Research Article
Anti-viral effect of a compound isolated from Liriope platyphylla against
hepatitis B virus in vitro
Tsurng-Juhn Huanga*, Yu-Chi Tsai h, Shang-Yu Chiang h, Guei-Jane Wang b, c, d,
Yu-Cheng Kuoe, f, Yi-Chih Changg, Yi-Ying Wug, Yang-Chang Wuh, i, j, k* a Department of Biological Science and Technology and Research Center for
Biodiversity, China Medical University, Taichung 404, Taiwan
b Graduate Institute of Clinical Medical Science, China Medical University, Taichung
404, Taiwan
cDepartment of Medical Research, China Medical University Hospital, Taichung 404,
Taiwan
dDepartment of Health and Nutrition Biotechnology, Asia University, Taichung 413,
Taiwan
e Department of Radiation Oncology, China Medical University Hospital, Taichung,
Taiwan
f Department of Biomedical Imaging and Radiological Science, China Medical
University, Taichung 404, Taiwan
g Department of Medical Laboratory Science and Biotechnology, China Medical
University, Taichung 404, Taiwan
h Graduate Institute of Natural Products, Kaohsiung Medical University, Kaohsiung
807, Taiwan
i School of Pharmacy, College of Pharmacy, China Medical University, Taichung
404,Taiwan
j Chinese Medicine Research and Development Center, China Medical University
Hospital, Taichung 404, Taiwan
k Center for Molecular Medicine, China Medical University Hospital, Taichung 404,
Taiwan
Correspondence should be addressed to: Tsurng-Juhn Huang, Ph.D TEL: +886 4 22053366 ext. 8105 FAX: +886 4 22071507 E-mail: tjhuang@mail.cmu.edu.tw 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38
Co-correspondence should be addressed to: Yang-Chang Wu, Ph.D TEL: +886 4 22053366 ext. 1012 FAX: +886 4 22060248 E-mail: yachwu@mail.cmu.edu.tw Abstract
The compound LPRP-Et-97543 was isolated from Liriope platyphylla roots and was 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61
observed to have potential anti-viral effects in HepG2.2.15 cells against hepatitis B virus (HBV). The antiviral mode was further clarified, and the HBV-transfected Huh7 cells were used as the platform. During viral gene expression, LPRP-Et-97543
treatment had apparent effects on the viral precore/pregenomic and S/preS RNA. Promoter activity analysis demonstrated that LPRP-Et-97543 significantly reduced Core, S, and preS but not X promoter activities. Further examination showed that putative signaling pathways were involved in this inhibitory effect, indicating that NF-B may serve a putative mediator of HBV gene regulation with LPRP-Et-97543. In addition, the nuclear expression of p65/p50 NF-B member proteins was
attenuated with LPRP-Et-97543 and augmented cytoplasmic IB protein levels but without affecting the expression of these proteins in HBV non-transfected cells during treatment. Moreover, LPRP-Et-97543 reduced the binding activity of NF-B protein to CS1 element of HBV surface gene in a gel retardation analysis and inhibited CS1 containing promoter activity in HBV expressed cells. However, HBV transfection significantly enhances CS1 containing promoter activity without compound treatment in cells. Finally, transfection of the p65 expression plasmid significantly reversed the inhibitory effect of LPRP-Et-97543 on the replicated HBV DNA level in HBV positive cells. In conclusion, this study suggests that the mechanism of HBV inhibition by LPRP-Et-97543 may involve the feedback regulation of viral gene 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80
expression and viral DNA replication by HBV viral proteins, which interferes with the NF-B signaling pathway.
