The antioxidant protein alkylhydroperoxide reductase
of
Helicobacter pylori switches from a peroxide
reductase to a molecular chaperone function
Ming-Hong Chuang*†‡, Ming-Shiang Wu‡§, Wan-Lin Lo*†, Jaw-Town Lin§, Chi-Huey Wong¶储**, and Shyh-Horng Chiou*†**
*Institute of Biochemical Sciences, National Taiwan University, Taipei 106, Taiwan;†Institute of Biological Chemistry, Academia Sinica, Taipei 115, Taiwan; §Division of Gastroenterology, Department of Internal Medicine, National Taiwan University Hospital, Taipei 100, Taiwan;¶Genomics Research Center, Academia Sinica, Taipei 115, Taiwan; and储The Scripps Research Institute, La Jolla, CA 92037
Contributed by Chi-Huey Wong, December 16, 2005
Helicobacter pylori, an oxygen-sensitive microaerophilic bacte-rium, contains many antioxidant proteins, among which alkylhy-droperoxide reductase (AhpC) is the most abundant. The function of AhpC is to protect H. pylori from a hyperoxidative environment by reduction of toxic organic hydroperoxides. We have found that the sequence of AhpC from H. pylori is more homologous to mammalian peroxiredoxins than to eubacterial AhpC. We have also found that the protein structure of AhpC could shift from low-molecular-weight oligomers with peroxide-reductase activity to high-molecular-weight complexes with molecular-chaperone func-tion under oxidative stresses. Time-course study by following the quaternary structural change of AhpC in vivo revealed that this enzyme changes from low-molecular-weight oligomers under nor-mal microaerobic conditions or short-term oxidative shock to high-molecular-weight complexes after severe long-term oxida-tive stress. This study revealed that AhpC of H. pylori acts as a peroxide reductase in reducing organic hydroperoxides and as a molecular chaperone for prevention of protein misfolding under oxidative stress.
peroxiredoxin兩 oxidative stress 兩 phylogenetic comparison 兩 dual functionality兩 quaternary structural change
C
hronic infection by Helicobacter pylori, a Gram-negative bacterium, can lead to various gastrointestinal diseases, including chronic gastritis, gastric and duodenal ulceration, and gastric cancer (1–4). After successful isolation and culture of H.pylori from patients with gastritis⬇20 years ago by Marshall and
Warren (5), the complete genome sequences of various H. pylori strains have been identified (6). However, the specific functions of most gene products remain to be defined. Previous investi-gations in this field focused mainly on the postinfection patho-genic events and more recently on the alteration of gene expression and protein modification under environmental stresses (7–10).
Infection by H. pylori involves adhesion and colonization to the epithelial cells of gastric mucosa (11, 12), resulting in the activation of the immune system and gastric inflammatory responses, including discharge of reactive oxygen species (ROS) from phagocytes (13, 14). To survive under the oxidative stress,
H. pylori expresses antioxidant proteins, including
alkylhydroper-oxide reductase (AhpC), superalkylhydroper-oxide dismutase, and catalase to reduce the toxic and reactive peroxides and oxygen radicals (15, 16).
AhpC, a member of the thiol-dependent 2-Cys peroxiredoxin family (17), is the most abundant antioxidant protein in H. pylori (18). Previous reports on bacterial AhpC indicated that the protein forms homodimers through an intersubunit disulfide linkage which can then be reduced by electron donors, as exemplified by AhpF in Salmonella typhimurium and AhpD in
Mycobacterium tuberculosis (19, 20). However, the electron
donor flavoprotein (AhpF) used by most bacteria to reduce
AhpC was shown to be inactive for the AhpC of H. pylori (21–24). In addition, some AhpC-deficient mutant strains have been found to have an increase in the expression of neutrophil-activating protein (NapA) (15, 18) or a decrease in the activity of catalase (25). Our previous study has also revealed that the transcription and expression of AhpC in H. pylori isolated from duodenal ulcer patients decrease under long-term oxidative stress (⬎8 h in 20% O2) (26). These results suggest that AhpC
in H. pylori, in addition to providing its anti-oxidative activity, may also play an important role in the regulation of other cellular functions. It was indeed observed that yeast peroxiredoxins (Prxs) can switch from an enzymatic activity to a molecular-chaperone function under oxidative stresses (27). However, there is no report on AhpC in prokaryotes regarding its struc-tural modification or functional switch under oxidative stresses. To investigate whether prokaryotic AhpC possesses functional properties similar to those of eukaryotic Prxs, we have cloned and expressed the AhpC gene of H. pylori from patients with duodenal ulcer and gastric cancer, and we have studied the structural and functional alterations of AhpC caused by oxida-tive stresses. We have demonstrated that the AhpC of H. pylori acts not only as a peroxide reductase but also as a stress-dependent molecular chaperone.
