.InuruulofNeurnchemi .rtry
Raven Press, Lld., New York
© 1995 International Society tor Neurochemistry
A Protease Inhibitor of the Serpin Family Is a Major Protein
in Carp Perimeningeal Fluid: II. cDNA Cloning, Sequence
Analysis, and Escherichia coli Expression
*Chang-Jen Huang, Ming-Shyue Lee, *Fore-Lien Huang, and Geen-Dong Chang
*Institute of Biological Chemistry, Academia Sinica ; and yGraduate Institute
of
Biochemical Sciences and $Departmentof
Zoology, National Taiwan University, Taipei, TaiwanAbstract : A cDNA clone, pCP9, has been isolated from a common carp liver cDNA library by immunoscreening with polyclonal antiserum raised against purified bighead carp a,-antitrypsin . This clone is 1,396 by in length and has an open reading frame encoding a protein of 410 amino acid residues. The deduced amino acid sequence shows moderate homology to human a,-antitrypsin (38%), guinea pig contrapsin (35%), human a,-antichy-motrypsin (34%), and human proteinase C inhibitor (31 %), all members of the serine protease inhibitor (ser-pin) family . To confirm further that the cDNA clone was derived from the authentic carp a,-antitrypsin gene, the presumptive mature protein of pCP9 was expressed in Escherichia coli. The molecular mass of the recombinant protein matched that predicted from the nucleotide se-quence . This recombinant protein, which was recognized by antiserum against native a,-antitrypsin, was capable of formation of serpn-enzyme complexes with trypsin, chymotrypsin, and elastase. Therefore, we conclude that the protein encoded by the pCP9 clone is indeed carp a,-antitrypsin . Expression of a,-antitrypsin in brain was confirmed by reverse transcription and polymerase chain reaction performed on mRNA derived from both common carp and bighead carp brain . Key Words: Carp-Prote-ase inhibitor-Serpin-Perimeningeal fluid .
J. Neurochem . 64, 1721-1727 (1995) .
The serpins (serine proteinase inhibitors) are a fam-ily of glycoproteins that include members involved in the control of blood coagulation, fibrinolysis, comple-ment activation, and inflammation processes (Carrell and Boswell, 1986) . Many of them are plasma protease inhibitors synthesized and secreted by the liver, such as a,-antitrypsin (Long et al., 1984), contrapsin (Su-zuki et al., 1991), al -antichymotrypsin (Chandra et al., 1983), complement C, inhibitor (Bock et al ., 1986), and protein C inhibitor (Suzuki et al., 1987) . However, a few of them are synthesized in other tis-sues, such as elastase inhibitor (Remold-O'Donnell et al ., 1992), a,-antitrypsin ( Perlmutter et al ., 1985; Perl-mutter and Punsal, 1988), and complement C,
inhibi-1721
for (Bensa et al., 1983) from monocytes, protease nexin from glial cells (Sommer et al., 1987; Van Nos-trand et al., 1989), and plasminogen activator inhibitor from endothelial cells (Ny et al ., 1986) .
We have purified a 62-kDa protease inhibitor (p62) from bighead carp perimeningeal fluid (PMF) (Huang et al., 1995 ) . p62 resembles mammalian a,-antitrypsin in many respects. The protein forms complexes with and inhibits bovine trypsin, bovine chymotrypsin, and porcine pancreatic elastase. In addition, the protease inhibitor is a glycoprotein whose carbohydrate moiety can be removed by endoglycosidase F. Because a,-antitrypsin in mammals is synthesized and secreted from the liver, attempts were made to isolate cDNA clones encoding this protease inhibitor from a carp liver cDNA library. We obtained two cDNAs whose expression products were recognized by a polyclonal antiserum against pb2 . Evidence is provided to support the proposal that one of these clones encodes func-tional p62.
MATERIALS AND METHODS Enzymes and chemicals
Restriction enzymes, the Klenow fragment of DNA poly-merase, and T4 DNA ligace were purchased from either Promega (Madison, WI, U .S.A.) or Boehringer Mannheim (Mannheim, Germany) . Radiolabeled compounds were ob-tained from Amersham (Amersham, U.K.) . Sources of other chemicals were described in our preceding article ( Huang et al ., 1995) .
