科技部補助專題研究計畫成果報告
(□期中進度報告/█期末報告)
克雷白氏肺炎桿菌之未知 Carbapenem 抗藥機轉研究
計畫類別:█個別型計畫 □整合型計畫
計畫編號:MOST 103-2320-B-006-018-MY3
執行期間: 104 年 8 月 1 日至 107 年 7 月 31 日
執行機構及系所:國立成功大學醫學院病理學科
計畫主持人: 張孔昭/顏經洲
共同主持人:
計畫參與人員:
本計畫除繳交成果報告外,另含下列出國報告,共 ___ 份:
□執行國際合作與移地研究心得報告
□出席國際學術會議心得報告
期末報告處理方式:
1. 公開方式:
█非列管計畫亦不具下列情形,立即公開查詢
□涉及專利或其他智慧財產權,□一年□二年後可公開查詢
2.「本研究」是否已有嚴重損及公共利益之發現:█否 □是
3.「本報告」是否建議提供政府單位施政參考 █否 □是, (請列舉提供
之單位;本部不經審議,依勾選逕予轉送)
中 華 民 國 106 年 10 月 4 日
目錄
一、目錄 I 二、中英文摘要 (1)中文摘要 II (2)英文摘要 III 三、報告內容 (1)前言與目的 1 (2)研究方法 2 – 4 (3)結果 4 – 11 (4)結論與建議 12 (5)成果報告自評表 13-14英文摘要
II. 中文摘要
克雷伯氏肺炎桿菌是重要的院內與社區感染病菌之一,目前對廣效性乙內醯胺具抗藥性菌株的散佈已 是全球性的問題,主要以碳青黴烯來治療其感染。碳青黴烯抗藥菌株已逐漸增加,成為治療上的問題, 此抗藥性的產生主要經由:(i)細菌細胞膜對抗生素通透性的改變加上 AmpC 酵素或廣效性乙內醯胺 酶的產生;(ii) 碳青黴烯酶的產生。我們過去的研究顯示,約有 1/4 的碳青黴烯抗藥菌株無法以已知 的抗藥機轉解釋,本計畫的主要目的在嘗試揭露未知的碳青黴烯抗藥機轉。在第一年計畫中,我們利 用多位點核酸序列分型技術建立一菌株資料庫,加上細胞外膜基因多形性分型以及脈衝場凝膠電泳, 可以快速篩選出適合的碳青黴烯敏感菌株以供次世代基因體定序比較分析,用以篩選出可能的抗藥基 因決定因子。另外,我們運用次世代基因體定序技術,比較來自同一病人、具有親緣關係、且具相同 細胞外膜型、但對碳青黴烯具有不同敏感性之不同分離菌株,以篩選出可能的抗藥基因決定因子。目 前我們建立的細胞外膜基因多形性分型方法將可用來分別篩選克雷伯氏肺炎桿菌的高毒性菌株以及 多重抗藥性的高危險菌株,未來亦可利用此技術,藉由比較不同 clone 但具有特定感染疾病或高抗藥 性關聯性的菌株,可找出細菌毒性或易形成抗藥性的決定因子。利用多位點核酸序列分型技術建立一 菌株資料庫,未來可作為研究高毒性菌株或多重抗藥性的基礎。另外利用次世代基因體定序已篩選出 可能的碳青黴烯抗藥基因,已經或正在將這些基因進行 cloning 或基因剔除,將進一步進行分析確定 其與碳青黴烯抗藥性的關聯性。目前我們已完成分屬兩個 clones 的 5 株菌株全基因解碼,將可儲存於 基因資料庫,作為日後高抗藥性高危險菌株的研究基礎。 關鍵詞:克雷伯氏肺炎桿菌、碳青黴烯、抗藥機轉、外胞膜、多位點核酸序列分型、次世代基因體定 序III. 英文摘要
Klebsiella pneumoniae is one of the most important pathogens of both community-acquired and nosocomial infections in Taiwan. The spread of extended-spectrum-resistant K. pneumoniae isolates has become a global problem, and carbapenems are usually used for the treatment of infections caused by such isolates. The emergence of carbapenem-resistant K. pneumoniae isolates has been recognized and is a potential serious threat. The most common resistance mechanisms responsible for carbapenem resistance in K. pneumoniae are: (i) carbapenemase production and (ii) porin loss coupled with AmpC or extended-spectrum β-lactamase production. About one fourth of carbapenem-resistant K. pneumoniae isolates had no such mechanisms in our previous studies. This project is proposed to elucidate carbapenem resistance mechanisms that have not been reported in K. pneumoniae. We also attempt to investigate the link of the identified resistance
mechanisms with bacterial adaptation or virulence, which may affect the persistence of such resistance mechanisms. In the first year of the project, genetic differences between genetically related isolates from the same patient were identified by comparative genomic analysis, and resistance determinants will be
investigated by analyzing the genetic differences. We are constructing a bacterial database using multilocus sequence tying, and a rapid test and strategy using ompK tying, multilocus sequence typing, and pulsed-field gel electrophoresis to select appropriate control strains rapidly for comparative genomic analysis have been developed. The genetically related strains with different carbapenem susceptibilities have been subjected to whole genome sequencing using Illumina Genome Analyzer IIx. Genetic differences among these isolates have been identified, which are being analyzed to identify resistance determinants. Complete genome sequences are achieved by de novo assembly, and contigs are further assembled by PCR and Sanger sequence. In conclusion, we have developed a novel typing method (ompK36 typing) for K. pneumoniae, which may be used for screening high-risk resistant or hypervirulent clones; candidate genes for carbapenem resistance have been obtained and need further investigation; a K. pneumoniae database based on MLST have been established, which can be used for the basis of studies on bacterial virulence and resistance. Moreover, isolates of the different clones associated with resistance or specific infections in this project can be used for comparison analysis to identify virulence or resistance determinants.