Results
Cloning and Nucleotide Sequencing. DNAs of two different H.
pylori strains (HD30 and HC28) isolated from patients with
duodenal ulcer and gastric cancer, respectively, were amplified by PCR and the products (597 bp in size) were cloned and sequenced. Through cloning and sequencing we obtained the complete cDNA sequences and deduced the amino acid se-quences of these two AhpC genes (sese-quences have been depos-ited in databanks).
Both AhpC proteins have the two conserved catalytic sites similar to 2-Cys peroxiredoxins of eukaryotic species. The catalytic sites contain Cys-49 and Cys-169 in the conserved tripeptide Val-Cys-Pro (VCP) and form an intramolecular di-sulfide bond essential for the catalytic activity (21, 27, 28). When we compared these two AhpC proteins, their amino acid se-quences were found to be almost identical, with only one point
Conflict of interest statement: No conflicts declared.
Abbreviations: AhpC, alkylhydroperoxide reductase; HMW, high-molecular-weight; EM, electron microscopy; LMW, low-molecular-weight; Prx, peroxiredoxin; Trx, thioredoxin; TrxR, thioredoxin reductase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY871104 (ahpC gene of HC28) and AY871312 (ahpC gene of HD30)].
‡M.-H.C. and M.-S.W. contributed equally to this work.
**To whom correspondence may be addressed. E-mail: shchiou@ntu.edu.tw or wong@ scripps.edu.
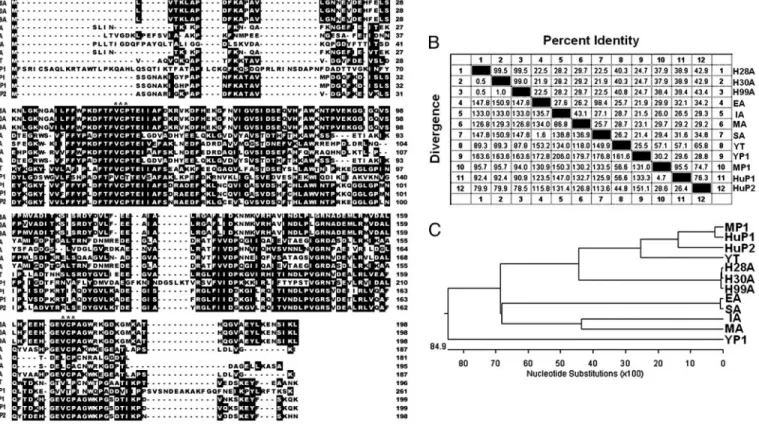
mutation, at residue 101, valine in the HD30 strain and methi-onine in the HC28 strain (data not shown). The biological significance of the point mutation is, however, unclear. Sequence Alignment and Comparison.We aligned 12 protein se-quences encompassing representative AhpC proteins and per-oxiredoxins of H. pylori and other prokaryotic and eukaryotic organisms (Fig. 1A). Distinct differences were found at the N-and C-terminal regions with VCP tripeptide (designated by three stars) being mostly conserved in these aligned sequences.
We have also performed pair-wise sequence homology and divergence comparison among these sequences and the result (Fig. 1B) showed only⬇20–30% sequence homology between H.
pylori AhpC and other bacterial AhpC. However, the sequence
homology between H. pylori AhpC and mammalian (human or mouse) peroxiredoxins is as high as 43%. On the other hand, the phylogenetic tree analysis similar to our previous report (29) showed that all of the three H. pylori AhpC proteins are more homologous to mammalian peroxiredoxins than to other bacte-rial AhpC proteins (Fig. 1C). Using computer modeling, we have also found that the AhpC protein of H. pylori isolates is more similar structurally to mammalian peroxiredoxins (data not shown). Taken together, these results suggested that the AhpC
proteins present in the H. pylori isolates of human origin are more similar to mammalian than bacterial peroxiredoxins. Con-ceivably, their functional or enzymatic characteristics may also be closer to eukaryotic peroxiredoxins.
