 2D). These data support the theory that cAMP can repress the expression of hfq from the P3hfq promoter, leading to a re-duced Hfq level.
Interaction between cAMP-CRP and P3hfq promoter. We tested the hypothesis that the cAMP-CRP complex binds to
FIG. 1. Effects of glucose and cAMP on the stability of E. coli
ompA mRNA as determined by Northern blot analysis. (A) Wild-type
cells (BW25113) were grown in LB medium supplemented with and without 40 mM glucose (Glc). (B) Wild-type cells (BW25113) were grown in LB with 40 mM glucose and 10 M cAMP. (C) cAMP-deficient cells (JW3778) were grown in LB with and without 40 mM glucose. After exponential phase was reached (OD600⫽ 0.3 to 0.5),
rifampin was added to the cultured cells to inhibit new synthesis of mRNA, and the cells were harvested at different time points from 0 to 26 min. Analysis of the ompA mRNA levels was performed by frac-tionation of total RNAs on 1.2% agarose-formaldehyde gel and trans-fer of the RNA onto a nylon membrane. The membrane was then hybridized with DIG-labeled ompA DNA. Each experiment was re-peated three times and subjected to statistical analysis.
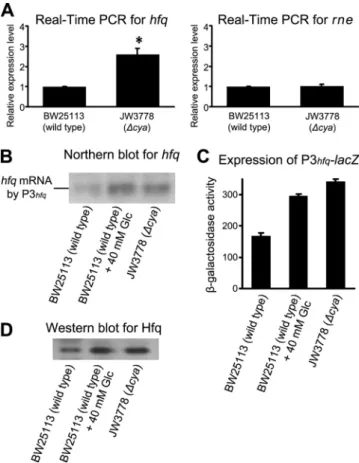
the P3hfqpromoter region of the hfq gene and acts as a tran-scriptional factor, by the use of an EMSA. First, we prepared a full-length recombinant CRP and incubated it with cAMP to form the cAMP-CRP complex. We then examined the binding of the cAMP-CRP complex with four sequential fragments spanning the P2hfqand P3hfqpromoter region (sequence⫺429 to 68) (Fig. 3A, top) and found that the sequence from⫺151 to⫺32 was sufficient for the binding (Fig. 3A, bottom). Sec-ond, we used the MATCH program prediction software to delineate a potential binding region over the P3hfqpromoter (⫺151 to ⫺32). The MATCH program is a weight matrix-based tool for searching the putative binding region for a transcription factor in DNA sequences (20). The cAMP-CRP complex binding region was putatively identified as being lo-cated between⫺104 and ⫺89 relative to the P3hfqpromoter. The binding profile of CRP was constructed based on similarity to the consensus sequences inferred from known CRP binding sites using RegulonDB, version 6.4. A fragment (⫺115 to ⫺78) was synthesized within the proposed sequence (⫺104 to ⫺89) for EMSA to test the binding of the cAMP-CRP complex. Figure 3B depicts that the cAMP-CRP complex bound to this
synthetic fragment in a CRP dose-dependent manner (0 to 200 nM), whereas cAMP or CRP alone (Fig. 3B, bottom) failed to bind to the proposed region. Finally, the specificity of the binding region was confirmed because the cAMP-CRP com-plex could not bind to the other synthetic fragment containing the mutated sequences, ACGCTAGGGGTAGAGT (Fig. 3B, top). The EMSA results demonstrated that the cAMP-CRP complex directly interacts with the hfq promoter region (⫺104 to⫺89) in a dose-dependent manner. Next, we used a DNase I footprinting assay to verify the cAMP-CRP complex binding site. A DNA fragment from positions⫺251 to ⫺32 was sub-jected to DNase I footprinting in the absence or presence of CRP (400 nM). The digested fragments were separated by capillary electrophoresis and peak heights on the chromato-grams. We detected only one CRP protected region located from⫺112 to ⫺87 (including a hypersensitive region) (Fig. 3C, bottom) when comparing sequence patterns in the absence or presence of CRP (Fig. 3C, top and middle). Therefore, the CRP protected region verified by the DNase I footprinting assay is similar to the binding site detected by EMSA. The results shown in Fig. 3 suggest that a binding region of the cAMP-CRP complex is located on the⫺429-to-68 fragment.