Key words: Klebsiella pneumoniae, multilocus sequence typing, carbapenem resistance, resistance mechanisms, porins, proteomics, next-generation sequencing, comparative geneomic analysis.
前言
Carbapenems are usually used to treat serious infections caused by multidrug-resistant Enterobacteriaceae isolates, those producing extended-spectrum β-lactamases (ESBLs) or AmpC β-lactamases in particular (1-3). The emergence of carbapenem resistance and carbapenemases (e.g. KPC, NDM) in Enterobacteriaceae has thus become a great worldwide concern (3-6). Carbapenemases mainly belong to three of four molecular classes of β-lactamases, Ambler’s class A (serine enzyme), class B, also called metallo-β-lactamases (MBLs), and class D (4). Despite the wide geographic distribution of carbapenemases, Enterobacteriaceae isolates more commonly develop resistance to carbapenems by producing AmpC enzymes or ESBLs in combination with the loss of major porins (7-10).
Klebsiella pneumoniae is one of the most important hospital- and community-acquired bacterial pathogens, and it is an important reservoir of drug resistance determinants in Taiwan. In addition to SHV- and CTX-M-type ESBLs that are worldwide distributed (4), the DHA-1 AmpC-type enzyme is common in K. pneumoniae in Taiwan (11). The first carbapenemase discovered in K. pneumoniae in Taiwan was the IMP-8 MBL (12), and the VIM-1 and NDM-1 MBLs and the KPC-type carbapenemase were detected recently (13). Outbreaks of KPC-producing K. pneumoniae isolates occurred recently in northern Taiwan (13).
Despite the increasing prevalence of carbapenemases in K. pneumoniae in Taiwan, impermeability due to the porin loss coupled with AmpC or ESBL production remains the major carbapenem resistance mechanism (10, 13). A previous study conducted by Chiu et al. (13) using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed that ~95% of carbapenem-resistant K. pneumoniae isolates exhibited either OmpK35 or OmpK36 porin loss or both. The lack of porin loss in ~5% of carbapenem-nonsusceptible isolates and variation in carbapenem susceptibilities among K. pneumoniae isolates with similar porin profiles, suggesting the presence of non-porin-mediated carbapenem resistance mechanisms (13). In this study, the DHA-1 AmpC enzyme was the most common β-lactamase followed by the CTX-M-14 ESBL. The prevalence of DHA-1 in Taiwan has thus become a clinical problem. It not only confers resistance to extended-spectrum cephalosporin resistance but also causes the selection of carbapenem resistance in K. pneumoniae.
The spread of antimicrobial resistance has been found to be accompanied by global dissemination of particular bacterial clones with high interhost transmission and colonization efficiency (14, 15). These clones were identified by multilocus sequence typing (MLST), a sequence-based technique that can be used to study population biology of microorganisms by polymorphism analysis of a set of conserved housekeeping genes (16). The technique is easy to perform, reproducible among laboratories, and reliable (17). High prevalence of clones ST258 and ST11 among carbapenemases-producing and non-carbapenemase-producing carbapenem-resistant K. pneumoniae isolates was noted recently (13, 15, 18-20). These findings suggest the presence of “high-risk” clones for antimicrobial resistance in K. pneumoniae (15, 21). In addition to the association of carbapenem resistance with K. pneumoniae ST11 (13), a predominance of the ST23 clone among K. pneumoniae isolates causing liver abscess was also revealed by MLST in Taiwan (22, 23).
which resistance can be manifest (24). Since classical measures of controlling the emergence and spread of antimicrobial resistance are becoming increasingly insufficient, the development of new strategies such as eco-evo drugs and strategies is urgently needed (21). Development of strategies that target and selectively block antibiotic resistance mechanisms to rescue antibiotic activity is one of the important research directions in this regard (21). The knowledge on the sources of resistance mechanisms, their evolution, and the ways in which they are spread in microbial communities is the basis for the development of new drugs and strategies and should be clearly understood. Since treatment against carbapenem-resistant K. pneumoniae and the prevention of their emergence and spread are challenging, it is very important to elucidate carbapenem resistance mechanisms.
目的
Since there were still many clinical K. pneumoniae isolates with unidentified resistance mechanisms in our previous studies, the present project was proposed to elucidate carbapenem resistance
mechanisms that have not been reported in K. pneumoniae. We also attempted to investigate the link of the resistance mechanisms with bacterial adaptation or virulence, which may affect the persistence of such resistance mechanisms.
研究方法
Bacterial isolates
(1) More than 10,000 K. pneumoniae isolates recovered at National Cheng Kung University Hospital between 1999 and 2011 were consecutively collected, and one isolate per patient were analyzed to determine the prevalence of various carbapenem-resistant mechanisms.
