INTRODUCTION
Infectious hypodermal and hematopoietic necrosis virus (IHHNV) is a virus that can have severe patho-genic effects in penaeid shrimp and presents a risk to commercial shrimp farming. When the virus was first discovered in 1981, it resulted in high mortalities (up to 90%) in infected Penaeus stylirostris (taxonomy used is according to Holthuis 1980) cultured in Hawaii (Lightner et al. 1983a,b). The virus has since been detected in a number of other penaeid species and from stocks around the world, including the Americas,
Oceania, and Asia (Lightner 1996, Flegel 1997). The effects of the virus vary among penaeid species. For example, IHHNV infection does not cause mortality in stocks of P. vannamei and P. monodon. However, it results in a disease called runt-deformity syndrome in both species (Bell & Lightner 1984, Kalagayan et al. 1991, Primavera & Quinitio 2000), and this can also cause substantial economic losses (Wyban et al. 1992).
Based on morphological and biochemical charac-teristics, IHHNV is taxonomically placed within the family Parvoviridae (Bonami et al. 1990). It is an
ico-© Inter-Research 2003 · www.int-res.com *Email: fengjyu@u.arizona.edu
Geographic variations among infectious hypodermal
and hematopoietic necrosis virus (IHHNV) isolates
and characteristics of their infection
Kathy F. J. Tang
1,*, Bonnie T. Poulos
1, Jun Wang
1, Rita M. Redman
1, Hsiu-Hui Shih
2,
Donald V. Lightner
11Department of Veterinary Science and Microbiology, University of Arizona, Tucson, Arizona 85721, USA 2Department of Zoology, National Taiwan University, Taipei, Taiwan
ABSTRACT: Nucleotide sequence variations of a 2.9 kb fragment of infectious hypodermal and hematopoietic necrosis virus (IHHNV) isolated from samples of Penaeus monodon were determined and compared with an isolate from Hawaii. The infection characteristics of these isolates were exam-ined by histology, in situ hybridization, and laboratory challenge studies with P. vannamei. Isolates of IHHNV were obtained from samples collected from the SE Asia region (the Philippines, Thailand, and Taiwan). Isolates of putative IHHNV were obtained from African samples (Tanzania, Madagascar, and Mauritius). The Philippine isolate had a very high nucleotide sequence identity (99.8%) to Hawaii IHHNV. The Thailand isolate showed a slightly lower identity (96.2%). The putative IHHNV sequences collected from Tanzania and Madagascar showed greater divergence from Hawaii IHHNV, 8.2% difference for Tanzania and 14.1% difference for Madagascar. A phylogenetic analysis showed that the Philippine IHHNV clustered with IHHNV found in the western hemisphere. This supports the theory that the Philippines was the origin of IHHNV that was first detected in Hawaii. In the laboratory infection study, both the Philippine and Thailand IHHNV were passed into P. vannamei, and the infected shrimp did not suffer any mortalities. In another laboratory infection, P. vannamei injected with a tissue homogenate of P. monodon from Madagascar, which tested posi-tive for IHHNV by PCR, did not demonstrate IHHNV infection, suggesting that this putaposi-tive IHHNV is not infectious to P. vannamei.
KEY WORDS: Infectious hypodermal and hematopoietic necrosis virus · IHHNV · Penaeus monodon · Genetic variations
sahedral, non-enveloped virus and contains a single-stranded, 4.1 kb DNA genome (Bonami et al. 1990, Mari et al. 1993). Nearly the entire IHHNV genome has been sequenced (Nunan et al. 2000, Shike et al. 2000). It contains 3 large open reading frames (ORF1, 2, and 3). ORF1 comprises approximately 50% of the genome encoding a polypeptide of 666 amino acids, which is predicted to be a non-structural protein 1 (NS1) based upon its degree of homology with 2 mosquito brevidensoviruses (Shike et al. 2000). ORF2, which starts 56 nucleotides upstream and overlaps ORF1, could encode a 343 amino acid putative non-structural protein 2 (NS2). The 5’ end of ORF3 also overlaps ORF 1, and the ORF3 encodes a 329 amino acid polypeptide. At least 4 structural proteins of Mrs
(74, 47, 39, and 37.5 kDa) are found in the purified viri-ons (Bonami et al. 1990). According to partial amino acid sequencing, the 47 kDa protein was part of ORF3 (B. T. Paulos and D. V. Lightner unpubl. data). Based on the gene structure and organization of other par-voviruses, it is likely that the ORF3 encodes structural proteins.
There is little information available on sequence variation among IHHINV isolates. However, a recent study on IHHNV from infected Penaeus stylirostris and P. vannamei from the western hemisphere indicated that the IHHNV genome is very stable (Tang & Light-ner 2002); less than 0.5% nucleotide sequence diver-gence was found among 14 isolates collected from Hawaii and the Americas from 1982 to 1997. In this report, we analyzed sequence variations of IHHNV from P. monodon collected from SE Asia and Africa, and characterized infection of these isolates through histology, in situ hybridization, and laboratory chal-lenge studies.