Modification of AhpC from a Peroxide Reductase to a Molecular Chaperone-Like Peroxiredoxinin Vitro.To study whether the AhpC protein from H. pylori has both the peroxide reductase and molecular chaperone activities, we performed a series of exper-iments under oxidative stress in vitro. First, we analyzed the peroxide reductase and chaperone activities of these cloned AhpC proteins from H. pylori isolates after exposure to 10 mM H2O2for 1 or 12 h (Fig. 2 A and B). Second, we checked their
molecular size by native and SDS兾PAGE. Both AhpC proteins treated with H2O2 for 12 h were shown to lose their enzyme
activity greatly and have a structural change from low-molecular-weight (LMW) oligomers (⬇75–125 kDa) to a HMW complex (⬎669 kDa) as revealed by native gradient-gel (4–12%) elec-trophoresis (Fig. 2C). In contrast to the result of native PAGE, SDS兾PAGE of both the normal and stressed AhpC showed clearly a single band of⬇26 kDa, with stressed AhpC showing a slightly larger size than normal AhpC (Fig. 2C Inset). The difference was probably due to the oxidation of some amino acid
Fig. 1. Sequence alignment and homology comparison of H. pylori AhpC with homologous AhpC and Prxs from other species. (A) Alignment of the amino acid sequences of AhpC and Prxs (2-Cys Prx) from several representative prokaryotes and eukaryotes. The encoded amino acid sequences were aligned, and gaps (dashes) were introduced to optimize sequence alignment. Identical and consensus amino acids in all sequences are shown in black boxes. The highly conserved tripeptide (VCP) related to the catalytic function is designated by stars. (B) Pairwise comparison of protein sequence identity and divergence. (C) Construction of phylogenetic tree based on sequence divergence between the AhpC sequences from clinical H. pylori isolates (HC28 and HD30) and AhpC homologues from other species in A. Sequence data were analyzed with theMEGALIGNprogram ofLASERGENE(DNASTAR, Madison, WI) as described previously (29). The percent divergence is calculated by comparing sequence pairs in relation to the relative positions in the phylogenetic tree, in contrast with the percent identity, which is estimated by comparing percent sequence identity directly without accounting for phylogenetic relationships. The tree was built by using theCLUSTAL Wprogram and weighted residue-weight table. The length in each branch represents the sequence distance between aligned pairs. The scale beneath the tree measures the distance between sequences (in nucleotide substitutions). It is noted that the AhpC proteins of clinical H. pylori isolates were more similar to mammalian Prxs (human and mouse) than eubacterial AhpC. The abbreviations for amino acid sequences of AhpC and Prx from various species are as follows: H28A and H30A, AhpC of two clinical strains of H. pylori studied in this report; H99A, AhpC of J99 strain of H. pylori; EA, AhpC of Escherichia coli; IA, AhpC of Idiomarina loihiensis; MA, AhpC of M. tuberculosis; SA, AhpC of S. typhimurium; YT, Trx peroxidase of yeast (Saccharomyces cerevisiae); YP1, Prx I of yeast (S. cerevisiae); MP1, Prx I of mouse; HuP1, Prx I of human; HuP2, Prx II of human.
residues in AhpC upon H2O2treatment. As shown in lines 2 and
3 of Fig. 2B, AhpC started to act as a chaperone protein upon exposure to H2O2treatment for⬎1 h, and it exhibited the full
chaperone activity after 12 h. To rule out the possibility that
protein aggregation induced by DTT in this chaperone-activity assay was due to the reduction of HMW complexes of AhpC, we determined the sulfhydryl contents in the native and oxidized forms of AhpC (Table 1). The results indicated that the native form of AhpC contained three disulfide bonds, and no disulfide bonds were detected in the oxidized HMW form of AhpC. It clearly suggests that all of the disulfide bonds in native AhpC were converted to sulfinic or sulfonic acid (–SO2H or –SO3H) in
the oxidized HMW form of AhpC by H2O2(28) and are thus
resistant to reduction by DTT. Therefore, the experimental results support the notion that the protein aggregation in the chaperone-activity assay is indeed due to the reductive unfolding of insulin induced by DTT, resulting in the accumulation of insulin aggregate upon refolding. It is noteworthy that HMW complexes of AhpC can indeed protect insulin from DTT-induced protein aggregation.
The gross structural change of AhpC under oxidative stress can also be detected by electron microscopy (EM). The EM images showed that AhpC appeared as small molecules of irregular shape under normal conditions. HMW AhpC com-plexes can be found as spherical particles after 10 mM H2O2
treatment at 37°C for 12 h (Fig. 2D), corroborating the size-estimation results based on native-gel electrophoresis. Collec-tively, these in vitro assays clearly suggested that AhpC possesses dual functions: i.e., it acts as peroxide reductase in the form of oligomers under normal physiological conditions and changes to HMW complexes with chaperone-like activity under oxidative stress.