Stability of ompA mRNA following the hfq knockout in cAMP-deficient mutant.Our data suggested that elevated ex-pression levels of hfq in response to low levels of cAMP were associated with the increased degradation of ompA mRNA. To further test this hypothesis, we used a cAMP-deficient mutant, which already displayed a high degradation level of ompA mRNA (Fig. 1A and C, top), to create a cya hfq double mutant using a phage P1-generalized transduction method. The cya hfq double mutant should have low levels of both cAMP and Hfq. Figure 4 shows, interestingly, that once the hfq gene was deleted, ompA mRNA became markedly more stable. The increases in stability were independent of the cAMP levels because both the hfq mutant and cya hfq double mutant exhib-ited similar increases in the stability of ompA mRNA. These findings confirm that high levels of hfq expression reduce the stability of ompA mRNA.
Roles of transformed hfq in ompA mRNA stability of hfq knockout mutant in the presence of glucose.In addition, we used a hfq knockout mutant to assess the role of hfq in the presence of glucose by transforming the hfq knockout mutant with a plasmid containing the P3hfqpromoter and wild-type hfq gene. The half-life of ompA mRNA was more stable in the hfq knockout mutant than in the wild-type strain (Fig. 4, bottom). When hfq levels were restored by transforming the mutant with a plasmid containing the wild-type hfq operon, the ompA mRNA was destabilized (Fig. 5A, top). However, the stability of the ompA mRNA was further reduced by the addition of glucose to the growth medium (Fig. 5A, bottom). This suggests that the presence of the hfq gene is necessary for the negative effects of glucose on ompA mRNA stability in response to the reduced cAMP levels. On the other hand, glucose did not have a negative effect when the P3hfqpromoter region was mutated (namely, PNCH, with a mutation of the binding region for the cAMP-CRP complex) (Fig. 5B). This finding reveals that, first, the predicted cAMP-CRP binding region of the P3hfq pro-moter is essential for glucose response, and second, the under-lying mechanisms involved in the negative regulation by glu-cose of ompA mRNA stability is associated with the activation
FIG. 2. Increases in hfq expression of cAMP-deficient mutant. (A) E. coli was grown in LB medium. Relative expression of hfq and
rne mRNA in wild type (BW25113) and cAMP-deficient mutant
(JW3778) measured by a real-time PCR (in triplicate).ⴱ, P ⬍ 0.05. (B) Expression of hfq mRNA measured by Northern blot analysis. BW25113 was grown in LB supplemented with or without 40 mM glucose (Glc). JW3778 was grown in LB. (C) Activity of -galacto-sidase was measured for BW25113 and JW3778 transformed with pRW50-P3hfq-lacZ. The cells were grown as described above for panel
of the hfq gene caused by the loss of the repressive effects of the decreased cAMP level (Fig. 1B).
In contrast to the repressive effects of the cAMP-CRP com-plex on the P3hfqpromoter, it is well known that the complex can positively activate the lacZ promoter. Next we replaced the P3hfqpromoter with the lacZ promoter and used it to address whether the negative effects of glucose on ompA mRNA sta-bility could be reversed by reducing hfq expression at low cAMP levels. We found that using the lacZ promoter resulted in a stabilization of ompA mRNA after adding glucose (Fig. 5C). This indicates that the decreased level of cAMP induced by glucose failed to activate the lacZ promoter, resulting in lower expression of hfq. Under such conditions, the trans-formed cells mimicked those for the hfq knockout mutant (Fig. 4, bottom). Western blot analysis also indicated that the sta-bility of ompA mRNA correlates with Hfq levels (Fig. 5D).
Positive effects of cAMP and negative effects of Hfq on fur mRNA stability.The relationship between cAMP and Hfq on mRNA stability was examined using another mRNA (fur) with a degradation rate that is regulated by the Hfq protein in the E. coli wild type (35). Compared to the wild type (BW25113) (Fig. 6A), fur mRNA was less stable in cAMP-deficient mu-tants (Fig. 6B). However, knockout of the hfq gene in the cya and hfq double mutant (CPT021) markedly stabilized the fur mRNA (Fig. 6C). These results were closely similar to those described in the legend to Fig. 4 for the stability of ompA mRNA.