Keywords: Liriope platyphylla, hepatitis B virus, NF-B, HepG2.2.15, Huh7
1. Introduction
Hepatitis B virus (HBV) is a causative agent that often leads to acute and chronic infections in humans (Gitlin, 1997). Liver damage occurs in approximately 80% chronic HBV carriers, which may lead to liver cirrhosis and hepatocellular carcinoma (HCC) (Park et al., 2006). Despite the development of a recombinant vaccine, which provides immunity against the HBV and the use of a specific Hepatitis B
immunoglobulin in susceptible populations, there are still more than 400 million chronic HBV carriers (Gish, 2005).While interferons and nucleotide analogs have been widely used to treat chronically infected patients, the rapid development of resistance and undesirable side effects associated with this treatment makes alternate treatments necessary. Recent studies have been performed to determine how the viral gene is regulated and controlled to ameliorate HBV. This is a crucial issue for
antiviral strategies (Huang et al., 2014a) because the focus is not on viral genome replication but on curing hepatitis B. Thus, continued development of new antiviral agents with novel targets and mechanisms are urgently required to eradicate HBV in chronic carriers. 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99
The large repertoire of herbal compounds may be the essential ingredients required for developing new drugs that relieve the threat of HBV and its associated liver diseases (Bent, 2008; Bent and Ko, 2004). At present, more that 80% of the population in developing countries uses alternative or traditional medical resources. Systematic methodologies have been developed to determine the active compounds in traditional Chinese herbal medicine because some of the herbal plants exhibit potent anti-viral effects (Li and Peng, 2013).
Plant species belonging to the Liriope genus were reported to have a broad spectrum of biological activities, such as anti-inflammatory (Hu et al., 2011),
antibacterial (Kim et al., 2002), anticancer (Sun et al., 2010; Wang et al., 2013; Wang et al., 2011), antidiabetic (Bai et al., 2009b), neuroprotective (Hur et al., 2004), oestrogenic, anti-platelet (Tsai et al., 2013), and hepatoprotective (Wu et al., 2001) functions. Recent studies showed the root extract from Liriope platyphylla reduces inflammation and alleviates symptoms from metabolic and neurodegenerative disorders, such as diabetes and pathological obesity (Bai et al., 2009a; Chen et al., 2009; Choi et al., 2011; Lee et al., 2005). Bioactivity-guided fractionation and
purification of L. platyphylla can be used to examine its antiviral effects. In this study, an active compound of L. platyphylla root part extract (LPRP), named LPRP-Et-97543, and its anti-hepatitis B virus activities was examined. Further evaluation of 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118
LPRP-Et-97543 was performed by transfecting it into the HBV genome using a hepatoma cell.
2. Materials and Methods
2.1. Materials
The anti-preSSclonesurface sc-23944, anti-HBcAg (13A9 clone) (sc-23946), anti-NF-B (p65/p50)(sc-109/sc-7178) and anti-IB (sc-371) antibodies were used as the HB antibodies and purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Proliferating cell nuclear antigen (PCNA)(PC10 clone)(#2586) antibody was purchased from Cell Signaling Technology Inc. (Danvers, MA, USA). The DIG high prime DNA labeling and detection starter kit was obtained from Roche (Mannheim, Germany) while the Power SYBR Green PCR master mix was purchased from Applied Biosystem (Foster City, CA, USA). The QIAquick Gel extraction kit was purchased from Qiagen (Valencia, CA, USA) and the dual-Luciferase reporter assay kit was purchased from Promega (Madison, WI, USA). The Trizoltotal RNA isolation solution and the Lipofectamine 2000 transfection reagent were from Invitrogen (Carlsbad, CA, USA). Lamivudine (2′, 3′-dideoxy-3′-thiacytidine, commonly known as 3TC) and an anti--tubulin antibody were purchased from Sigma (St Louis, MO, USA). pNF-B-Luc, p-activator protein (AP)-1-Luc, pAP2-Luc and p-interferon-sensitive response element (ISRE)-pAP2-Luc promoter-luciferase 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137
reporter constructs from Stratagene (La Jolla, CA, USA) were provided by Dr. Cheng-Wen Lin (Department of Medical Laboratory Science and Biotechnology, China Medical University, Taichung, Taiwan). The pCMV-p65 expression plasmid was provided by Dr. Chen-Kung Chou (Department of Biomedical Sciences, Chang Gung University, Tao-Yuan, Taiwan)
2.2. Plant material
In August 2009, Liriope platyphylla was collected from Taichung County, Taiwan, and the plant was identified by Dr. Ming-Hong Yen from the Graduate Institute of Natural Products (GINP), Kaohsiung Medical University, Kaohsiung, Taiwan. A voucher specimen (LP002) was deposited at GINP. The roots were air-dried at room temperature and crushed using a mill (Rong Tsong, Taichung, Taiwan) before extraction.