Exposure ofH. pylori to Oxidative Stress in Vivo Induces Functional Change of AhpC. A previous report showed that the enzyme activity of Prxs was rapidly decreased by bursts of intracellular peroxide production (30), in agreement with our previous result (26) showing that the amount of monomeric AhpC in H. pylori was greatly decreased after long-term oxidative stress. Herein we have further performed a time-course analysis of AhpC under long-term oxidative (20% O2) conditions by native PAGE and
Western blotting to probe the structural changes of the AhpC of
H. pylori isolates under oxidative stress in vivo.
Crude extracts prepared from cell cultures of H. pylori isolates incubated under normal or oxidative stress conditions for 8 and 16 h were subjected to native PAGE. The AhpC proteins obtained from normal H. pylori cells consisted of multiple forms of LMW oligomers in a size range of 75–125 kDa (stained with Coomassie blue) similar to that shown in Fig. 2C. A time-course study on native PAGE followed by immunoblotting with anti-bodies to H. pylori AhpC (Fig. 3) revealed that normal or short-term stress (0–8 h) AhpC formed approximately LMW trimers or higher oligomers. Almost all AhpC protein molecules were, however, converted to HMW complexes after long-term oxidative stress (⬎16 h). Consistent with the in vitro analysis (Fig. 2 A–C), these in vivo analyses also suggest that H. pylori AhpC switched from the peroxide reductase activity (LMW oligomers) under normal conditions to chaperone-like activity (HMW complexes) under oxidative stress.
Fig. 2. The dual-function activity of recombinant H. pylori AhpC from HC28. (A) Peroxide reductase activity assays of recombinant AhpC from HC28 were measured in the absence of Trx system (line 1). Peroxide reductase activity assays of AhpC pretreated with 10 mM H2O2(12 h) were measured in the presence of Trx system (line 2). The same activity assay of AhpC was carried out in the presence of Trx system without pretreatment with H2O2(line 3). Recombinant AhpC from HC28 and HD30 possesses similar peroxide reductase activity under short-term oxidative treatment (⬍1 h) as shown by the rapid decrease of NADPH absorbance at 340 nm (line 3). However AhpC lost most of the peroxide reductase activity when treated with H2O2for 12 h (line 2). (B) The chaperone activity of recombinant AhpC from HC28 or HD30 (data not shown) was measured by the turbidity change at 360 nm of DTT-induced aggregation of insulin at 25°C. The activity was measured in the absence of H2O2(line 1) or in the presence of 10 mM H2O2for 1 h (line 2) and 12 h (line 3), respectively. The protein exhibited its full chaperone activity after 12 h. (C) Structural change of purified AhpC was analyzed by native PAGE (4 –12% gradient polyacrylamide gel) after treatment with 10 mM H2O2for 12 h. The proteins were converted to high-molecular-weight (HMW) complexes with molecular mass⬎669 kDa as estimated by gel electrophoresis with molecular mass markers. (Inset) All of the different forms of AhpC isolated from normal or stressed conditions showed a 26-kDa single band on SDS兾15% PAGE. N, normal condition; O, oxidative stress condition. Gels were stained with Coo-massie blue and molecular mass markers are shown on the left. (D) Electron micrographs showing structures of recombinant purified AhpC protein from HC28 under normal condition (Left) or after treatment with 10 mM H2O2for 12 h (Right). Electron micrographs were obtained by negative staining with uranyl acetate. (Scale bar, 100 nm.) Arrows indicate the presence of HMW complex particles under oxidative stress. Similar EM images were obtained with recombinant AhpC of HD30.
Table 1. Determination of sulfhydryl contents of normal and stressed recombinant AhpC proteins from H. pylori with or without dithiothreitol (DTT) by Ellman’s reagent
AhpC DTT No. of Cys per protein Normal ⫺ 0.12⫾ 0.09 Normal ⫹ 5.98⫾ 0.32 Stressed ⫺ 0.08⫾ 0.15 Stressed ⫹ 0.03⫾ 0.02
Discussion
To combat the reactive oxygen species released from the host immune system and survive in the gastric mucosa, pathogenic H.
pylori is equipped with a number of detoxifying proteins (25, 31).
Among these, the most abundant one is AhpC. Several func-tional studies of H. pylori AhpC have shown that the AhpC-deleted mutants have increased sensitivity to oxygen (15), over-express neutrophil-activating protein (18), and accelerate inactivation of catalase (25).
As a member of 2-Cys peroxiredoxins (17, 21, 31), AhpC has been shown to be Trx-dependent (17, 27, 32, 33). The native structure of AhpC in S. typhimurium was shown to exist as a dimer (34, 35). Although the structure of bacterial AhpC was shown to be similar to that of eukaryotic Prxs, the cofactors for AhpC and Prxs were different. The peroxide reductase activity of AhpC from S. typhimurium was shown to be flavin-dependent, whereas the AhpC in M. tuberculosis was lipoamide and AhpD-dependent (19, 20). All these data indicated that the bacterial AhpC proteins were more heterogeneous in functional charac-teristics than eukaryotic Prxs.