MATERIALS AND METHODS
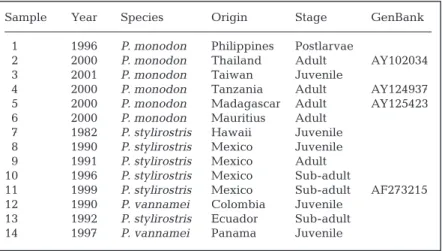
Origin of penaeid shrimp. The samples of Penaeus monodon, P. vannamei, and P. stylirostris were sent to our laboratory as diagnostic materials at various times from 1990 to 2001. The samples were archived and stored frozen at –70°C. We selected isolates to repre-sent the major geographic areas where the shrimp are either farmed or found in the wild. We included one of the earliest samples from Hawaii, which was collected during the first reported outbreak of IHHNV. (Data on the origins, life stages, and dates of collection of the samples are summarized in Table 1.)
DNA extraction and PCR amplification. DNA was extracted from either hemolymph or pleopods of shrimp with a high-pure DNA template preparation kit (Roche Molecular Biochemicals), and amplified by PCR. The reaction mixture contained 10 mM Tris-HCl, pH 9.0;
50 mM KCl; 0.1% Triton-X; 200 µM each dNTP: 2 mM MgCl2; 0.3 µM primers, extracted DNA (1 µl), and
0.625 U Taq DNA polymerase (Promega) in a final volume of 50 µl. Amplification was performed for 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, 3 min at 72°C, with a final extension at 72°C for 7 min. Primers IHHN3065F (5’-GAC GAC GAA GAA TGG ACA GA-3’) and IHHN3065R (5’-TGC CTG GGT AGC TGG TAT GTA TA-3’) were used to amplify a 3.0 kb frag-ment comprising Nucleotides 792 to 3856. The nucleo-tide positions refer to the published IHHNV sequence (GenBank AF218266). The amplified DNA was puri-fied with a QIAquick PCR purification kit (Quiagen). The amplified products were directly sequenced with an automated DNA sequencer 377 (Applied Biosys-tems) at the sequencing facility of the University of Arizona. Sequences obtained from isolates of Thai-land, Tanzania, and Madagascar have been assigned GenBank Accession Nos AY102034, AY124937, and AY125423, repectively.
DNA sequence analysis. The nucleotide sequence of 2.9 kb from Nucleotides 816 to 3744 was compared among these IHHNV isolates. Distances among these sequences were calculated using the Jukes-Cantor method, and the un-rooted tree was calculated using the neighbor-joining method (Saitou & Nei 1987) with the GCG package. The un-rooted tree was displayed with the Treeview program (Page 1996). A boot-strapped tree was generated with the PAUP software (Version 4.0 in GCG package); distance analysis was implemented with a heuristic algorithm. The tree was generated with tree-bisection-reconnection. The data were re-sampled by 1000 bootstrap replicates to deter-mine the confidence index within the phylogenetic tree. Bootstrap values greater than 70 (as percentage of 1000 replications) were considered to be related with a significance of 95% (Hillis & Bull 1993).
Digoxigenin-labeled IHHNV probe and in situ hybridization. A DNA probe for IHHNV was previ-ously developed in our laboratory (Mari et al. 1993, Lightner 1996). The probe was labeled with digoxi-genin-11-dUTP in a PCR reaction as described by Lightner (1996). Postlarval and juvenile Penaeus mon-odon from the Philippines and Taiwan, respectively, were fixed with Davidson’s AFA (alcohol-formalin-acetic acid) fixative, processed, embedded in paraffin and sectioned (4 m thick) according to standard meth-ods. The procedures used for routine hematoxylin & eosin (H&E) staining and in situ hybridization were described by Lightner (1996). Tissue sections from IHHNV-infected P. stylirostris and specific-pathogen-free (SPF) P. vannamei were used, respectively, as pos-itive and negative controls for in situ hybridization.
Laboratory infection. Small juveniles of SPF Penaeus vannamei (10 shrimp, 0.3 g) were fed minced
tissue from either the Philippine or Thailand IHHNV-infected P. monodon at 10% of their body weight once daily for 3 to 10 d in a 90 l glass tank. For the following 2 to 4 wk, the shrimp were maintained on a pel-letized ration. After an incubation period of 2 to 4 wk, challenged shrimp were fixed in Davidson’s AFA for his-tological and in situ hybridization analyses.
For challenge studies with Madagas-car Penaeus monodon, injection was used because there was limited shrimp tissue available, and injection is rou-tinely used for laboratory infections (Lightner 1996). These shrimp were found to be IHHNV-positive by PCR; then the tissue was homogenized in a
buffer (0.02 M Tris-HCl, pH 7.4, 0.4 M NaCl; 1 g/10 ml), and centrifuged at 5000 ×g for 30 min. The super-natant was diluted (1:20, v:v) with sterile 2% saline to generate an inoculum. A volume of 50 µl of inoculum was injected into each SPF P. vannamei (10 shrimp, 1 g). The shrimp were maintained on pelletized ration for 1 mo, then fixed in Davidson’s AFA for in situ hybridization analysis.