Received April 15, 1994; revised manuscript received August 17, 1994; accepted August 31, 1994.
Address correspondence and reprint requests to Dr. C.-D. Chang al Graduate Institute of Biochemical Sciences, National Taiwan Uni-versity, P.O. Box 23-106, Taipei 10098, Taiwan.
The sequence data in this article have been submitted to the EMBL/Genbank Data Libraries under the accession numbers of L08689 and L27172.
Abbreviations u.sect. PBS, phosphate-buffered saline; PCR, poly-merase chain reaction; PMF, perimeningeal fluid; RT, reverse tran-scription ; SDS, sodium dodecyl sulfate.
1722
General methods in molecular biology
Standard procedures in molecular biology (Sambrook et al., 1989) were used for preparation of planmid DNA, restric-tion enzyme digesrestric-tion, DNA agarose gel electrophoresis, DNA ligation, and bacterial transformation.
lmmunoscreening of a carp liver cDNA library A carp liver (Cyprinus carpio) cDNA library prepared from poly(A)-enriched RNA by unidirectional insertion of cDNA into A-ZAP II ( Short et al., 1988 ) was purchased from Stratagene (La Jolla, CA, U.S .A.) . To screen the library, A-ZAP II phages were plated at a density of 5 X IOa plaques per agar plate (150 mm i .d.) . A total of 10 plates was initially screened . After incubation for 3 h at 42°C, the plates were overlaid with nitrocellulose filters (pore size, 0.45 p,m; Mi-cron Separations, Westboro, MA, U.S.A.) that had been im-pregnated with 10 mM isopropyl-ß-~-thiogalactopyrano-side. Incubation was continued overnight at 37°C. The filters were then removed, washed wíth phosphate-buffered saline (PBS) at room temperature, and blocked in 3% skim milk in PBS for 1 h at room temperature. Following blocking, filters were probed with a polyclonal antiserum specific for carp a,-antitrypsin (p62) at a 1:500 dilution in PBS con-taining 3 mg/ml of bovine serum albumin, 1 mM EDTA, and 0.4% Triton X-100 at 4°C for 16 h. The filters were then washed three times at room temperature in PBS and incubated with horseradish peroxidase-conjugated anti-guinea pig IgG (Sigma, St. Louis, MO, U.S.A.) for 1 h at room temperature. After three washes with PBS, the immune complexes were incubated in PBS containing 0.2 mg/ml of diaminobenzamidine and revealed by adding HZO: (10 ~I in 10 ml of PBS) . Phages demonstrating stronger signals were isolated for secondary screening . Five positive clones were isolated after screening 5 x 105 plaque-forming units, and pBluescript plasmids containing cDNA inserts were obtained by in vivo excision according to the protocols provided by Stratagene .
DNA sequencing and sequence analysis
Plasmids carrying cDNA inserts were sequenced in both directions using T7 polymerase (U.S . Biochemical Corp., Cleveland, OH, U.S .A.) and the dideoxy chain termination method (Sanger et al ., 1977) . Programs from IntelliGenetics (Mountain View, CA, U.S.A.) were used to analyze the nucleotide sequences, and the SEQ program was used to deduce the amino acid sequence. A search for related se-quences in GenBank, EMBL, SWISS-PROT, and Protein Identification Resource was carried out with an IF1ND pro-gram using the FASTA algorithm of Pearson and Lipman (1988) . Alignment of the deduced amino acid sequences with those of mammalian serpins was accomplished with the GENALIGN program using the FASTP algorithm ofLipman and Pearson (1985 ) .
Expression and purification of recombinant cr,-antitrypsin
To express carp a,-antitrypsin in Fscherichia cnli, the polymerase chain reaction (PCR) was carried out to amplify the DNA fragment containing the putative mature protein using two primers: F1, (5'-GGAT000ATCACTACCAC-CATCTCCAC-3') and R1 (5'-TCTGTTTGGAGTTGG-ACCTCA-3') . PCR was performed on the planmid DNA (pCP9) with the two primers and Tay polymerase by stan-dard procedures (Mullis and Faloona, 1987) . An amplified DNA fragment of 1 .2 kb was isolated from an agarose gel, J. Neurocheru., Vol. 64, No . 4, t 995
C.-,l. HUANG FT AL.