We have previously demonstrated that the enzyme activity of AhpC in H. pylori of clinical isolates was suppressed under long-term oxidative stress (26). We have further compared the sequences of AhpC and Prxs from different species, including bacteria, yeasts, and mammals (Fig. 1 A–C), and found that the AhpC of H. pylori is more homologous to human Prx than to eubacterial AhpC. These results suggest that a long-term infec-tion of H. pylori may facilitate the recombinainfec-tion of its AhpC gene with human Prx genes to form a human-like AhpC (36–38). The AhpC of H. pylori is closer to eukaryotic Prx than to other prokaryotic AhpC phylogenetically (Fig. 1C), and it was found that H. pylori AhpC also acts as a molecular chaperone as does yeast Prx. Herein we propose a likely mechanism underlying the structural and functional changes of AhpC in H. pylori under short-term and long-term oxidative stresses (Fig. 4). When H.
pylori encounters intracellular attack by reactive oxygen species,
the AhpC will react to ameliorate peroxides and oxygen radicals generated under short-term oxidative stress. However, some AhpC will be converted to HMW chaperones to prevent the misfolding or unfolding of proteins under long-term stress conditions. If the oxidative stress is too severe because of the presence of excessive damaged proteins (perhaps including some essential transcription factors for AhpC transcription), all ex-pressed AhpC may be switched to molecular chaperones for salvage of unfolded proteins. Based on the full genome se-quences of H. pylori (6), it is of interest to note that H. pylori does not appear to contain all small heat-shock proteins (sHSPs) used as potential molecular chaperones by most eukaryotes. There-fore, it is suggested that AhpC or other as-yet-unidentified proteins in H. pylori may possess stress-induced chaperone property like sHSPs (39). Under various environmental stresses, these proteins may act as potent stress sensors and chaperones for H. pylori to survive and persist in the extreme environment of human stomachs.
Materials and Methods
Materials.The H. pylori strains used in this study were isolated from the gastric biopsy specimens in two patients, one with duodenal ulcer and one with gastric cancer, and were abbrevi-ated by HD30 (from human duodenal ulcer) and HC28 (from human gastric cancer), respectively. The Escherichia coli strains used for sequencing, genetic manipulation and expression were
Fig. 3. Oxidative stress-dependent switching of AhpC structures in vivo. Crude proteins extracted from two clinical H. pylori isolates HC28 and HD30 under atmospheric oxidative stress were separated by native 4 –12% PAGE and subjected to immunoblotting with anti-H. pylori-AhpC polyclonal antibodies. The results of a time-course study indicated that the proteins formed oli-gomers (lower arrow) under short-term (8 h) stress; however, all AhpC were converted to HMW complexes (higher arrow) with molecular chaperone activity after long-term stress (16 h).
Fig. 4. A schematic representation of the oxidative stress-dependent structural and functional switching of AhpC in H. pylori. Two aggregate forms of AhpC in H. pylori with distinct quaternary structures are formed in two independent pathways, i.e., under antioxidant homeostasis (A) and severe hyperoxia stress conditions (B). When H. pylori grows in a microaerobic environment or short-term oxidative shock, their AhpC proteins may exist as LMW oligomeric forms to effect the inherent peroxide reductase function for reducing toxic reactive oxygen species (ROS). AhpC proteins could be converted to HMW complexes with chaperone activity for prevention of unfolded proteins from aggregation under severe long-term stress conditions.
ECOS101 (DH5␣) and ECOS21 (BL21), respectively. These H.
pylori strains were grown on Centers for Disease Control and
Prevention (CDC) anaerobe blood agar plates (BD) at 37°C in a modular atmosphere-controlled system (5% O2兾10% CO2兾
85% N2) (Don Whitley Scientific, Shipley, U.K.), and confirmed
to be H. pylori because of their urease activity and helical morphology as determined by phase-contrast microscopy. Luria– Bertani (LB) agar and media supplemented with ampicillin (100 g兾ml) were used for growing E. coli strains at 37°C. Protein concentrations were determined with the bicinchoninic acid (BCA) protein assay kit (Pierce). Protein standards used in PAGE were purchased from Amersham Pharmacia. Insulin, Ellman’s reagent,L-cysteine, and the bacterial thioredoxin (Trx) system used in AhpC activity assays including thioredoxin, thioredoxin reductase, NADPH, and H2O2were obtained from
Sigma.