RESULTS
Comparisons of IHHNV and putative IHHNV sequences to Hawaii IHHNV
The IHHNV from Hawaii, the best characterized iso-late among those studied here, has been completely sequenced (GenBank AF218266). It was used in this study as a reference genotype to assess the variations of IHHNV (and putative IHHNV) sequences of Penaeus monodon collected from 6 different regions (Table 1, Samples 1 to 6). The 2.9 kb fragment that is compared contains approximately 70% of the entire viral genome including the full length ORF1 and ORF3. The results showed that the Philippine isolate was most similar to Hawaii IHHNV, with a difference of only 0.2% in the nucleotide sequence (Table 2). The Thailand isolate showed a higher variation from Hawaii IHHNV, with an identity of 96.2%. The Taiwan isolate was very sim-ilar (99.7% identity) to the Thailand isolate, differing only in 9 nucleotides. The extracted DNA from P. mon-odon from Tanzania, Madagascar, and Mauritius con-tained putative IHHNV sequences. Among them, the Tanzania sequence had an identity of 91.8% with the Hawaii IHHNV. The Madagascar sequence had a lower identity, 85.9%. The sequence from Mauritius
displayed only 1 nucleotide difference from the Mada-gascar sequence.
Alignment of these sequences showed that all the nucleotide changes were substitutions and that no new termination codons were introduced. Both ORF1 and ORF3 had the same number of amino acids (aa) in their predicted polypeptides, i.e. 666 aa from ORF1 and 329 aa from ORF3. A length of 352 aa was predicted from the portion of ORF2 that overlaps ORF1. We compared these nucleotide (nt) sequences to that of the Hawaii isolate. The sequence identities ranged from 86 to 100% for ORFl (nt 816 to 2816), and 91 to 99.8% for ORF3 (nt 2758 to 3747) (Table 2). In each case, the Philippine isolate was most similar to the Hawaii iso-late and the Madagascar isoiso-late was the least similar. The comparison of amino acid similarity followed the same pattern; and in most cases the amino acid sequence similarities were very close to the corre-sponding nucleotide sequence identities. However, the ORF3 amino acid sequence similarities for the Tanza-nia and Madagascar isolates were higher than their corresponding nucleotide sequence identities.
Table 1. Penaeus spp. Year, origin and related information on the IHHNV
isolates used in the study
Sample Year Species Origin Stage GenBank
1 1996 P. monodon Philippines Postlarvae
2 2000 P. monodon Thailand Adult AY102034
3 2001 P. monodon Taiwan Juvenile
4 2000 P. monodon Tanzania Adult AY124937
5 2000 P. monodon Madagascar Adult AY125423
6 2000 P. monodon Mauritius Adult
7 1982 P. stylirostris Hawaii Juvenile
8 1990 P. stylirostris Mexico Juvenile
9 1991 P. stylirostris Mexico Adult
10 1996 P. stylirostris Mexico Sub-adult
11 1999 P. stylirostris Mexico Sub-adult AF273215
12 1990 P. vannamei Colombia Juvenile
13 1992 P. stylirostris Ecuador Sub-adult
14 1997 P. vannamei Panama Juvenile
Table 2. Comparison of nucleotide and predicted amino acid sequences of IHHNV (and putative IHHNV) isolated from samples of Penaeus monodon in this study with IHHNV
isolated from Hawaii
Isolate Nucleotide sequence Amino acid origin identity (%) similarity (%)
2.9 kb ORF1 ORF3 ORF1 ORF2 ORF3 Philippines 99.8 100 99.8 100 100 99.1 Thailand 96.2 96.8 98.3 96.4 96.3 97.9 Tanzania 91.8 92.3 94.7 92.8 93.2 98.2 Madagascar 85.9 85.8 91.0 86.0 85.8 95.1
Sequence motifs of ORF1
As shown in Table 2, the putative IHHNV sequences from Africa had more substitutions, particularly in ORF1, than the IHHNV isolates from the western hemisphere (Tang & Lightner 2002). The amino acid
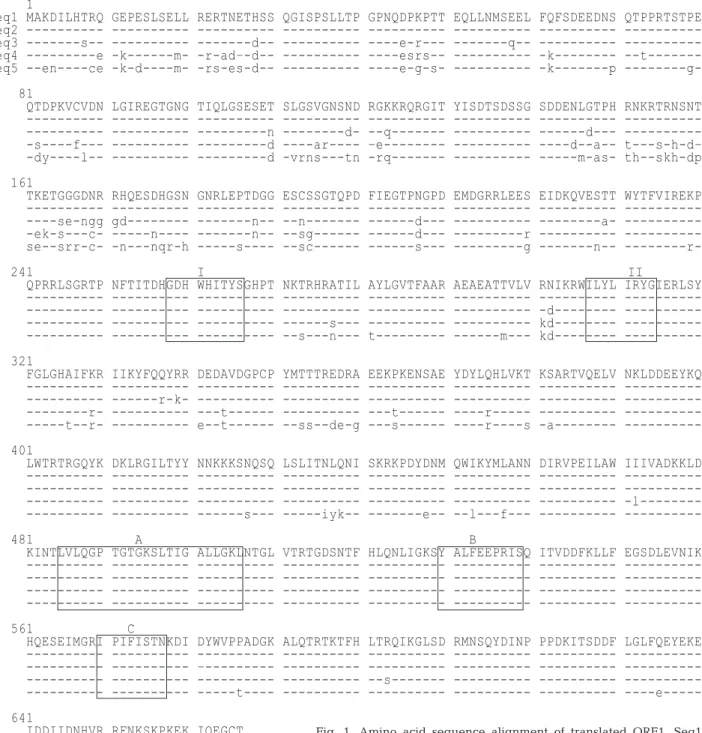
sequences predicted for ORF1 were examined to determine whether or not they contained the charac-teristic motifs found in IHHNV. Although the amino acid differences were distributed throughout the se-quence, a higher percentage of substitutions was found among the first 200 amino acids (Fig. 1).