(2) All isolates collected in 2008 have been investigated for carbapenem resistance, and 7 isolates that were found to have higher levels of imipenem resistance (MIC, >4 μg/ml) and intact ompC in our previous studies were used for screening for “as-yet-unknown” carbapenem-resistant determinants.
(3) E. coli K12 was used as the competent cell strain for transformation experiments.
Identify candidate carbapenem-resistant determinants from the mutant library.
(1) We screened for carbapenem-resistant mutants using 1 μg/ml ertapenem and 64 mg/L of chloramphenicol in Luria-Bertani (LB) agar from the mutant library established previously.
(2) DNA sequences adjacent to the transposon insertion sites were determined by inverse PCR and DNA sequencing. The genomic DNA of mutants was extracted and subjected to complete digestion by restriction endonuclease BglII followed by circularization. Sequence analysis of DNA flanking the transposon was carried out using the transposon-specific primer (5'-CGGATTACAGCCGGATCCCCG-3') and by gene walking with custom sequencing primers. Nucleotide and amino acid sequences were analyzed and compared by use of the BLAST computer program (National Center for Biotechnology Information) and the FASTA
program (Genetics Computer Group).
Establish a bacterial database for comparative analysis.
In addition to carbapenem-resistant isolates with known STs obtained by MLST previously, 100 isolates susceptible to all cephalosporins were selected for MLST. These isolates are the basis for the bacterial database, and the isolates analyzed later in this project were enrolled in the database. MLST was performed with 7 housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB) (http://www.pasteur.fr/recherche/genopole/PF8/mlst/primers_Kpneumoniae.html) according to the protocol of Diancourt et al. (27). The database includes STs and drug-resistant profiles of isolates, and the bacterial isolates, their drug-resistant profiles, and the data obtained in this project were used in the future for other principal investigators who study on drug resistance or virulence of K. pneumoniae in Taiwan.
Develop rapid tests for bacterial strain typing and a strategy for rapid selection of appropriate control strains for comparative genomic analysis.
(1) ompK typing. Nucleotide sequences of ompK for isolates with known ST types were determined by PCR
(10). Nucleotide comparisons were performed using the GAP program of the Genetics Computer Group sequence analysis package (University of Wisconsin). Multiple sequence alignments were generated using the PILEUP program of GCG. Phylogenetic tree was constructed using the Growtree program of GCG. PCR primers specific for target STs based on ompK35 and ompK36 sequences were designed.
(2) Strategy for rapid selection of control strains. STs of study isolates were determined by MLST. If there
were cephalosporin-susceptible isolates with the same STs, the study isolates and controls were subjected to pulsed-field gel electrophoresis (PFGE) to confirm their genetic relationship, and those with similar PFGE profiles were subjected to comparative analyses. If no cephalosporin-susceptible isolate with the same ST was found, we screened other isolates using PCR with primers specific for the ST. The PCR-positive isolates were subjected to MLST to identify isolates with the same STs. PFGE using XbaI-digested genomic DNA was performed on a CHEF-DR 3 apparatus (Bio-Rad Laboratories, Hercules, CA, USA) according to the instruction manual. PFGE profiles were compared using the BioNumerics program (Bio-Rad). Genetic relatedness was calculated based on the Dice coefficient, and isolates were considered genetically related if they showed ≥80% similarity. The PFGE profiles were included in the database.
Screen for carbapenem-resistant determinants by whole genome sequencing and comparative genomic analysis between genetically related carbapenem-resistant and –susceptible isolates.
(1) The genetically related carbapenem-sensitive and -resistant strains were subjected to whole genome sequencing. Paired-end library with average distance of 300 bp was constructed and Illumina Genome Analyzer IIx (Illumina, CA, USA) was used for determination of the sequences. This was performed in Yourgene Bioscience (Taipei, Taiwan). de novo assembly was performed with CLC genomic workbench (CLC Bio, Aarhus, Denmark). Gap between each contig was amplified by PCR and sequence was performed by Sanger method. The coding regions wereanalyzed and annotated by using the CLC genomic workbench and BLAST.
(2) Resequence analysis was performed with CLC genomic workbench to identify the single nucleotide polymorphisms and indels. The genome comparison was performed with the Artemis Comparison Tool (ACT) and progressive Mauve to identify the genome rearrangement, including recombination, segmental duplication, gain, and loss (28, 29). PCR specific primers were designed to confirm the identified genetic differences.
結果
(1) Identification a hypervirulent group and a high-risk resistant group in K.
pneumoniae by ompK36 typing
(A) Development of a novel phylogenetic typing method for K. pneumoniae
MLST was performed firstly using 35 previously characterized K. pneumoniae isolates with decreased carbapenem susceptibilities and intact ompK36 sequences. Twelve STs were obtained, of which ST11 was the most common (16 isolates, 45.7%). Nucleotide sequences of ompK36 were compared using the GCG SeqWeb software (University of Wisconsin Biotechnology Centre, Madison, WI, USA), and multiple sequence
alignments and a phylogenetic tree were generated using the Pileup and Growtree programs, respectively. Thirteen ompK36 alleles were obtained; they were divided into 4 clusters, designated groups A, B, C, and D (Fig. 1a). All isolates of high-risk clones ST11, ST15, and ST37 belonged to group A. Pulsed-field gel electrophoresis was performed, and the result was consistent with the MLST analysis and showed genetic unrelatedness among isolates of different CGs with similar ompK36 sequences (Fig. 1b).