Cloning and Sequencing ofahpC from Clinical H. pylori.Genomic DNA was prepared from confluent cultures on CDC agar plates with a QIAprep DNA minikit (Qiagen, Chatsworth, CA). The primers used for PCR (40) and sequencing are listed in Table 2. Briefly, specific PCR was carried out in 50-l mixtures contain-ing 10 ng of DNA, 0.2M deoxyribonucleoside triphosphates (dNTPs), 1M each primer, 1 M cDNA, and 1 unit of Taq DNA polymerase (Promega) in a standard PCR buffer for 35 cycles under the following conditions: 94°C for 1 min, 60°C for 1 min, and 72°C for a time period chosen based on the size of expected fragments (1 min兾kb). The PCR products were then analyzed by 1.5% agarose gel electrophoresis using standard protocols. After PCR amplification, the products (⬍650 bp) were collected and purified by using an EasyPure PCR clean-up kit (Bioman, Taipei, Taiwan), treated with T4 DNA ligase (Promega), and cloned into the yT&A vector to give yAhpC-D30 and yAhpC-C28, which were then transformed into E. coli DH5␣ cells. These constructs were sequenced in both directions by using a Taq DyeDeoxy Terminator Cycle Sequencing kit
(Ap-plied Biosystems Prism) on a 373A sequencing system (Perkin– Elmer).
Expression and Purification of Recombinant AhpC.For expression of the AhpC in E. coli, the yAhpC-D30 and yAhpC-C28 plasmids were doubly digested with the restriction endonucleases XhoI and NdeI, and the products were ligated into pET21b (Novagen), a His-taq protein expression vector bearing the T7 promoter and ampicillin resistance. The plasmids with the correct gene se-quences were then transformed into E. coli strain BL21 (DE-3). The His6-fused AhpC was purified by using a native Ni-NTA
column (Qiagen, Chatsworth, CA). The enzyme activity of the recombinant AhpC containing the added six His residues at the C terminus was measured according to previous reports (21, 27). The purity of the purified recombinant AhpC was determined to be⬎99% based on SDS兾PAGE. The structural and functional changes of recombinant AhpC proteins from HD30 or HC28 after treatment with 10 mM H2O2and Trx system for 1 or 12 h
were analyzed by molecular size measurement on native PAGE and SDS兾PAGE and peroxide reductase and chaperone-activity assays plus electron microscopy as described below.
Gel Electrophoresis and Western Blot Analysis ofH. pylori AhpC Under Oxidative Stress.Plate-grown H. pylori cells of HC28 and HD30 strains incubated under normal (5% O2) condition for 48 h were
transferred to oxidative-stress (20% O2) conditions.
Subse-quently, the cells were harvested at various time intervals (0, 8, or 16 h) and suspended in PBS. For native PAGE, 50g of total soluble proteins extracted from cells disrupted by lysozyme and sonication was dissolved in a native buffer without SDS and then applied to native gel electrophoresis. After electrophoresis, the proteins were stained directly with Coomassie blue or trans-ferred to poly(vinylidene difluoride) (PVDF) membranes under a voltage of 392 V for 30 min. After transfer, the membranes were saturated with 5% (wt兾vol) nonfat dry milk powder in PBS兾0.1% Tween 20 at room temperature for 2 h, followed by incubation with polyclonal antibodies against H. pylori-AhpC (anti-HPAhpC) for 3 h. After three washes with PBS兾0.1% Tween 20, the membranes were incubated with a solution of anti-rabbit Ig conjugated with alkaline phosphatase. After 1-h incubation at room temperature, the membranes were washed three times with PBS兾0.1% Tween 20 and the membrane blots were developed by using color-forming nitroblue tetrazolium (NBT)兾5-bromo-4-chloro-3-indolyl phosphate (BCIP) reagents. EM and Imaging. The AhpC proteins of normal or 10 mM H2O2-treated (12 h) forms were absorbed to glow-discharged
carbon-coated copper grids by incubation at 25°C for 5 min, then the grids were rinsed with deionized water and stained with 2% (wt兾vol) uranyl acetate. The EM images were recorded with a JEOL JSM-1200EX II transmission electron microscope at an accelerated voltage of 80 kV and a magnification of 50,000-fold. Light-optical diffractograms were used to select micrographs to avoid defocus and verify that no drift or astigmatism was present (27). For image processing, the negative image scanner (Kodak) and EM software package (IMAGIC) were used.
Sequence Database Analysis.We have performed sequence com-parison and alignment of the two AhpC homologues from two
H. pylori clinical isolates (HC28 and HD30) and the 10 reported
amino acid sequences obtained from bacterial and eukaryotic AhpC proteins in the National Center for Biotechnology Infor-mation (NCBI) sequence database. The alignment, phylogenetic analysis, and estimation of sequence divergence were carried out by usingMEGALIGNandPROTEANprograms (DNASTAR, Mad-ison, WI).