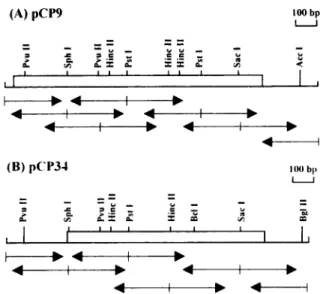
FIG. 1 . Restriction enzyme maps of cDNA clones pCP9 and pCP34 and DNA sequencing strategy . The coding regions are indicated by the open boxes. Arrows indicate the direction and lengths of determined DNA sequences.
digested with Barn HI, and cloned into the BamHl-SmaI sites of the expression vector pQE30, which was purchased from QIAGEN (Chatsworth, CA, U .S.A.) . The sequence of the PCR product was verified by DNA sequencing. This construction introduced 12 amino acids (MRGSHHHHH-HGS) into the amino-terminus of the mature protein . The resulting planmid was transformed into F. c.~oli strain JM109, and the resulting transformants were grown at 37°C over-night. The overnight culture was diluted 50-fold and regrown at 37°C for 1 h. Isopropyl-ß-t>-thiogalactopyranoside was then added to a final concentration of 1 mM, and the culture was continued for 5 h longer. The culture medium was cen-trifuged at 4,000g for 10 min, and the pellet was stored at -70°C for subsequent purification . Purification of His-tagged recombinant a,-antitrypsin by Ni - ' -nitrilotriacetate resin (QIAGEN) was accomplished according to the
proce-dures provided by the supplier. Cyanogen bromide cleavage
Purified bighead carp a,-antitrypsin ( 100 ug ) was reduced by l0 mM dithiothreitol in 8 M urea/50 mM Tris-HCl (pH 8.0) for 60 min at 37°C. Neutralized iodoacetate was added to a final concentration of 15 mM, and the sample was incubated for an additional 20 min. The sample was then dialyzed against distilled water, concentrated, and treated with Cyanogen bromide (10 mghnl ) in 100 N,l of 0.1 M HCl for 24 h at 25°C. Residual Cyanogen bromide wan evaporated in a Speed-Vac concentrator. The peptides were then dis-solved in 0.1 % trifluoroacetic acid and separated on a C,n HPLC column with a linear gradient of CH,CN. Three peaks were chosen for amino acid sequencing. Automated amino acid sequencing was performed in a gas vapor sequenator (model 471 A; Applied Biosystems, Forest City, CA, U .S.A.) equipped with an on-line phenylthiohydantoin analyzer. Reverse transcription (RT) and PCR
Total RNA was isolated from the whole brain of common carp and bighead carp (Ar-istichthos nobili.s) using a rapid acid-guanidinium procedure (Chomczynski and Sacchi,
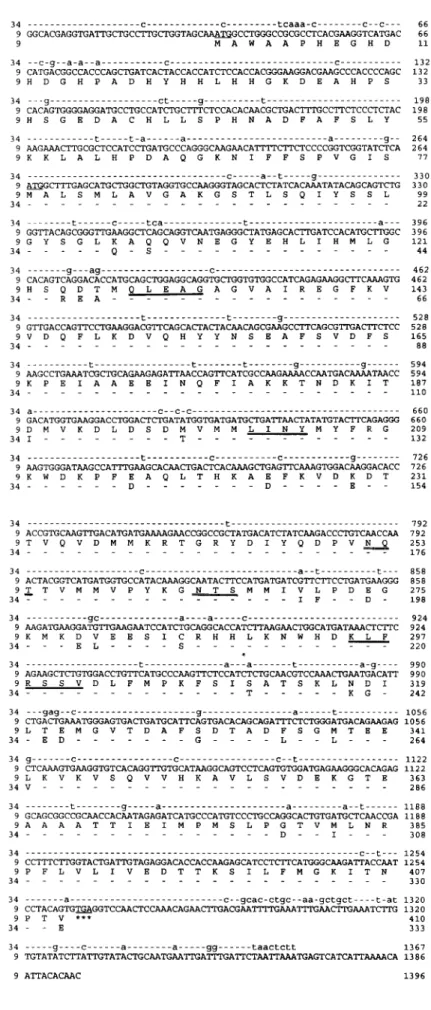
FIG. 2. Nucleotide and deduced amino acid se-quences of cDNA clones pCP9 and pCP34. The nucleotide sequences of both clones are shown in the upper two lines, whereas deduced amino acid sequences are shown in the one-letter code below the nucleotide sequences. ForpCP34, only nucleotides (in lowercase letters) and amino acid residues different from those of pCP9 are indi-cated. The initiation and termination codons are underlined. Two potential N-glycosylation sites are double underlined. Amino acid residues that match peptide sequences derived from purified p62 are underlined with thick lines.