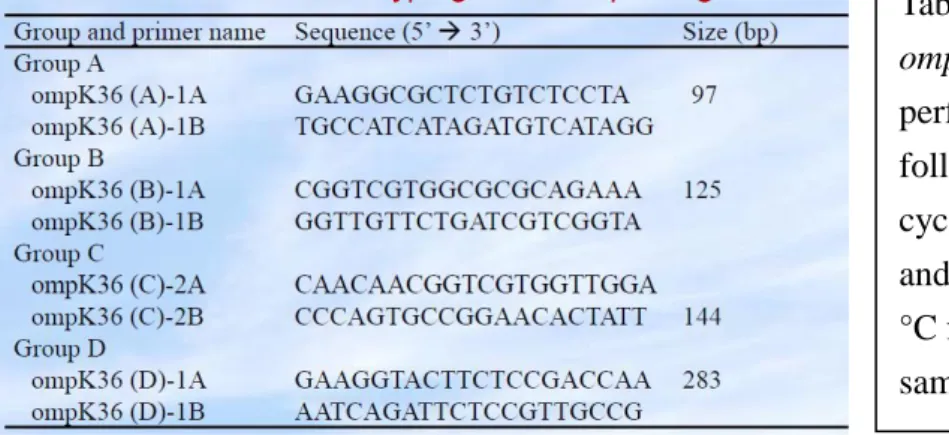
A polymerase chain reaction (PCR)–based method using ompK36-targeted group-specific primer pairs was then developed, and 69 previously characterized extended-spectrum β-lactamase (ESBL)–producing and/or AmpC–producing K. pneumoniae isolates from 7 medical centres were examined. The primer sequences and amplicon sizes are shown in Table 1.
Of the 69 isolates, 35 (50.7%), 6 (8.7%), 23 (33.3%), and 5 (7.2%) isolates were assigned to groups A, B, C, and D, respectively. Twelve ST11, 7 ST15, 3 ST37, 1 ST273, 3 ST395, 2 ST709, and 2 ST833 isolates were included in group A. The ST11, ST15, and ST37 isolates were collected from 5, 4, and 2 hospitals, respectively. Group B comprised 3 ST17, 1 ST414, 1 ST1465, and 1 ST1535 isolates. In group C, 10 STs (ST1, ST23, ST25, ST35, ST39, ST42, ST48, ST101, ST163, and ST416) were identified, of which each included 1 or 2 isolates. Group D comprised 1 ST321, 2 ST378, 1 ST528, and 1 ST655 isolates. Currently available alleles or STs were not obtained for 7 isolates.
The prevalence of each ompK36 group among 81 K. pneumoniae bloodstream isolates collected at the National Cheng Kung University Hospital in 2010 was determined by the PCR-based ompK36 typing tests. Phenotyping and genotyping of β-lactamases were performed. Fourteen (17.3%), 9 (11.1%), 39 (48.1%), and 17 (21.0%) of the 81 isolates belonged to groups A, B, C, and D, respectively, and 2 isolates cannot be typed. The MLST analysis showed 41 STs and 13 unassignable types (Table 2). The major international clones (ST11, ST15, ST37, and ST147) were uncommon (6 isolates, 7.4%). The virulent clone ST23 in group C was the most common ST (n = 13). Seventeen (21.0%) of the 81 isolates carried blaESBL, blaDHA-1 (6 isolates), or
both (4 isolates), and groups A, B, C, and D accounted for 8 (47.1%), 1 (5.9%), 5 (29.4%), and 3 (29.4%) of the 17 isolates, respectively.
Table 2. Results of ompK36 typing, β-lactamase typing and multilocus sequence typing for 81 K. pneumoniae isolates collected from the National Cheng Kung University in 2010
Table 1. Primers used for PCR typing of ompK36. Simplex PCR experiments were performed under the PCR conditions as follows: 1 cycle of 94 °C for 10 min; 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 60 s; a final extension at 72 °C for 10 min. PCR conditions were the same for all primer pairs.
In summary, a total of 59 currently available STs were identified, among which the major international high-risk clones (29 ST11, 13 ST15, 7 ST37, and 1 ST147 isolates) were grouped together by ompK36 typing. Six STs reported to be associated with pyogenic liver abscess were detected, and of note, all isolates of these STs (15 ST23, 2 ST65, 3 ST86, 1 ST163, 1 ST373, and 2 ST375 isolates) were assigned to the other ompK36 allele group.