Never-1
Seq1 MAKDILHTRQ GEPESLSELL RERTNETHSS QGISPSLLTP GPNQDPKPTT EQLLNMSEEL FQFSDEEDNS QTPPRTSTPE Seq2 Seq3 ---s-- ---d-- ----e-r--- ---q-- ---Seq4 ---e -k---m- -r-ad–-d-- --- ----esrs-- --- -k--- --t---Seq5 --en----ce -k-d----m- -rs-es-d-- --- ----e-g-s- --- -k---p
---g-81
QTDPKVCVDN LGIREGTGNG TIQLGSESET SLGSVGNSND RGKKRQRGIT YISDTSDSSG SDDENLGTPH RNKRTRNSNT ---n ---d- --q--- ---d--- ---s----f--- --- ---d ----ar---- –e--- --- ----d--a-- t---s-h-d– -dy----l-- --- ---d -vrns---tn -rq--- --- ---m-as- th--skh-dp 161
TKETGGGDNR RHQESDHGSN GNRLEPTDGG ESCSSGTQPD FIEGTPNGPD EMDGRRLEES EIDKQVESTT WYTFVIREKP ----se-ngg gd--- ---n-- --n--- ---d--- ---a- -ek-s---c- ---n---- ---n-- --sg--- ---d--- ---r ---se--srr-c- -n---nqr-h ---s---- --sc--- ---s--- ---g ---n--
---r-241 I II
QPRRLSGRTP NFTITDHGDH WHITYSGHPT NKTRHRATIL AYLGVTFAAR AEAEATTVLV RNIKRWILYL IRYGIERLSY -d--- ---s--- kd--- --s---n--- t--- ---m--- kd--- ---321
FGLGHAIFKR IIKYFQQYRR DEDAVDGPCP YMTTTREDRA EEKPKENSAE YDYLQHLVKT KSARTVQELV NKLDDEEYKQ ---r-k- ---r- ---t--- ---t--- ----r--- ---t--r- e--t--- --ss--de-g ---s--- ----r----s -a--- ---401
LWTRTRGQYK DKLRGILTYY NNKKKSNQSQ LSLITNLQNI SKRKPDYDNM QWIKYMLANN DIRVPEILAW IIIVADKKLD --- --- --- --- --- --- --- -l--- -l--- ---s--- ---iyk-- ---e-- --l---f--- -l--- -l--- ---481 A B
KINTLVLQGP TGTGKSLTIG ALLGKLNTGL VTRTGDSNTF HLQNLIGKSY ALFEEPRISQ ITVDDFKLLF EGSDLEVNIK ---561 C
HQESEIMGRI PIFISTNKDI DYWVPPADGK ALQTRTKTFH LTRQIKGLSD RMNSQYDINP PPDKITSDDF LGLFQEYEKE –--- --s--- --- --- ---t---- --- --- --- --- ----e---641
IDDIIDNHVR RFNKSKPKEK IQEGCT ---- ---- ---- ----i--- --e---s--- ----g----t ---e---sy-- ----g----t -e---a
Fig. 1. Amino acid sequence alignment of translated ORF1. Seq1: Hawaii IHHNV (GenBank AF218266); Seq2: Philippine IHHNV; Seq3: Thailand IHHNV (GenBank AY102034); Seq4: Tanzania tive IHHNV sequence (GenBank AY124937); Seq5: Madagascar puta-tive-IHHNV sequence (GenBank AY125423). Dashes (---) indicate the same amino acids as those in Hawaii IHHNV. I, II: replication initiator protein motifs. A, B, C: NTP-binding and helicase domains
theless, the replication Initiator Protein Motifs I (aa 258 to 266) and II (aa 307 to 314) (Ilyina & Koonin 1992, Shike et al. 2000) were conserved, as were the NTP-binding and helicase domains A, B, C (aa 460 to 640), which comprise an ATP/GTP-binding site motif (A/G)-X4-GK(S/T) (aa 489-496) (Walker et al. 1982).