SERPIN IN CARP PERIMENINGEAL FLUID
1723
1724
1987) . Total cellular RNA (50 Ng) was incubated at 65°C for 5 thin in a buffer containing 50 mM Tris-HCI, 75 mM KCI, 3 mM MgC12, 10 mM dithiothreitol, 2 units of RNasin, and 1 .25 mM deoxynucleotide triphosphates (dGTP, dATP, dTTP, and dCTP) and then kept on ice. Two hundred units of Superscript Moloney murine leukemia virus reverse tran-scriptase (GIBCO BRL, Gaithersburg, MD, U.S.A.) and 1 pg of oligo-dT,~ ,H primer were added and incubated at 37°C for 1 h. The reaction was then stopped by incubation at 95°C for 5 min.
Two sets of degenerated primers were designed according to the highly conserved amino acid sequence of the serpin family (see Fig. 3) . They wcre 5'CA(T/C)AA(T/C)-GCNGA(T/C)TT(T/C)GCNTT (F1 ), 5'GGNAA(A/G)-TGGGANAA(A/G)CCNTT (F2), 5'AANGG(T/C)TTN-TCCCA(T/C)TTNCC (Rl), and 5'GCNGC(T/C)TC-NGTNCC(T/C)TT(T/C)TC . PCR was carried out by addition of 20 pl of the heal-treated RT mixture and 80 pl of PCR buffer containing 25 mM Tris-HCI, 37.5 mM KCI, 1 .5 mM MgCI=, 5 mM dithiothreitol, 0.25 mM deoxynucleo-tide triphosphates, 0.25 NM primers FI/RI or F2/R2, and 2 .5 units of Tay polymerise (Promega) . Forty cycles of reaction was performed in a thermocylcer (Hybaid Ltd., Middlesex, U.K.) by the following program: 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min. The PCR products of 510 (for primers Fl/R1) and 475 by (for primers F2/R2) were isolated from an agarose gel and subcloned into the Smni site of the pUC 19 vector (Yanisch-Perron et al., 1985 ) . ,l. Neurochent., Vol. 6~ . No. 4, 1995
C.-J. HUANC ET AL.
FIG. 3. Comparison of the deduced amino acid sequence of clone pCP9 with other members of the serpin family. Known signal peptides are underlined. Gaps introduced for maximal align-ment are indicated with dots. Amino acid posi-tions are indicated at the end of each line. Shaded residues indicate amino acids that are identical in at least four proteins. The numbers at the end of each sequence show the percentage of iden-tity between a given serpin and carp a,-antitryp-sin .
Other analytical methods wcre essentially the same as de-scribed in our preceding article ( Huang et al., 1995 ) .
RESULTS
cDNA cloning
After screening 5 X 105 plaques with polyclonal antiserum raised against purified carp a,-antitrypsin, flue positive clones were isolated. On the basis of their restriction enzyme maps and partial sequences, two clones, pCP9 and pCP34, were chosen for further char-acterization. The restriction map and sequencing strat-egy of the two clones are shown in Fig . 1 .
The nucleotide sequences and the deduced amino acid sequences of both eDNA clones, pCP9 and pCP34, are shown in Fig. 2. pCP9 and pCP34 have inserts of 1,396 and 1,367 bp, respectively . The pCP9 clone contains 33 by of the 5'-untranslated region, an open reading frame of 1,233 bp, and 130 by of the 3'-untranslated region. A putative initiating ATG codon, which agrees with Kozak's rule (Kozak, 1987), is lo-cated at nucleotide 34. The open reading frame is pre-dicted to encode a proteïn of 410 amino acids with a calculated molecular mass of 46,330 Da.
