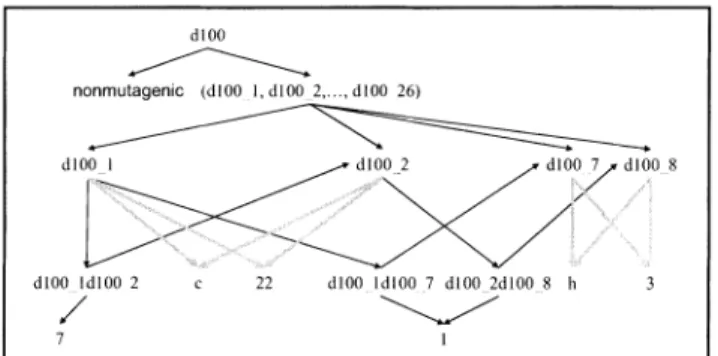
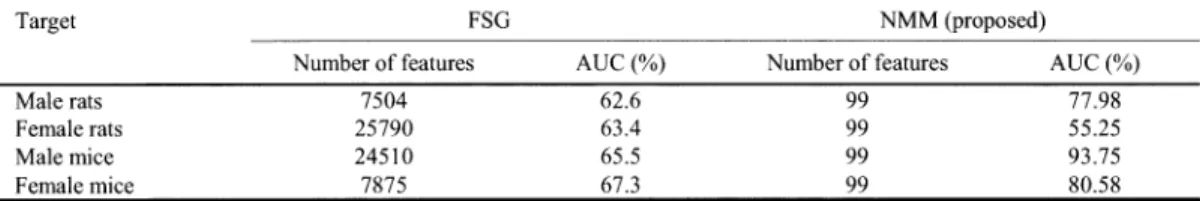
Network Motif Model: An Efficient Approach to Extract Features from Relational Data
全文
數據

相關文件
Is end-to-end congestion control sufficient for fair and efficient network usage. If not, what should we do
Responsible for providing reliable data transmission Data Link Layer from one node to another. Concerned with routing data from one network node Network Layer
This option is designed to provide students an understanding of the basic concepts network services and client-server communications, and the knowledge and skills
In the work of Qian and Sejnowski a window of 13 secondary structure predictions is used as input to a fully connected structure-structure network with 40 hidden units.. Thus,
3. Works better for some tasks to use grammatical tree structure Language recursion is still up to debate.. Recursive Neural Network Architecture. A network is to predict the
To solve this problem, this study proposed a novel neural network model, Ecological Succession Neural Network (ESNN), which is inspired by the concept of ecological succession
This study proposed the ellipse-space probabilistic neural network (EPNN), which includes three kinds of network parameters that can be adjusted through training: the variable
It is always not easy to control the process of construction due to the complex problems and high-elevation operation environment in the Steel Structure Construction (SSC)