Table 2. Strains, plasmids, and primers
Strains, plasmids
and primers Description and source Strains
H. pylori
HC28 Clinical strain isolated from a patient with gastric cancer
HD30 Clinical strain isolated from a patient with duodenal ulcer
E. coli
DH5␣ Cloning strain from commercial ECOS101 (Yeastern Biotech, Taipei, Taiwan) BL21 Expression strain from commercial ECOS21
(Yeastern Biotech) Plasmids
yT&A Cloning vector for sequencing (Yeastern Biotech) yAhpC-C28 ahpC of HC28 inserted into yT&A
yAhpC-D30 ahpC of HC30 inserted into yT&A
pET21b Cloning vector for expression (in our laboratory stock)
pAhpC-C28 ahpC of HC28 inserted into the Xhol and Ndel
sites of pET21b
pAhpC-D30 ahpC of HD30 inserted into the Xhol and Ndel
sites of pET21b Primers
ahpCF 5⬘-CCATATGTTAGTTACAAAACTTGCC-3⬘ ahpCR 5⬘-CTCGAGAAGCTTAATGGAATTTTC-3⬘
Peroxide Reductase and Chaperone-Like Activity Assays.The perox-ide reductase activity of recombinant AhpC was monitored with an Ultrospec 4000 spectrophotometer (Amersham Pharmacia) at 25°C by following the decrease in A340within 12 min due to
NADPH oxidation. Each assay was performed by using 20M each AhpC in a total volume of 1.0 ml of 50 mM potassium phosphate buffer (pH 7.0)兾0.1 M ammonium sulfate兾0.5 mM EDTA containing Trx system (5 M thioredoxin, 0.5 M thioredoxin reductase, and 150M NADPH) and 1 mM H2O2
as described (21). Chaperone-like activity of recombinant AhpC was studied based on the DTT-induced insulin reductive un-folding and chaperone-assisted reun-folding (29, 41). The reaction mixture was prepared in a total volume of 1.0 ml of PBS buffer containing insulin and DTT. The insulin substrate stock solution consisted of 8.5 mg of insulin dissolved in 1 ml of 0.1 M NaOH, 1.7 ml of 0.5 M NaH2PO4兾Na2HPO4buffer (pH 6.8), and 1.7 ml
of 1 M NaCl, and the total volume was adjusted to 17 ml with distilled H2O. The AhpC-dependent chaperone assay was
car-ried out at 25°C by recording the turbidity change of OD360
within 42 min upon the initiation of DTT-induced insulin aggregation or until the turbidity curve reaches a plateau (29). Determination of the Sulfhydryl Contents.The sulfhydryl contents were determined by using Ellman’s reagent and DTT according to a published method (28). The protein concentrations of purified recombinant AhpC and oxidative HMW complexes induced by 10 mM H2O2 plus their reduced proteins in the
presence of 10 mM DTT were determined by the BCA protein assay kit as described above. The sulfhydryl concentrations in these proteins were determined from the calibration curve with known concentrations of standardL-cysteine solutions.
We thank Hwei-Yuan Chang for her preparing and providing the clinical isolates of H. pylori in this study. This work was supported in part by Academia Sinica and National Science Council Grants 93-2311-B-002-033 and 94-2311-B-002-027 (to S.-H.C.) and a grant from National Taiwan University Hospital (to M.-S.W. and J.-T.L.).
1. Hopkins, R. J., Girardi, L. S. & Turney, E. A. (1996) Gastroenterology 110, 1244–1252.
2. Kreiss, C. & Blum, A. L. (1995) Curr. Opin. Gastroenterol. 11, 25–31. 3. Sugiyama, T. & Asaka, M. (2004) Med. Electron Microsc. 37, 149–157. 4. Stoicov, C., Saffari, R., Cai, X., Hasyagar, C. & Houghton, J. M. (2004) Gene
341,1–17.
5. Marshell, B. J. & Warren, J. R. (1984) Lancet. 1, 1311–1315.
6. Tomb, J. F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., et al. (1997) Nature 388, 539–547.
7. Wen, Y., Marcus, E. A., Matrubutham, U., Gleeson, M. A., Scott, D. R. & Sachs, G. (2003) Infect. Immun. 71, 5921–5939.
8. Park, A. M., Li, Q., Nagata, K., Tamura, T., Shimono, K., Sato, E. F. & Inoue, M. (2004) Free Radical Biol. Med. 6, 1126–1133.
9. Barnard, F. M., Loughlin, M. F., Fainberg, H. P., Messenger, M. P., Ussery, D. W., Williams, P. & Jenks, P. J. (2004) Mol. Microbiol. 51, 15–32. 10. Kawakubo, M., Ito, Y., Okimura, Y., Kobayashi, M., Sakura, K., Kasama, S.,
Fukuda, M. N., Fukuda, M., Katsuyama, T. & Nakayama, J., (2004) Science 305, 1003–1006.