The nucleotide sequence of pCP34 is similar to that of pCP9 except for several point mutations . One point
FIG. 4. SDS-polyacrylamide gel electrophoresis and western blot of recombinant and native a,-antitryp-sin. Coomassie Brilliant Blue R-250 staining of the gel is shown in lanes 1 and 2 (0.5 Ng), whereas the west-ern blot using an antiserum against a,-antitrypsin is shown in lanes 3 and 4 (0.1 Ng) . M, markers of 94, 75, 45, 28, 22, and 18 kDa; lanes 1 and 3, purified recombinant protein ; lanes 2 and 4, purified native p62.
mutation, T to C, occurs at the presumptive initiation ATG colon. As a result, the pCP34 clone contains 264 by of the 5'-untranslated region, an open reading frame of 1,002 bp, and 101 by of the 3'-untranslated region. The open reading frame encodes a protein of only 333 amino acids with a predicted molecular mass of 37,629 Da. The deduced sequence of pCP34 corresponds to amino acid residues 78-410 of pCP9 with 93%n iden-tity at the amino acid level and 90% ideniden-tity at the nucleotide level. Two potential N-glycosylation sites were identified in the deduced amino acid sequences of both cDNA clones.
Sequence homology
The deduced amino acid sequence of pCP9 shows moderate sequence homology (31-38%) with other members in the serpin family such as human cx,-anti-trypsin (Long et al., 1984), guinea pig contrapsin (Su-zuki et al ., 1991), human a,-antichymotrypsin (Chan-dra et al., 1983), and human proteinase C inhibitor (Suzuki et al., 1987) (Fig. 3) . The amino-terminal regions, including the signal peptide, are most variable. Stretches of highly conserved sequences were found, especially in the carboxy-terminal regions. A putative junction for the signal peptide and mature protein is located at a similar position as that in human a,-anti-trypsin and guinea pig contrapsin.
Characterization of recombinant protein
Most of the His-tagged recombinant protein was found in inclusion bodies and was thus solubilized with 8 M urea. The presence of 8 M urea did not affect the binding of recombinant protein to the metal column. Figure 4 shows sodium dodecyl sulfate (SDS)-gel electrophoresis and western blot analysis of the puri-fied recombinant cr,-antitrypsin. A major protein band of ~46 kDa was detected (His-p46) . `The major band represented ^-5% of the total cellular protein (data not shown) . A band of lower molecular mass was also observed . This protein appears to be the degradation product of His-p46 because its amount increased dur-ing storage (data not shown) . To demonstrate that the recombinant a,-antitrypsin was active, the ability of recombinant protein to form complexes with serine proteases was examined. Figure 5 shows that this re-combinant protein formed SDS-resistant complexes not only with trypsin but also with chymotrypsin and elastase . Polyclonal antiserum was also raised against
SERPIN IN CARP PERIMENINGEAL FLUlD
FIG. 5. Complex formation between re-combinant a,-antitrypsin and prote-ases. Recombinant His-p46 (0.3 Ng) and protease (0.2 Ng) in 5 NI of PBS were mixed and incubated at 4°C for 1 min. The mixtures were then analyzed by SDS-gel electrophoresis and visual-ized by silver staining. Lane 1, His-p46; lane 2, p46/trypsin ; lane 3, His-p46/chymotrypsin ; lane 4, His-p46/ elastase . The positions of complexes
are indicated by arrows .
DISCUSSION
mes
this recombinant a,-antitrypsin . This antiserum recog-nized native a,-antitrypsin in PMF and in other body fluids with the exception of ovarian fluid (Fig. 6) . RT-PCR of brain mRNA
Two sets of primers were designed to perform RT-PCR on mRNA derived from common carp and big-head carp brain. PCR products from both species were identical in length, and only the products from bighead carp were sequenced . The derived amino acid sequence was 84°In identical to that of the pCP9 clone (Fig. 7) .