(B) Identification of a hypervirulent group and a high risk resistant group of K.
pneumoniae by ompK36 typing
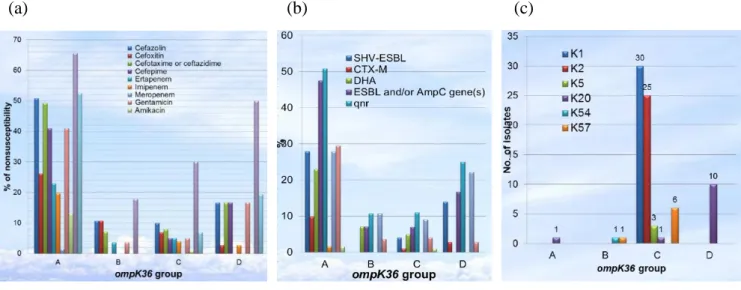
The four groups of K. pneumoniae separated by ompK36 typing were further characterized using 226 nonduplicate K. pneumoniae bloodstream isolates collected at a Taiwanese hospital in 2011. Statistical analysis showed significantly higher rates of antimicrobial resistance (all tested antibiotics except
meropenem), extended-spectrum β-lactamases or DHA-1 (47.5% together), Qnr-type quinolone resistance determinants (50.8%), and IncFIIA-type plasmids (49.2%) in group A than in others (Fig. 2a and 2b). Seventeen isolates were identified as belonging to 3 international high-risk clones (4 sequence type [ST] 11, 10 ST15, and 3 ST147 isolates); all but 1 ST15 isolate were classified in group A. The significant
characteristics of group C were hypermucoviscosity (62.0%) and higher virulence gene content (Table 3). This group included all serotype K1 (n=30), K2 (n=25), and K5 (n=3) isolates, 6 of 7 K57 isolates, all isolates of major clones associated with pyogenic liver abscess (28 ST23, 11 ST65, 5 ST86, 7 ST373, and 1 ST375 isolates), and 16 (94.1%) of 17 isolates causing bacteremic liver abscesses (Fig. 2c and Table 4) . Twelve (42.9%) of the group B isolates were responsible for bacteremic biliary tract infections (Table 4). Group D was predominant (83.3%) among 12 K20 isolates (Fig. 2c). The present data suggests that clinical K. pneumoniae isolates can be separated into four groups with distinct characteristics based on ompK36 types.
(a) (b) (c)
(
2) Screen for carbapenem-resistant determinants by whole genome sequencing and
comparative genomic analysis
(A) Selection of test isolates
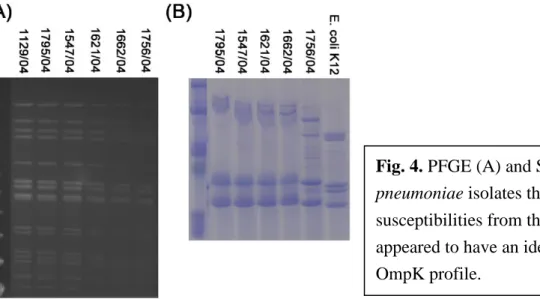
An isolate (isolate 1756/04) was from a patient with multiple K. pneumoniae isolates that were susceptible or intermediately susceptible to carbapenems, and these isolates also had the same β-lactamase content (Table 5). These isolates were suggested to be from the same clone by pulsed-filed gel electrophoresis and had the similar porin profiles revealed by SDS-PAGE (Fig. 4) and the same porin ompK35 and ompK36 nucleotide sequences. These results suggest that the occurrence of the stepwise accumulation of genetic alterations in non-porin genes led to the stepwise decrease in carbapenem susceptibility in a K. pneumoniae strain.
Table 5. Isolates of the same clone with different carbapenem susceptibilities from the same patient. Fig. 2. Comparison among four groups of K. pneumniae determined by ompK36 typing in: (a) antimicrobial resistance, (b) extended-spectrum β-lactamases and the DHA-1 enzyme, and (c) capsular types.
Table 3. Hypermucoid phenotype and
virulence genes among four groups of K.
pneumoniae Table 4. Types of infections due to four groups of K.
Disk diffusion testa MIC (μg/ml)a
Isolate CMZ CAZ CRO CPO ETP IPM MEM ETP IPM MEM β-lactamases
1129 R R R S S S S 1547 R R R I R S S 6 0.5 0.25 CMY-2, SHV-11, TEM-1 1621 R R R S R S S 1662 R R R S R S S 1756 R R R R R R R >32 4 8 CMY-2, SHV-11, TEM-1 1795 R R R S S S S 1.5 0.094 0.25 CMY-2, SHV-11, TEM-1 a
CAZ, ceftazidime; CMZ, cefmetazole; CPO, cefpirome; CRO, ceftriaxone; ETP, ertapenem; I, intermediately susceptible; IPM, imipenem; MEM, meropenem; S, susceptible; R, resistant. MICs were determined by Etest.
(B) Comparative genomic analysis
Screening for genetic alterations leading to carbapenem resistance was performed by comparative genomic analysis with whole genome sequencing on Illumina Genome Analyzer IIx. Flowchart of the next generation sequence data analysis is shown in Figure 5. Complete genome sequences will be achieved by de novo assembly and contigs are being further assembled by PCR and Sanger sequence. Three representative isolates of the same clone with different carbapenem susceptibilities were subjected to reference sequence mapping (K. pneumoniae NTUH-K2044, accession no. NC_012731.1). When we mapped these reads to the reference genome, gapped alignment and soft clip strategy were used. This method is essential for variant discovery because the sequence reads may contain insertion-deletion polymorphism (INDEL). Without this alignment approach, a read may still be mapped onto the correct position but with consecutive mismatches at INDEL locations. A read was mapped onto the reference if it has at least 90% similarity in matched region, and the matched region was at least 100% of the read length fraction. The SNP detection was performed based on the Neighborhood Quality Standard (NQS) algorithm. A SNP was called if a variant has at least 35% frequency at a location with a minimum coverage of 4. The SNP has a minimum quality score of 20, and its surrounding 10 bases had quality scores greater than 15. Fraction of reference sequence covered was 0.92 for each isolate, and numerous INDEL and SNP sites are found. Numbers of genetic differences between the test isolates are shown in Table 6.