Further-more, BLAST analysis found no matches for these putative IHHNV sequences in the GenBank other than IHHNV.
Phylogenetic analyses
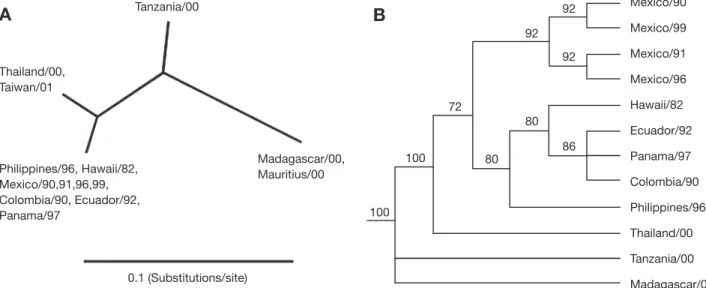
The relationship of these IHHNV (and putative IHHNV) sequences were examined in a phylogenetic analysis with 8 representative IHHNV isolates found in the western hemisphere; 1 Hawaii isolate, 4 isolates collected from the Gulf of California, Mexico, and 3 from Central/South America (Table 1: Samples 7 to 14). The un-rooted tree showed that the distance between the Philippine IHHNV and those from the western hemisphere is only 0.2% (i.e. 0.002 substitu-tions/site) and they clustered together (Fig. 2A). The Mauritius isolate had the same genotype as the isolate from Madagascar. The Taiwan isolate was grouped with the Thailand isolate. The Madagascar sequence displayed approximately the same distance (14%) from either the Philippine or the Thailand IHHNV. The distance between Thailand and the Philippine IHHNV was 3.8% (0.038 substitutions/site). Two sequences from Africa had a rather large distance between each other, 10% (0.1 substitution/site).
With the brevidensovirus Aedes densovirus (Gen-Bank M37899) as an outgroup in a bootstrapped tree, there were 3 main lineages corresponding to the origins of isolates: Thailand/the Philippines/western hemisphere, Tanzania, and Madagascar (Fig. 2B). Philippine and Thailand IHHNV are grouped together with a bootstrap value of 100. The Philippine isolate was closely related to the 4 IHHNV isolates originating from Hawaii and Central/South America, with a boot-strap value of 80. Three isolates from Central/South America were in the same group with a bootstrap value of 86. Four Mexico isolates were grouped to-gether with a bootstrap value of 92 and appeared sep-arate from Hawaii and Central/South America isolates.
In situ detection of IHHNV in Philippine and Thailand Penaeus monodon
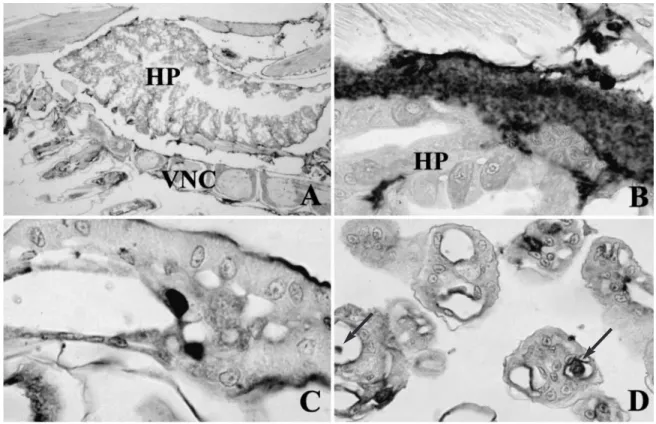
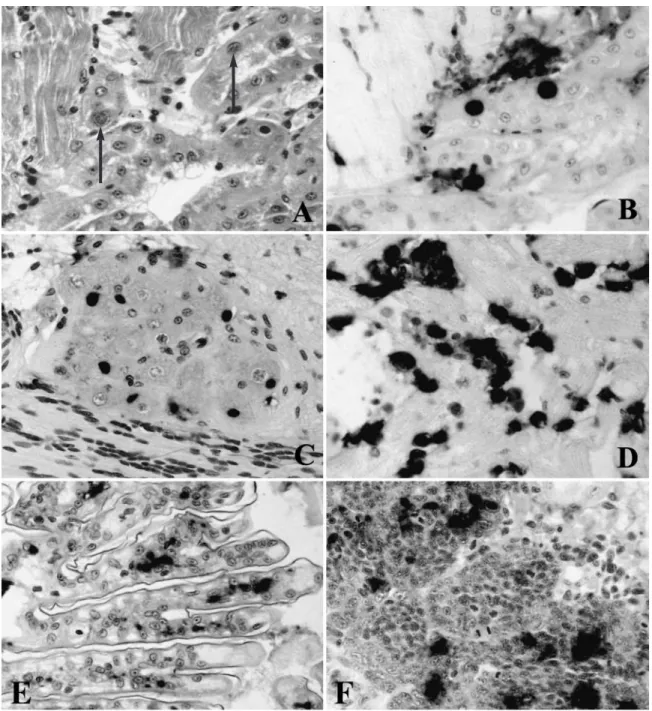
For the Philippine IHHNV-infected Penaeus mono-don, histological examination indicated that the hall-mark Cowdry A intranuclear inclusion bodies were rare. However, in situ hybridization showed that the postlarvae had systemic infections (Fig. 3A), and IHHNV was detected in various tissues including the hemolymph of the coelomic cavity (Fig. 3B). Strong intranuclear staining was found in the epithelial cells of the antennal gland (Fig. 3C). Circulating hemocytes and the epithelial cells of gills were also highly infected (Fig. 3D).