We have isolated two cDNA clones, pCP9 and pCP34, encoding common carp a,-antitrypsin . A me-thionine colon with a flanking Kozak sequence ( Ko-zak, 1987) is located at position 34 in pCP9. Because this methionine precedes the first two consensus se-yuences found in all serpins, FAFSLY in an a-helix hA domain and NIFFSPV in a ß-sheet s6B domain, it is likely that this colon represents the translation initiation site. Therefore, the cDNA clone pCP9 has an open reading frame encoding a protein of 410 amino acids with a calculated molecular mass of 46,330 Da, which is close to the deduced molecular mass of most members of the serpin family . Stretches of sequences
FIG. 6. Body fluid distribution of carp ~x,-antitrypsin. Proteins from various body fluids as well as purified ce,-antitrypsin (0.1 Ng) were electrophoresed in a SDS-polyacrylamide gel and sub-jected to western blotting using an antiserum against purified recombinant His-p46. Lanes 1 and 2, two isoforms of carp rx,-antítrypsin (Mono-P-a and Mono-P-b) ; lanes 3-6, 0.1 NI of se-rum, 0 .1 NI of PMF, 1 NI of ovarian fluid, and 1 pl of milt.
1726
that are conserved in all serpins, including pCP9,
corre-spond to the structural features analyzed in detail in
a,-antitrypsin (Huber and Carrell, 1989) .
The reactive center of the serpins is located near
their carboxy-terminus with well-defined motifs
(Car-rell and Travis, 1985 ) . Comparison of the sequence
of this region also suggests that pCP9 belongs to the
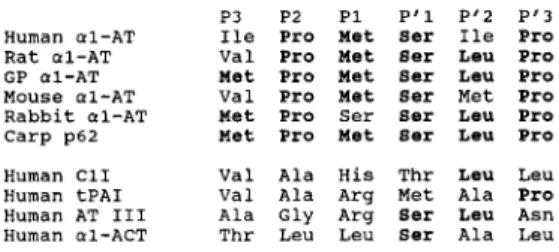
a,-antitrypsin subfamily (Fig. 8) . The amino acid of
p62 is homologous with a,-antitrypsin from other
spe-cies within this six-amino-acid region (P3-P'3) . The
above structural similarities strongly suggest that pCP9
encodes a carp a,-antitrypsin.
The cDNA clone pCP9 was originally isolated by
immunoscreening a carp liver cDNA library with
anti-serum against purified p62. We suggest that the gene
product encoded by the pCP9 is identical or similar to
p62 based on the following observations . First,
ex-pressed protein encoded by the pCP9 clone can be
recognized by the same antiserum used for
immuno-screening . Second, this recombinant protein is capable
of forming complexes not only with trypsin but also
with chymotrypsin and elastase. Tn addition, polyclonal
FIG. 8. Comparison of the amino acid sequence of the reactive site region of carp a,-antitrypsin (p62) and other serpins . a1-AT, a,-antitrypsin ; C11, complement C, inhibitor ; tPAI, tissue plasminogen activator inhibitor ; AT III, antithrombin III; a1-ACT, ce,-antichymotrypsin . Sequence data of other serpins were taken from Huber and Carrell (1989) and from Suzuki et al. (1991) . J. Neurochena., Vol. 64, Nn. 4, 1995
C .-J.
HUANG ET AL.
FIG. 7. Comparison of deduced partial amino acid sequence of pCP9 and PCR products from bighead carp brain. Identical amino acid residues are indi-cated by dashes. Amino acids that are identical to the peptide sequence derived from purified p62 are marked with double lines above the sequence. The positions of PCR primers are indicated by arrows.
antiserum raised against this recombinant protein
rec-ognized native p62. Finally, stretches of amino acid
residues in positions 128-132, 201-204, and
295-301 in pCP9 were identical to the sequence of peptides
derived from purified p62 by cyanogen bromide
cleav-age . However, two discrepancies arose during
compar-ison of the structures of purified p62 and the pCP9
clone . First, one peptide fragment, KLFRSSV, was
not preceded with a methionine residue. Second, the
deglycosylated form of p62 has an apparent molecular
mass of 53 kDa, which is 7 kDa larger than the value
predicted from the nucleotide sequence.
Gene duplication occurs frequently in teleost fish.
As a result, some species are tetraploid, including
gold-fish and common carp ( Risinger and Larhammar,
1993 ) . Goldfish and common carp have ti 100
chro-mosomes, twice the number of chromosomes in other
cyprinidae fish (Ohno et al., 1968) . Should gene
dupli-cation occur, the two resulting genes may
accommo-date different degrees of mutations. One gene may
retain the original function, whereas the other is free
to accumulate mutations, resulting in gene products
with null or different functions. In this study, we have
cloned two genes in carp liver encoding a,-antitrypsin.