Fig. 4. PFGE (A) and SDS-PAGE (B) of K.
pneumoniae isolates that varied in carbapenem susceptibilities from the same patient. They appeared to have an identical PFGE pattern and OmpK profile.
Fig. 5. Flowchart of next generation sequence data analysis.
Table 6. No. of genetic differences obtained by comparative genomic analysis between isolates of the same
clone with different carbapenem susceptibilities
1756/04 versus 1795/04 1547/04 versus 1795/04 No. of SNP sitesa 10 345
No. of INDEL sites 119 1286
a
SNP, single-nucleotide polymorphism; INDEL, insertion or deletion.
Genetic differences among the carbapenem-nonsusceptible and carbapenem-resistant isolates were confirmed by PCR and nucleotide sequencing. Predicted encoded proteins of these genes were obtained by BLAST analysis, and 20 genes that might be associated with carbapenem nonsusceptibility were then selected (Table 7).
Among the 20 candidate resistance determinants, 5 are associated with membrane molecule transport,
including 2 porins (OmpC and LamB), 1 putative permease (membrane transport protein) (KP1_1478), and 2 ion channel proteins (potassium channel protein Kch and sodium/panthothenate symporter); 1 is supposed to be the target of carbapenems (MrsA, a penicillin-binding protein 1A); 1 may be one constituent of outer membrane (KP1_0410, putative fimbrial biogenesis outer membrane usher protein); and 2 may be associated with outer membrane synthesis (KP1_5236, putative cellulose synthase; Glf, UDP-galactopyranose mutase) . The wild-type and mutants of these genes have been cloned and will be analyzed firstly to validate their association with carbapenem resistance.
Table 7. Candidate genes that might contribute to carbapenem nonsusceptibility obtained by whole
genomic comparison between carbapenem-nonsusceptible isolates 1756/04 and 1547/04 and carbapenem-susceptible isolate 1795/04
Abbreviations: Del, deletion; Ins, insertion; NSPM, nonsense point mutation; MSPM, missense point mutation; S, carbapenem-susceptible; I, intermediate susceptible to carbapenems; R, carbapenem resistant; presence of
genetic change.
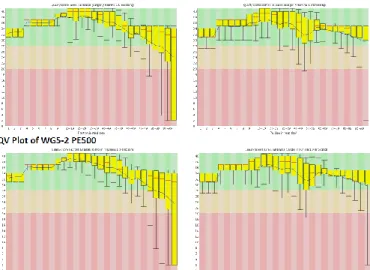
Another one carbapenem-susceptible and one carbapenem-resistant isolates were subjected to whole genome sequencing on Illumina Genome Analyzer IIx this year (Fig. 6) and were mapped by comparison with K. pneumoniae NTUH_K2044 (Fig. 7).
Fig. 6. Quality values (QVs) of one carbapenem-susceptible (WG-1) and one carbapenem-resistant (WG-2)
isolates.
Fig. 7. Analysis pipeline and mapping of one carbapenem-susceptible (WG-1) and one carbapenem-resistant
結論與建議
The present results demonstrated the association of the major high-risk resistant clones and virulent STs with specific ompK36 allele groups. The association might result from the occurrences of convergent evolution driven by the selective pressure created by antimicrobial use and host factors. Active surveillance of the epidemic resistant clones followed by appropriate infection control measures is needed to prevent the spread of antimicrobial resistance, and the ompK36-targeted PCR method developed in this study may be useful for screening of epidemic clones. The major high-risk resistant clones were uncommon. The use of the PCR screening method should therefore be cost-effective despite the lack of specificity, and subsequent sequencing of one or two housekeeping genes for MLST should reduce the number of isolates for complete MLST
analysis further. Further large scale studies are needed to confirm the association identified by ompK36 typing and the usefulness of the typing method.
We are constructing a bacterial database using multilocus sequence tying, and a rapid test and strategy using ompK tying, multilocus sequence typing, and pulsed-field gel electrophoresis to select appropriate control strains rapidly for comparative genomic analysis have been developed. The ompK36 typing method for K. pneumoniae can be used for screening high-risk resistant or hypervirulent clones. The K. pneumoniae
database based on MLST can be used for the basis of studies on bacterial virulence and resistance. Moreover, isolates of the different clones associated with resistance or specific infections in this project can be used for comparison analysis to identify virulence or resistance determinants.
Genetic differences between genetically related isolates from the same patient were identified by comparative genomic analysis, and resistance determinants will be investigated by analyzing the genetic differences. Genetic differences among these isolates have been identified, which are being analyzed to identify resistance determinants. Complete genome sequences are achieved by de novo assembly, and contigs are further
assembled by PCR and Sanger sequence. Candidate genes for carbapenem resistance have been obtained and validation is being carried out.
參考文獻
1. Oteo J, Pérez-Vázquez M, Campos J. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis 2010; 23: 320-6.
2. Dalhoff A, Janjic N, Echols R. Redefining penems. Biochem Pharmacol. 2006; 71: 1085-95.
3. Ramphal R, Ambrose PG. Extended-spectrum β-lactamases and clinical outcomes: current data. Clin Infect Dis 2006; 42 (Suppl 4): S164-72.