For Thailand IHHNV, the tissue distribution was determined using Penaeus monodon from Taiwan
Tanzania/00 Thailand/00, Taiwan/01 0.1 (Substitutions/site) Philippines/96, Hawaii/82, Mexico/90,91,96,99, Colombia/90, Ecuador/92, Panama/97 Madagascar/00, Mauritius/00 Mexico/90 Mexico/99 Mexico/91 Mexico/96 Hawaii/82 Ecuador/92 Panama/97 Colombia/90 Philippines/96 Thailand/00 Tanzania/00 Madagascar/00 92 86 92 80 80 72 100 100 92
Fig. 2. Phylogenetic analysis of representative IHHNV nucleotide sequences collected from western hemisphere and sequences obtained in the current study. (A) Un-rooted tree generated by neighbor-joining method; bar = 0.1 substitutions per site. (B) Boot-strapped tree generated by PAUP analysis. The tree was rooted using nucleotide sequence of Aedes densovirus (GenBank
M37899) as an outgroup; numbers indicate the percentages of bootstrap support from 1000 replicates; branches with values of < 70 were collapsed. Name of sequence is indicated by the origin and year of isolation in both (A) and (B)
that were naturally infected with Thailand IHHNV (Table 1, Sample 3). H&E staining showed Cowdry A intranuclear inclusion bodies in the antennal gland (Fig. 4A), ganglia, nerve tract, and gills (data not shown). With in situ hybridization, IHHNV was found essentially in all the tissues including the antennal gland (Fig. 4B), nerve ganglia (Fig. 4C), heart (Fig. 4D), gills (Fig. 4E), hematopoietic tissue (Fig. 4F), lymphoid organ, and connective tissues (data not shown). Hybridization was found in the cytoplasmic area of infected cells as well as in intranuclear in-clusion bodies.
Laboratory infections
To determine if the Philippine IHHNV can infect other species of penaeid shrimp, a laboratory infection was carried out in a bioassay using Penaeus vannamei as indicator shrimp: 2 wk after exposure to IHHNV, there were no mortalities among the indicator shrimp. Hemolymph was drawn from the challenged P. van-namei for PCR analysis through the use of IHHNV-specific primers (Nunan et al. 2000), and the samples tested positive for IHHNV.
Another laboratory infection performed with Thai-land IHHNV was carried out through feeding Penaeus vannamei with minced tissue of IHHNV-infected P. monodon: 4 wk after exposure to IHHNV, no mor-talities were observed. The P. vannamei were fixed and analyzed by in situ hybridization. The Thailand IHHNV-infected P. vannamei showed strong reactions within the antennal gland, hematopoietic nodules, nerve tract, heart, gill filaments, and connective tissues (data not shown). The majority of reaction sites were in cytoplasmic areas with a few intranuclear stainings in the nerve tract. When a 2.9 kb IHHNV fragment from these experimentally infectedP. vannamei was ampli-fied by PCR and sequenced for comparison with that from P. monodon, the results showed that there was no change in nucleotide sequence after passage through P. vannamei.
In the challenge study using injection of tissue homogenate from Madagascar Penaeus monodon, the indicator shrimp showed no mortality 1 mo after injec-tion. Hemolymph drawn from the challenged shrimp was negative for IHHNV when tested with primers designed from the Madagascar IHHNV sequence. The result of in situ hybridization of tissue sections from challenged shrimp was negative.