The open reading frame of pCP34 encodes a protein
of only 333 amino acids, corresponding to the amino
acid residues 78-410 of pCP9. The gene product of
pCP34 lacks a signal peptide, the helix domain hA,
the sheet domain s6B, and half of the helix domain
hB, structural elements associated with a,-antitrypsin.
Some natural a,-antitrypsin variants such as antitrypsin
I and antitrypsin M Procida (Huber and Carrell, 1989)
have single substitution in the hA domain, leading to
predisposition for emphysema. Another variant,
anti-trypsin I Malton, has one phenylalanine deletion in the
s6B domain and is also nonfunctional . By analogy with
these human a,-antitrypsin variants, the gene product
ofpCP34 is expected to be a null mutant. In addition,
the product of the pCP34 gene was not detected in
body fluid. Therefore, it appears that only one a,-anti-trypsin gene(pCP9) isfunctional. We have previously cloned two types of cDNA encoding the a (al and a2) subunitofcarp gonadotropin (Chang et al., 1988) .
These twocDNAsencode proteins that differ by seven amino acids (three in the signal peptide and tour in the mature polypeptide) . Both recombinant aI and a2 subunits, expressed in insect Sf21 cells, are able to associate with theßsubunit, but only the cx1 /ß hetero-dímer displays biological activity (Huang et al.,1991) .
Therefore, tetraploid fish provide rich sources for the isolation of gene variants and provide valuable duos
for the study of the structure-function relationship.
Acknowledgment : We thank Drs. L. S. Kao and S . R. Roffler for critically reading the manuscript. This study was supported by grants from the National Science Council and hrom Academia Sinica, Taiwan, R.O.C.
REFERENCES
Bensa J . C., Reboul A., and Colornb M. G. ( 1983) Biosynthesis in vitro of complement subcomponents Clq, Cls and Cí inhibitor by resting and stimulated human monocytes. Biachern . .I. 216, 385-392.
Bock S. C., Skriver K ., Metsen E., Thógersen H.-C., Wiman R., Donaldson V . H., Eddy R. L., Marrinan J., Radziejcwska E., Huber R., Shows T. B., and Magnusson S. (1986) Human Cl inhibitor : primary structure, cDNA cloning, and chromosomal localization. Biochemistry 25, 4292-4301 .
Carrell R. W. and Boswell D. R. ( 1986) Serpins : the superfamily of plasma serine proteinase inhibitors, in Yroteinasc Inhibitors (Barren A. J. and Salvesen G ., eds), pp. 405-419. Elsevier Biomedical Press, Amsterdam .
Carrell R. W. and Travis J . (1985) a,-Antítrypsin and the serpins: variation and countervariation . Trends Bindrem. Sci. 10,
20-24.
Chandra T., Stackhouse R., Kidd V . J ., Robson K. J. H., and Woo S. L. C. ( 1983) Sequence homology between human a,-anti-chymotrypsin, a,-antitrypsin, and antithrornbin Ill . Biochemis-try 22, 5055-5060 .
Chang Y.-S., Huang C.-J., Huang F.-L., and Lo T.-B. (1988) Pri-mary structure of carp gonadotropin subunits deduced from cDNA nucleotide sequences . Irrt. ,I. Pept. Protein Rca. 32,
556-564.
Chomczynski P. and Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biocherry. 162, 156-159.
Huang C.-J., Huang F.-L., Chang G.-D., Chang Y.-S ., Lo C.-F., Fraser M . J ., and Lo T.-B. ( 1991 ) Expression of two forms of carp gonadotropin a subunits in insect cells by recombinant baculovirus . Proc. Natl. Acaad. Sci. USA 88, 7486-7490. Huang C.-J., Chen C.-C., Chen H.-J., Huang l'.-L., and Chang
G.-D. ( 1995) A protease inhibitor of the serpin family is a major protein in carp perimeningeal fluid: 1. Protein purification and characterization. J. Neuroc/rem. 64, 1715-1720 . Huber R. and Carrell R. W. ( 1989) Implications of the
three-dimen-SF.'RPIN IN CARP PERIMENINCEAL FLUID 1727
sional structure of a,-antitrypsin for structure and function of serpins . Biochemisstry 28, 8951-8966 .