DISCUSSION
While the decay rate of ompA mRNA in E. coli has been reported (29), our finding that glucose negatively regulates the stability of ompA mRNA has not been reported previously according to the best of our knowledge. The growth rate-dependent stability of ompA mRNA is probably due to the fact that glucose is a preferred carbon source for E. coli for most of
FIG. 3. Delineation of hfq promoter region interacted with cAMP-CRP complex and its specificity. (A) (Top) Four DNA fragments of the P3hfqpromoter region ranging from sequence position⫺429 to 68
were used to narrow down the potential binding region of the cAMP-CRP complex. The numbers indicate the DNA sequences counted from the P3hfqpromoter. (Bottom) The band shift resulted from the
binding of cAMP-CRP complex by EMSA. (B) The MATCH program predicted and screened the putative binding region (hfq fragment). The hfq and NCH fragments were synthesized and used for EMSA. NCH indicates non-CRP binding region mutant. (Top) EMSA results as shown by32P-labeled hfq or NCH fragment in the presence of 200
uM cAMP and different concentrations of CRP as indicated. (Bottom) EMSA results as shown by32P-labeled hfq fragment. The
concentra-tions of cAMP and CRP are indicated above the panels. (C)
Full-length electropherograms of DNase I footprinting assays showing the protected pattern of the⫺251-to-⫺32 fragment in the absence (blue, top) or presence (red, middle) of CRP. The red square shows a sig-nificant difference in the peak pattern. The square was expanded to demonstrate where the region was protected by aligning the results for 0 nM and 400 nM CRP (bottom). The red sequence indicates the protected region.
FIG. 4. Stability of ompA mRNA following the hfq knockout in cAMP-deficient mutant. Northern blot analysis was used to examine the effect of hfq deletion on the stability of ompA mRNA in two E. coli strains. CPT021 was a double mutant with both hfq and cAMP defi-ciency. JW4130 was the mutant with hfq defidefi-ciency.
its nutrients, based on carbon catabolite repression (9). It is therefore expected that the uptake of glucose results in the enhanced growth rate of the cells, and in turn, the increased growth rate results in the increase in the stability of ompA mRNA (25, 36). However, our study has shown that the
glu-cose supplement did not make a significant difference in the growth rates when E. coli was growing in LB medium. These findings are consistent with those of the previous study showing that the growth rate of E. coli is barely affected when glucose is added to LB media (31). In this study, the doubling times were similar: 41 min when wild-type E. coli was grown in LB and 37 min when it was grown in LB supplemented with glu-cose (40 mM). Therefore, the data presented reflect the effects of glucose on degradation of ompA mRNA that is independent of the growth rate. The half-life (⬃11.5 min) for ompA mRNA in the wild type (Fig. 1) is in close agreement with those found in previous studies (25, 36).
It is well known that cAMP levels are low in E. coli growing in the presence of glucose (23). We have shown that changing cAMP levels in E. coli regulates ompA mRNA stability. In-creasing cAMP levels by the addition of exogenous cAMP to LB increased the stability of ompA mRNA (Fig. 1B). Subse-quently, we showed that the half-life of ompA mRNA was significantly attenuated in the cAMP-deficient mutant, sug-gesting that the regulation of ompA mRNA stability was cAMP dependent (Fig. 1C).
The mechanism(s) involved in the negative effects of glucose on ompA mRNA in the present study seems to be complicated. A schematic drawing to simplify our working hypothesis is depicted in Fig. 7. There are several lines of evidence in the present study, which have not been reported previously, sug-gesting that cAMP is associated with the expression of hfq. First, hfq was derepressed in the cAMP-deficient mutant when assessed by real-time PCR, Northern blot analysis, and West-ern blot analysis (Fig. 2). Second, cells grown in the presence
FIG. 5. Roles of transformed hfq in ompA mRNA stability. (A) OmpA mRNA stability was measured in hfq-deficient cells (JW4130) transformed with pSP73-Phfq-hfq plasmid when growing in
LB medium supplemented without (top) or with (bottom) 40 mM glucose (Glc). (B) OmpA mRNA stability was measured in hfq-defi-cient cells transformed with a negative-control pSP73-PNCH-hfq
plas-mid. (C) OmpA mRNA stability was measured in hfq-deficient cells transformed with a hybrid plasmid containing lacZ promoter and hfq gene (pSP73-PlacZ-hfq). All the experiments were conducted by
North-ern blotting in triplicate. (D) Hfq protein expression in response to glucose supplement was analyzed by Western blotting.
FIG. 6. Stability of fur mRNA following the hfq knockout in cAMP-deficient mutant. Northern blot analysis was used to examine the effects of hfq deletion on the stability of fur mRNA in the three E. coli strains. (A) Wild type. (B) Mutant with cAMP deficiency. (C) Double mutant with both cya and hfq deficiency.