Fig. 3. Penaeus monodon. In situ hybridization of Philippine IHHNV-infected postlarvae with a digoxigenin-labeled IHHNV
DNA probe. (A) Mid-sagittal section of cephalothorax including tissues of hepatopancreas (HP) and ventral nerve cord (VNC); total magnification 20×. (B) Coelomic cavity and circulating hemocytes; total magnification 200×. (C) Antennal gland; total
DISCUSSION
Our data support the theory that Philippine stocks could be the origin of IHHNV that is currently found in the western hemisphere, a conclusion reached by Lightner (1999) based on inference from knowledge of the introductions of Penaeus monodon stocks into the Americas. We found a high degree of identity of the nucleotide sequences between isolates from the
Philip-pines and Hawaii. Also, in the phylogenetic analysis, the Philippine IHHNV isolate clustered with the iso-lates found in the western hemisphere.
Based on similar evidence, it is likely that IHHNV may have been introduced to Taiwan from Thailand. Sequence from the Taiwan IHHNV samples had 99.7% identity with those from Thailand, and it is known that both live and frozen shrimp are frequently imported from Thailand to Taiwan.
Fig. 4. Penaeus monodon. H&E histology and in situ hybridization of Thailand IHHNV-infected shrimp. (A) H&E staining of
antennal gland (arrows indicate) Cowdry A intranuclear inclusion body; (B) in situ hybridization of serial section of antennal
gland with a digoxigenin-labeled IHHNV probe; (C), (D), (E), (F)in situ detection of IHHNV in nerve ganglia, heart, gills and
The 4 Mexico isolates clustered together and were separated from isolates from Hawaii, Central and South America. These 4 Mexico isolates were collected along the northern Gulf of California, Mexico, and may have the same lineage, as it is thought that the spread of IHHNV in this region may have started from 1 farm in 1987 (Lightner et al. 1992).
In general, genetic variation among parvoviruses is only up to 4% (Erdman et al. 1996). We found, how-ever, that IHHNV sequences isolated from Penaeus monodon have unexpectedly high variation, up to 14%. This is in contrast to our previous finding of less than 0.5% variation among IHHNV isolates from pop-ulations of P. stylirostris and P. vannamei (Tang & Lightner 2002). The lower variation in P. stylirostris and P. vannamei may be because of the short time available for variation to develop. These species became infected after exposure to IHHNV from infected stocks of P. monodon imported from Asia. Stocks of P. monodon were first introduced into the west during the mid to late 1970s, so if this hypothesis is correct, the IHHNV found in the western hemi-sphere has been circulating in this region for only 2 to 3 decades.
Three putative IHHNV sequences were found in the shrimp collected in Tanzania, Madagascar, and Mauri-tius. The Mauritius nucleotide sequence essentially has the same genotype as that of Madagascar, reflect-ing the close proximity of these geographic regions. There is a 10% difference between nucleotide se-quences from Tanzania and Madagascar. The reason for this rather distinct sequence difference from 2 rela-tively close areas is not known. Three ORFs that are characteristic of IHHNV can still be found in these sequences. The sequence motifs of replication initiator protein and NTP-binding/helicase in ORF1 are con-served. But there is a higher percentage of variations in ORF1/2 (full length ORF1 and overlapped 95% of ORF2) than in ORF3, which is rather unusual for a small virus with a considerable overlap of ORFs. This is also different from Hawaii IHHNV and most other parvoviruses, in which the structural protein coding region (ORF3) is usually more variable than the non-structural coding region (ORF1/2) (Truyen et al. 1995, Hemauer et al. 1996, Tang & Lightner 2002). ORF1 is thought to encode NS1, which is thought to be respon-sible for the majority of enzymatic activities involved in viral transcription and replication. ORF2 could encode NS2, the function of which is less clear but also known to be required for viral multiplication. Greater varia-tion in ORF1 and 2 may indicate weaker selecvaria-tion pressure on these ORFs, suggesting that they do not encode functional NS1 and NS2. If ORF1 and 2 are non-functional, then these putative IHHNV sequences may not be from infectious IHHNV. If so, this could
explain the fact that they are not infectious to Penaeus vannamei. Further challenge experiments with their natural host, P. monodon, are planned to determine if these putative IHHNV sequences are not infectious. The histopathology of Thailand IHHNV is very simi-lar to that of Philippine IHHNV, indicating that the 3.8% difference in nucleotide sequence does not affect its characteristic pathology. Both Thailand and Philip-pine IHHNV are infectious to Penaeus vannamei in laboratory trials. The amplified 2.9 kb portion of the IHHNV genome was sequenced from these infected P. vannamei and was shown to be identical to the orig-inal IHHNV sequences, indicating that the IHHNV sequence was not modified by passage through P. van-namei.
The emergence of IHHNV in the western hemi-sphere 2 to 3 decades ago may have been from the movement of IHHNV-infectedPenaeus monodon. Be-cause IHHNV infection is asymptomatic in P. mon-odon, the presence of the virus was not noticed until stocks of other, more susceptible, species became in-fected.