11. Petersen, A. M. & Krogfelt, K. A. (2003) FEMS Immunol. Med. Microbiol. 36, 117–126.
12. Suerbaum, S. & Michetti, P. (2002) N. Engl. J. Med. 347, 1175–1186. 13. Bagchi, D., Bhattacharya, G. & Stohs, S. J. (1996) Free Radical Res. 24,
439–450.
14. Monack, D. M., Mueller, A. & Falkow, S. (2004) Nat. Rev. Microbiol. 2, 747–765.
15. Olczak, A. A., Olson, J. W. & Maier, R. J. (2002) J. Bacteriol. 184, 3186–3193. 16. Comtois, S. L., Gidley, M. D. & Kelly, D. J. (2003) Microbiol. 149, 121–129. 17. Wood, Z. A., Schroder, E., Harris, J. R. & Poole, L. B. (2003) Trends Biochem.
Sci. 28, 32–40.
18. Olczak, A. A., Seyler, R. W., Olson, J. W. & Maier, R. J. (2003) Infect. Immun. 71,580–583.
19. Reynolds, C. M. & Poole, L. B. (2001) Biochemistry 40, 3912–3919. 20. Koshkin, A., Knudsen, G. M. & Ortiz De Montellano, P. R. (2004) Arch.
Biochem. Biophys. 427, 41–47.
21. Baker, L. M. S., Raudonikiene, A., Hoffman, P. S. & Poole, L. B. (2001) J. Bacteriol. 183, 1961–1973.
22. Lundstrom, A. M. & Bolin, I. (2000) Microb. Pathol. 29, 257–266.
23. Yan, J., Kumagai, T., Ohnishi, M., Ueno, I. & Ota, H. (2001) Helicobacter 6, 274–282.
24. Owen, R. J. & Xerry, J. (2003) J. Med. Microbiol. 52, 515–524.
25. Wang, G., Conover, R. C., Benoit, S., Olczak, A. A., Olson, J. W., Johnson, M. K. & Maier, R. J. (2004) J. Biol. Chem. 279, 51908–51914.
26. Chuang, M.-H., Wu, M.-S., Lin, J.-T. & Chiou, S.-H. (2005) Proteomics 5, 3895–3901.
27. Jang, H. H., Lee, K. O., Chi, Y. H., Jung, B. G., Park, S. K., Park, J. H., Lee, J. R., Lee, S. S., Moon, J. C., Yun, J. W., et al. (2004) Cell 117, 625–635. 28. Britto, P. J., Knipling. L., McPhie, P. & Wolff, J. (2005) Biochem. J. 389,
549–558.
29. Yu, C.-M., Chang, G.-G., Chang, H.-C. & Chiou, S.-H. (2004) Exp. Eye. Res. 79,249–261.
30. Wood, Z. A., Poole, L. B. & Karplus, P. A. (2003) Science 300, 650–653. 31. Yang, K. S., Kang, S. W., Woo, H. A., Hwang, S. C., Chae, H. Z., Kim, K. &
Rhee, S. G. (2002) J. Biol. Chem. 277, 38029–38036.
32. Wang, G., Olczak, A. A., Walton, J. P. & Maier, R. J. (2005) Infect. Immun. 73,378–384.
33. Chae, H. Z., Chung, S. J. & Rhee, S. G. (1994) J. Biol. Chem. 269, 27670–27678. 34. Woo, H. A., Jeong, W., Chang, T. S., Park, K. J., Park, S. J., Yang, J. S. & Rhee,
S. G. (2005) J. Biol. Chem. 280, 3125–3128. 35. Poole, L. B. (1996) Biochemistry 35, 65–75.
36. Ellis, H. R. & Poole, L. B. (1997) Biochemistry 36, 13349–13356.
37. Aspholm-Hurtig, M., Dailide, G., Lahmann, M., Kalia, A., Ilver, D., Roche, N., Vikstro¨m, S., Sjo¨stro¨m, R., Linde´n, S., Ba¨ckstro¨m, A., et al. (2004) Science 305, 519–522.
38. Hennig, E. E., Mernaugh, R., Edl, J., Cao, P. & Cover, T. L. (2004) Infect. Immun. 72, 3429–3435.
39. Hendrick, J. P. & Hartl, F. U. (1993) Annu. Rev. Biochem. 62, 349–384. 40. Ikenoue, T., Maeda, S., Ogura, K., Akanuma, M., Mitsuno, Y., Imai, Y.,
Yoshida, H., Shiratori, Y. & Omata, M. (2001) Clin. Diagn. Lab. Immunol. 8, 181–186.
41. Sanger, F. (1949) Biochem. J. 44, 126–128.