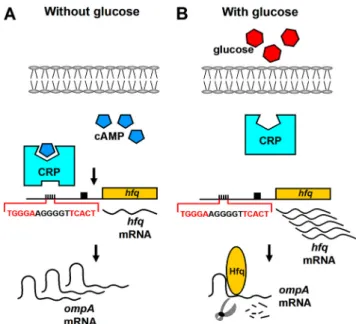
FIG. 7. Schematic drawing for the cascade effect of cAMP and Hfq on ompA mRNA stability. (A) In LB medium without the addition of glucose, cAMP forms a complex with CRP and specifically binds to the
hfq promoter to lower the transcription of hfq gene. The reduced Hfq
protein attenuates the ompA mRNA degradation. (B) In contrast, after the addition of glucose the cAMP levels are reduced, preventing the cAMP-CRP complex from binding to the hfq promoter. This re-sults in an increase in hfq transcription, and then Hfq protein promotes
of glucose (a condition that lowers the cellular cAMP levels) caused an increase in hfq expression (Fig. 2B and D). Third, only the cAMP-CRP complex, but not cAMP or CRP alone, could directly bind to the promoter of the hfq gene (Fig. 3B). The identified bound region (sequence⫺104 to ⫺89) for the cAMP-CRP complex appears to be specific and essential, as mutation of this region abolishes the binding of the complex in an EMSA and DNase I footprinting assay (Fig. 3B and C). In many cases, the cAMP-CRP complex binding site is positioned at a relatively long distance from the transcription start site (38). Downregulation of the guaB promoter by the cAMP-CRP complex has been reported at position⫺117.5 (14). In this study, the results shown in Fig. 3 show that only one binding site for the cAMP-CRP complex existed in the ⫺429-to-68 fragment. Fourth, ompA mRNA was destabilized in cAMP-deficient mutants, but the mRNA stability was markedly re-stored by deletion of the hfq gene in the cya hfq double mutant (Fig. 4). Our results suggest that the cAMP-CRP complex acts as a transcriptional factor to repress the expression of hfq. Fifth, subsequent transformation of the wild-type hfq operon back to the hfq mutant substantially destabilized ompA mRNA. Then, the regulation of the wild-type hfq operon for ompA mRNA stability in the presence of glucose was the same as that of the wild-type strain (BW25113) (Fig. 1A and 5A). In con-trast, substituting the P3hfq promoter with a lacZ promoter (that is positively regulated by cAMP-CRP) increased the sta-bility of the ompA mRNA in the presence of glucose. This is because a glucose-induced reduction in cAMP levels only min-imally activates PlacZ-hfq, and hfq expression was not sufficient to promote the ompA mRNA degradation (Fig. 5C).
Hfq has been proposed to control posttranscriptional or translational regulation by targeting small RNAs and mRNAs (4). In addition to forming base pairs of small RNA and mRNA, Hfq associates with the C-terminal scaffold region of RNase E to activate ptsG mRNA degradation (17, 24). Re-cently, Hfq is regarded as a limiting factor in vivo for small RNA signaling (15). The base-pairing of MicA small RNA (also named SraD) to the ompA mRNA in the vicinity of the ribosome-binding site is enhanced by the presence of Hfq, leading to mRNA decay and subsequent translational inhibi-tion. MicA participates in the downregulation of ompA mRNA in the stationary phase (OD600values of 1.5 to 2.0), and the
MicA mutant does not affect the stability of ompA mRNA during exponential growth (OD600 ⫽ 0.4) or the conditions
used in our experiments (data not shown) (27, 33). Further-more, CyaR (also named RyeE, the CRP-activated small RNA) negatively regulated the expression of ompX but had no effects on ompA mRNA (7). Therefore, the small RNA, which is already known to affect CRP or ompA mRNA stability, would not play a role for glucose regulation of ompA stability in this study.
Taken together, our data reveal that glucose can destabilize the ompA mRNA by reducing the cAMP-CRP complex-medi-ated activation of hfq gene expression. We also identified a binding site for the cAMP-CRP complex in the P3hfqpromoter, which may be responsible for the repression. Our findings provide new insights into the transcriptional regulation of hfq and posttranscriptional regulation of ompA by glucose. Since Hfq is an important RNA binding protein, the changes in cAMP levels induced by glucose appear to indirectly modulate
the stability of mRNA, such as ompA and fur, by an Hfq-dependent mechanism. This mechanism may play an important role in regulating the stability of other mRNA in E. coli.
ACKNOWLEDGMENTS
This work was supported by the National Council of Science (grant NSC 98-2311-B-009-003-MY2).
We thank Simon J. T. Mao of National Chiao Tung University for his critical review of the manuscript.
REFERENCES
1. Arnold, T. E., J. Yu, and J. G. Belasco. 1998. mRNA stabilization by the ompA 5⬘ untranslated region: two protective elements hinder distinct path-ways for mRNA degradation. RNA 4:319–330.
2. Baba, T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 3. Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes.
Mi-crobiol. Rev. 56:100–122.
4. Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10:125–133.
5. Chng, C., A. M. Lum, J. A. Vroom, and C. M. Kao. 2008. A key develop-mental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc. Natl. Acad. Sci. U. S. A. 105:11346– 11351.
6. Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromo-somal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645.
7. De Lay, N., and S. Gottesman. 2009. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191:461–476.
8. Emory, S. A., and J. G. Belasco. 1990. The ompA 5⬘ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol.
172:4472–4481.
9. Gorke, B., and J. Stulke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613– 624.
10. Gosset, G., Z. Zhang, S. Nayyar, W. A. Cuevas, and M. H. Saier, Jr. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expres-sion in Escherichia coli. J. Bacteriol. 186:3516–3524.
11. Hansen, M. J., L. H. Chen, M. L. Fejzo, and J. G. Belasco. 1994. The ompA 5⬘ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol. Microbiol. 12:707–716.
12. Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932.
13. Hollands, K., S. J. Busby, and G. S. Lloyd. 2007. New targets for the cyclic AMP receptor protein in the Escherichia coli K-12 genome. FEMS Micro-biol. Lett. 274:89–94.
14. Husnain, S. I., S. J. Busby, and M. S. Thomas. 2009. Downregulation of the Escherichia coli guaB promoter by upstream-bound cyclic AMP receptor protein. J. Bacteriol. 191:6094–6104.
15. Hussein, R., and H. N. Lim. 2011. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. U. S. A. 108:1110–1115. 16. Karr, E. A. 2010. The methanogen-specific transcription factor MsvR
regu-lates the fpaA-rlp-rub oxidative stress operon adjacent to msvR in Metha-nothermobacter thermautotrophicus. J. Bacteriol. 192:5914–5922. 17. Kawamoto, H., T. Morita, A. Shimizu, T. Inada, and H. Aiba. 2005.
Impli-cation of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 19:328–338.
18. Lion, T., and O. A. Haas. 1990. Nonradioactive labeling of probe with digoxigenin by polymerase chain reaction. Anal. Biochem. 188:335–337. 19. Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad
host range plasmids carrying the Escherichia coli lactose and galactose oper-ons. FEMS Microbiol. Lett. 74:271–276.
20. Matys, V., et al. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31:374–378.
21. Miller, J. H. 1972. Experiments in molecular genetics, p. 352–355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
22. Moll, I., T. Afonyushkin, O. Vytvytska, V. R. Kaberdin, and U. Blasi. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9:1308–1314.
23. Morita, T., W. El-Kazzaz, Y. Tanaka, T. Inada, and H. Aiba. 2003. Accu-mulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278:15608–15614.
24. Morita, T., H. Kawamoto, T. Mizota, T. Inada, and H. Aiba. 2004. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54:1063–1075.
25. Nilsson, G., J. G. Belasco, S. N. Cohen, and A. von Gabain. 1984. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature
312:75–77.
26. Peekhaus, N., and T. Conway. 1998. Positive and negative transcriptional regulation of the Escherichia coli gluconate regulon gene gntT by GntR and the cyclic AMP (cAMP)-cAMP receptor protein complex. J. Bacteriol. 180: 1777–1785.
27. Rasmussen, A. A., et al. 2005. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Micro-biol. 58:1421–1429.
28. Samarasinghe, S., et al. 2008. Autoregulation of the Escherichia coli melR promoter: repression involves four molecules of MelR. Nucleic Acids Res.
36:2667–2676.
29. Smith, S. G., V. Mahon, M. A. Lambert, and R. P. Fagan. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Micro-biol. Lett. 273:1–11.
30. Takayama, K., and S. Kjelleberg. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2:355–365. 31. Tseng, C. P., C. C. Yu, H. H. Lin, C. Y. Chang, and J. T. Kuo. 2001.
Oxygen-and growth rate-dependent regulation of Escherichia coli fumarase (FumA, FumB, and FumC) activity. J. Bacteriol. 183:461–467.
32. Tsui, H. C., G. Feng, and M. E. Winkler. 1996. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region
protease genes of Escherichia coli K-12 from clustered Esigma32-specific promoters during heat shock. J. Bacteriol. 178:5719–5731.
33. Udekwu, K. I., et al. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19:2355–2366.
34. Vecerek, B., M. Beich-Frandsen, A. Resch, and U. Blasi. 2010. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 38:1284–1293.
35. Vecerek, B., I. Moll, T. Afonyushkin, V. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897– 909.
36. Vytvytska, O., et al. 1998. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. U. S. A. 95:14118–14123.
37. Vytvytska, O., I. Moll, V. R. Kaberdin, A. von Gabain, and U. Blasi. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14:1109–1118.
38. Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874–5893.
39. Zianni, M., K. Tessanne, M. Merighi, R. Laguna, and F. R. Tabita. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17:103–113.