Acknowledgements. This work was supported by Gulf Coast
Research Laboratory Consortium Marine Shrimp Farming Program, CSRS, USDA, under Grant No. 99-38808-7431. We thank Mr. Glen Bieber and Dr. Dallas Weaver for providing
Penaeus monodon collected from Madagascar, Mauritius and
Tanzania. We thank Dr. Stephen Nelson for his assistance in editing this manuscript.
LITERATURE CITED
Bell TA, Lightner DV (1984) IHHN virus: infectivity and path-ogenicity studies in Penaeus stylirostris and Penaeus van-namei. Aquaculture 38:185–194
Bonami JR, Trumper B, Mari J, Brehelin M, Lightner DV (1990) Purification and characterization of the infectious hypodermal and hematopoietic necrosis virus of penaeid shrimps. J Gen Virol 71:2657–2664
Erdman DD, Durigon EL, Wang QY, Anderson LJ (1996) Genetic diversity of human parvovirus B19: sequence analysis of the vp1/vp2 gene from multiple isolates. J Gen Virol 77:2767–2774
Flegel TW (1997) Major viral diseases of the black tiger prawn (Penaeus monodon) in Thailand. World J Microbiol
Biotechnol 13:433–442
Hemauer A, Poblotzki A, Gigler A, Cassinotti P. Siegl G, Wolf H, Modrow S (1996) Sequence variability among different parvovirus B19 isolates. J Gen Virol 77:1781–1785 Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as
a method for assessing confidence in phylogenetic anal-ysis. Syst Biol 42:182–192
Holthuis LB (1980) FAO species catalogue. FAO Fisheries Syn-opsis No 125, Vol 1. Shrimps and prawns of the world: an annotated catalogue of species of interest to fisheries. Food and Agricultural Organization of the United Nations, Rome Ilyina TV, Koonin EV (1992) Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20:3279–3285
Kalagayan G, Godin D, Kanna R, Hagino G, Sweeney J, Wyban J, Brock J (1991) IHHN virus as an etiological factor in runt-deformity syndrome of juvenile Penaeus vannamei cultured
in Hawaii. J World Aquacult Soc 22:235–243
Lightner DV (1996) A handbook of shrimp pathology and diagnostic procedures for diseases of cultured penaeid shrimp. World Aquaculture Society, Baton Rouge, LA Lightner DV (1999) The penaeid shrimp viruses TSV, IHHNV,
WSSV, and YHV: current status in the Americas, available diagnostic methods, and management strategies. J Appl Aquacult 9:27–52
Lightner DV, Redman RM, Bell TA, Brock JA (1983a) Detec-tion of IHHN virus in Penaeus stylirostris and P. vannamei
imported into Hawaii. J World Maricult Soc 14:212–225 Lightner DV, Redman RM, Bell TA (1983b) Infectious
hypo-dermal and hematopoietic necrosis, a newly recognized virus disease of penaeid shrimp. J Invertebr Pathol 42: 62–70
Lightner DV, Williams RR, Bell TA, Redman RM, Perez A LA (1992) A collection of case histories documenting the intro-duction and spread of the virus disease IHHN in penaeid shrimp culture facilities in northwestern Mexico. ICES Mar Sci Symp 194:97–105
Mari J, Bonami JR, Lightner DV (1993) Partial cloning of the genome of infectious hypodermal and hematopoietic necrosis virus, an unusual parvovirus pathogenic for penaeid shrimps; diagnosis of the disease using a specific probe. J Gen Virol 74:2637–2643
Nunan LM, Poulos BT, Lightner DV (2000) Use of polymerase chain reaction for the detection of infectious hypodermal and hematopoietic necrosis virus in penaeid shrimp. Mar Biotechnol 2:319–328
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Primavera JH, Quinitio ET (2000) Runt-deformity syndrome in cultured giant tiger prawn Penaeus monodon. J Crustac
Biol 20:796–802
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shike H, Dhar AK, Burns JC, Shimizu C, Jousset FX, Klimple KR, Bergoin M (2000) Infectious hypodermal and hema-topoietic necrosis virus of shrimp is related to mosquito brevidensoviruses. Virology 277:167–177
Tang KFJ, Lightner DV (2002) Low sequence variation among isolates of infectious hypodermal and hematopoietic necrosis virus (IHHNV) originating from Hawaii and the Americas. Dis Aquat Org 49:93–97
Truyen U, Gruenberg A, Chang SF, Obermanier B, Veijalani-nen P, Parrish CR (1995) Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J Virol 69:4702–4910
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO (Eur Mol Biol Organ) J 1:945–951
Wyban JA, Swingle JS, Sweeney JN, Pruder GC (1992) De-velopment and commercial performance of high health shrimp using specific pathogen free (SPF) broodstock
Penaeus vannamei. In: Wyban JA (ed) Proc Special
Ses-sion on Shrimp Farming. World Aquaculture Society, Baton Rouge, LA, p 254–260
Editorial responsibility: Timothy Flegel, Bangkok, Thailand
Submitted: April 23, 2002; Accepted: August 2, 2002 Proofs received from author(s): January 27, 2003