Kozak M. ( 1987) Compilation and analysis of sequences upstream from the translation start site in eukaryotic rnRNAs. Nucleic Acids Re.s. 15, 8125-8148 .
Lipman D. J. and Pearson W. R. ( 1985 ) Rapid and sensitive prcxein similarity searches. Science 227, 1435-1441 .
Long G. L., Chandra T., Woo S. L. C., Davie E. W., and Kurachi K. (1984) Complete sequence of the cDNA for human a,-antitrypsin and the gene for the S variant. Biochemisn_t~ 23, 4828-4837.
Multis K. B. and Faloona F. A. ( 1987) Specific synthesis of DNA in vitro via a polyrnerase-catalyzed chain reaction. Methads En~ymnl. 155, 335-350.
Ny T., Sawdey M., Lawrence D., Millan J. L., and Loskutoff D. J. (1986) Cloning and sequence of a cDNA coding for the human ß-migrating endothelial-cell-type plasminogen activator inhibi-tor. Proc. Natl. Acad. Sci. USA 83, 6776-6780.
Ohno S., Wolf U ., and Atkin N . B. ( 1968) Evolution from fish to mammals by gene duplication . Heredito.s 59, 169-187 . Pearson W. R. and Lipman D. J . ( 1988) Improved tools for
biologi-cal sequence comparison. Proc. Nat/. Acad. Sci. USA 85, 2444-2448.
Perlmuher D. H. and Punsal P. I. ( 1988 ) Distinct and additive effects of elastase and endotoxin on expression ofa, proteinase inhibi-tor in mononuclear phagocytes. J. Binl. Chem. 263,
16499-16503.
Perlmutter D. H., Cole F. S ., Kilbridge P., Bossing T. H., and Colten H. R. ( 1985 ) Expression of the cx,-proteinase inhibitor gene in human monocytes and macrophages . Proc. Natl. Acad. Sci. USA 82, 795-799.
Remold-O'Donnell E., Chin J., and Athens M. ( 1992) Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor. Proc. Nat/. Acad. Sci. USA 89, 5635-5639. Risinger C. and Larhammar D. (1993) Multiple loci for synapse protein SNAP-25 in the tetraploid goldfish . Proc. Natl. Acad. Sci. USA 90, 10598-10602 .
Sambrook .i., Fritsch E. F., and Maniatis T. ( 1989) Molecular Clon-ing: A Laboratory Mameal. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
Sanger F., Nickten S., and Coulson A. R . (1977) DNA sequencing with chain terminating inhibitors . Proc. Natl. Acad. Sci. USA 74, 5463-5467 .
Short J. M., Fernanden J . H., Sorge J. A., and Huse W. D. ( 1988) Lambda 7_AP: a bacteriophage lambda expression vector with in vivo excision properties. Nuckis Acids Res. lfi, 7583-7600. Sommer J. S., Gloor S . M., Rovelli G . F., Hofsteenge J., Nick H., Meier R., and Monard D. ( 1987) cDNA sequence coding for rat glia-derived nexin and its homology to members of the serpin superfamily . Biochemistry 26, 6407-6410.
Suzuki K Deyashiki Y ., Nishioka J., Kurachi K., Akira M., Yama-rnoto S ., and Hashimoto S . ( 1987) Characterization of a cDNA for human protein C inhibitor. J. Biol. Chenr. 262, 61 I-616. Suzuki Y., Yoshida K., Honda E., and Sinohara H. ( 1991 ) Molecular
cloning and sequence analysis of cDNAs coding for guinea pig a,-antiproteinase S and F and contrapsin. J. Biol. Chern. 266, 928-932.
Van Nostrand W. E., Wagner S. L., Suzuki M., Choi B. H., Farrow J. S., Geddes J. W ., Cotman C. W., and Cunningham D. D. ( 1989) Protease nexin-II, a potent antichymotrypsin, shows identity to arnyloid /i-protein precursor. Nature 341, 546-549. Yanisch-Perron C., Vieira J., and Messing J. ( 1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUCl9 vectors. Gene 33, 103-119 .