4. Livermore DM, Woodford N. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006; 14: 413-20.
5. Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010; 10: 597-602.
6. Bratu S, Landman D, Haag R, et al. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 2005; 165: 1430–5.
7. Doumith M, Ellington MJ, Livermore DM, et al. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 2009; 63: 659-67.
8. Liu YF, Yan JJ, Ko WC, et al. Characterization of carbapenem-non-susceptible Escherichia coli isolates from a university hospital in Taiwan. J Antimicrob Chemother 2008; 61: 1020-3.
9. Yan JJ, Wu JJ, Lee CC, et al. Prevalence and characteristics of ertapenem-nonsusceptible Escherichia coli in a Taiwanese university hospital, 1999 to 2007. Eur J Clin Microbiol Infect Dis. 2010; 29: 1417-25.
10. Wu JJ, Wang LR, Liu YF, et al. Prevalence and characteristics of ertapenem-resistant Klebsiella pneumoniae isolates in a Taiwanese university hospital. Microb Drug Resist. 2011; 17: 259-66.
11. Yan JJ, Hsueh PR, Lu JJ, et al. Extended-spectrum β-lactamases and plasmid-mediated AmpC enzymes among clinical isolates of Escherichia coli and Klebsiella pneumoniae from seven medical centers in Taiwan. Antimicrob Agents Chemother. 2006; 50:1861-4.
12. Yan JJ, Ko WC, Chuang CL, Wu JJ. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying blaIMP-8 in a university medical center in Taiwan. J. Clin. Microbiol. 2001; 39:
4433–9.
13. Chiu SK, Wu TL, Chuang YC, et al. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One. 2013; 8: e69428.
14. Willems RJ, Hanage WP, Bessen DE, Feil EJ. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011; 35: 872-900. 15. Woodford N, Turton JF, Livermore DM. Multi-resistant Gram-negative bacteria: the role of high-risk
clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011; 35: 736-55.
16. Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998; 95: 3140-5.
17. Brehony C, Jolley KA, Maiden MC. Multilocus sequence typing for global surveillance of meningococcal disease. FEMS Microbiol Rev. 2007; 31: 15-26.
18. Li JJ, Sheng ZK, Deng M, et al. Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese University Hospital. BMC Infect Dis. 2012; 12:373. 19. Pereira PS, de Araujo CF, Seki LM, et al. Update of the molecular epidemiology of
KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340). J Antimicrob Chemother. 2013; 68: 312-6.
20. Kim SY, Shin J, Shin SY, Ko KS. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis. 2013; 76: 486-90.
21. Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob Agents Chemother. 2011; 55: 3649-60. 22. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive
syndrome. Lancet Infect Dis. 2012; 12: 881-7.
23. Siu LK, Fung CP, Chang FY, et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 2011; 49: 3761-5.
24. Committee on New Directions in the Study of Antimicrobial Therapeutics: New Classes of Antimicrobials, Committee on New Directions in the Study of Antimicrobial Therapeutics:
Immunomodulation, National Research Council. Treating Infectious Diseases in a Microbial World: Report of Two Workshops on Novel Antimicrobial Therapeutics. 2006. National Academy of Sciences.
科技部補助專題研究計畫成果自評表
請就研究內容與原計畫相符程度、達成預期目標情況、研究成果之學術或應用價
值(簡要敘述成果所代表之意義、價值、影響或進一步發展之可能性)
、是否適
合在學術期刊發表或申請專利、主要發現(簡要敘述成果是否具有政策應用參考
價值及具影響公共利益之重大發現)或其他有關價值等,作一綜合評估。
1. 請就研究內容與原計畫相符程度、達成預期目標情況作一綜合評估
█ 達成目標
□ 未達成目標(請說明,以 100 字為限)
□ 實驗失敗
□ 因故實驗中斷
□ 其他原因
說明:
2. 研究成果在學術期刊發表或申請專利等情形(請於其他欄註明專利及技轉之
證號、合約、申請及洽談等詳細資訊)
論文:█已發表□未發表之文稿 □撰寫中 □無
專利:□已獲得□申請中 █無
技轉:□已技轉□洽談中
█無
其他:
(以 200 字為限)
3. 請依學術成就、技術創新、社會影響等方面,評估研究成果之學術或應用價
值(簡要敘述成果所代表之意義、價值、影響或進一步發展之可能性,以 500
字為限)。
(1) 我們建立一種全新的克雷伯氏肺炎桿菌分型方法 (細胞外膜基因多形性分型
法),將可用來分別篩選克雷伯氏肺炎桿菌的高毒性菌株以及多重抗藥性的高危
險菌株,未來亦可利用此技術,藉由比較不同 clone 但具有特定感染疾病或高抗
藥性關聯性的菌株,篩選細菌毒性或易形成抗藥性的決定因子。
(2) 我們利用多位點核酸序列分型技術建立一菌株資料庫,未來可作為研究高毒
性菌株或多重抗藥性的基礎。
(3) 我們利用次世代基因體定序已篩選出可能的碳青黴烯抗藥基因,將可進一步
進行分析。全基因的解碼完成後,亦可儲存於基因資料庫,作為日後高抗藥性高
危險菌株的研究基礎。
4. 主要發現
本研究
具有政策應用參考價值:
█否 □是,建議提供機關_______
(勾選「是」者,請列舉建議可提供施政參考之業務主管機關)
本研究具影響公共利益之重大發現:█否 □是
科技部補助計畫衍生研發成果推廣資料表
日期: 年 月 日科技部補助計畫
計畫名稱: 計畫主持人: 計畫編號: 領域:研發成果名稱
(中文) (英文)成果歸屬機構
發明人
(創作人)
技術說明
(中文) (200-500 字) (英文)產業別
技術/產品應用範圍
技術移轉可行性及預期
效益
註:本項研發成果若尚未申請專利,請勿揭露可申請專利之主要內容。科技部補助專題研究計畫成果彙整表
計畫主持人: 計畫編號:MOST 103-2320-B-006-018-MY3 計畫名稱:克雷白氏肺炎桿菌之未知 Carbapenem 抗藥機轉研究 成果項目 量化 單位 質化 (說明:各成果項目請附佐 證資料或細項說明,如期刊 名稱、年份、卷期、起訖頁 數、證號...等) 國 內 學術性論文 期刊論文 0 篇 請附期刊資訊。 研討會論文 0 專書 0 本 請附專書資訊。 專書論文 0 章 請附專書論文資訊。 技術報告 0 篇 其他 0 篇 智慧財產權 及成果 專利權 發明專利 申請中 0 件 請附佐證資料,如申請案 號。 已獲得 0 請附佐證資料,如獲證案 號。 新型/設計專利 0 商標權 0 營業秘密 0 積體電路電路布局權 0 著作權 0 品種權 0 其他 0 技術移轉 件數 0 件 收入 0 千元 1. 依「科技部科學技術研 究發展成果歸屬及運用 辦法」第 2 條規定,研 發成果收入係指執行研 究發展之單位因管理及 運用研發成果所獲得之 授權金、權利金、價金、 股權或其他權益。 2. 請註明合約金額。 國 外 學術性論文 期刊論文 2 篇 1. Genome Announc. 2017; 5(13). 2. J Clin Microbiol. 2015; 53 (10):3256-63. 研討會論文 0 專書 0 本 請附專書資訊。 專書論文 0 章 請附專書論文資訊。 技術報告 0 篇 其他 0 篇智慧財產權 及成果 專利權 發明專利 申請中 0 件 請附佐證資料,如申請案 號。 已獲得 0 請附佐證資料,如獲證案 號。 新型/設計專利 0 商標權 0 營業秘密 0 積體電路電路布局權 0 著作權 0 品種權 0 其他 0 技術移轉 件數 0 件 收入 0 千元 1. 依「科技部科學技術研 究發展成果歸屬及運用 辦法」第 2 條規定,研 發成果收入係指執行研 究發展之單位因管理及 運用研發成果所獲得之 授權金、權利金、價金、 股權或其他權益。 2. 請註明合約金額。 參 與 計 畫 人 力 本國籍 大專生 0 人次 碩士生 博士生 博士後研究員 專任助理 非本國籍 大專生 碩士生 博士生 博士後研究員 專任助理 其他成果 (無法以量化表達之成果如辦理學術活動、獲得獎項、 重要國際合作、研究成果國際影響力及其他協助產業 技術發展之具體效益事項等,請以文字敘述填列。)
科技部補助專題研究計畫執行國際合作與移地研究心得報告
日期: 年 月 日一、執行國際合作與移地研究過程
二、研究成果
三、建議
四、本次出國若屬國際合作研究,雙方合作性質係屬:(可複選)
□分工收集研究資料
□交換分析實驗或調查結果
□共同執行理論建立模式並驗証
□共同執行歸納與比較分析
□元件或產品分工研發
□其他 (請填寫) _______
五、其他
計畫編號
MOST - - - - -
計畫名稱
出國人員
姓名
服務機構
及職稱
出國時間
年 月 日至
年 月 日
出國地點
出國研究
目的
□實驗 □田野調查 □採集樣本 □國際合作研究 □使用國外研究設施
24
科技部補助專題研究計畫出席國際學術會議心得報告
日期: 年 月 日一、參加會議經過
二、與會心得
三、發表論文全文或摘要
四、建議
五、攜回資料名稱及內容
六、其他
計畫編號
MOST - - - - -
計畫名稱
出國人員
姓名
服務機構
及職稱
會議時間
年 月 日至
年 月 日
會議地點
會議名稱
(中文)
(英文)
發表題目
(中文)
(英文)
25
科技部補助專題研究計畫執行出國參訪及考察心得報告
日期: 年 月 日一、參訪及考察過程
二、心得
三、建議
四、其他
計畫編號
MOST - - - - -
計畫名稱
出國人員
姓名
服務機構
及職稱
出國時間
年 月 日至
年 月 日
出國地點
26
科技部補助專題研究計畫國外學者來臺訪問成果報告
日期: 年 月 日一、訪問過程
二、對本項專題計畫產生之影響、貢獻或主要成果
三、建議
四、其他
計畫編號
MOST - - - - -
計畫名稱
邀訪學者
姓名
服務機構
及職稱
國籍
來臺時間
年 月 日至 年 月 日
來訪目的
(可複選)
□技術指導 □實驗設備設立 □計畫諮詢/顧問 □學術演講 □國際會
議主講員 □其他
27