2.3. Extraction and isolation of bioactive compound
The roots of L. platyphylla (3.5 kg) were extracted with 95% ethanol (EtOH) at room temperature for 24 h; this process was repeated two more times so that the roots were extracted at room temperature for a total of 72 h. The ethanolic extract (654.6 g) was concentrated in vacuo, suspended in water and partitioned into n-hexane, 70% EtOH in an aqueous solution and n-butanol (BuOH), respectively. The aqueous EtOH layer (28.5 g) was subjected to silica gel column chromatography (CC) and successively 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157
eluted with n-hexane:CH2Cl2 (90:10 – 0:100) and CH2Cl2:MeOH (100:0 – 80:20)
mobile phase system to afford 19 fractions. Fraction nine (1140.6 mg) was subjected to silica gel CC and eluted with CH2Cl2:MeOH (98:2) to yield nine subfractions.
Compound LPRP-Et-97543 (6.8 mg) was purified using a HPLC with MeOH:H2O
(75:25) solvent system. The structure of LPRP-Et-97543 was established as (3R)-3-(4'-hydroxybenzyl)-5,7-dihydroxy-6-methyl-chroman-4-one by spectroscopic analysis and was compared with the reported physical data (Nguyen et al., 2006; Tsai et al., 2013). Prior to treatment, LPRP-Et-97543 was dissolved in dimethyl sulfoxide (DMSO). The final DMSO concentration did not exceed 0.5%, and it did not cause any detectable effects during treatment.
2.4. Cell culture
HBV stably expressed cells were generated (in 1987) by transfecting HepG2 cells with the head-to-tail dimer of HBV to generate a stable cell line (HepG2.2.15) (Sells et al., 1987) and were maintained in MEM with heat-inactivated 10% fetal bovine serum (FBS) , 1% antibiotics and G418 (300 g/ml). In parallel experiments, human Huh7 hepatoma cells were maintained in DMEM supplemented with heat-inactivated 10% FBS and 1% antibiotics. HepG2.2.15 and Huh7 cells were both grown at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176
2.5. Cell viability assay
The cytotoxic effects of LPRP-Et-97543 was determined by a CellTiter 96 AQueous
one solution cell proliferation assay kit (MTS) (Promega, Madison, WI, USA) to pinpoint the non-toxic test compound concentration in HepG2.2.15 and Huh7 cells. In brief, HepG2.2.15 or HBV-transfected Huh7 cells were plated into 96-well plates at a density of 4 104cells /ml for 24 h. The cells were then treated with serial dilutions of
LPRP-Et-97543, which ranged from 2.5–320 g/ml, and the mixture was incubated for 3 days. Cell toxicity was measured according to the manufacturer’s protocol. All measurements were performed in four replicates and the results are presented as relative percentages over that of the control group.
2.6. Determination of HBsAg and HBeAg
After treating the HepG2.2.15 cells or HBV-transfected Huh7 cells, the levels of the viral surface (HBsAg) and e (HBeAg) antigens were measured in the culture media using an enzyme immunoassay (EIA) kit (Johnson and Johnson, Skillman, NJ, USA) according to the manufacturer’s instructions.
2.7. Plasmid construction
All of the constructs were produced using standard recombinant DNA techniques 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196
(Sambrook et al., 1989). The pHBV1.2 plasmid containing the 1.2-fold of HBV adw2 serotype genome (nt 2186-1986) was cloned into the DraI and BglII site of pGEM-7Zf (+) (Promega, Madison, WI, USA) (Blum et al., 1991) which was kindly provided by Dr. Cheng-Chan Lu (Department of Pathology, National Cheng-Kung University). The four viral promoter-reporter plasmids were constructed by
amplifying the HBV genomic fragments that corresponded to the Core
(nt1636~1851), S (nt3114~220), PreS (nt2438–2855) and X (nt1071–1357) gene promoter regions with a polymerase chain reaction (PCR) that used the pHBV1.2 plasmid as a template and was subsequently inserted into the SacI/XhoI sites of the pGL4.17 luciferase-reporter expression vector (Promega) as previously described (Huang et al., 2014a). In addition, the reporter plasmid, p5CS1-Luc, which contains five copies of the CS1 site (nt 207-216) within HBV surface gene coding sequence (adw2 serotype), was constructed from the pGL4.17 expression vector. All of the DNA sequences were verified with the appropriate restriction enzyme digestion and direct sequencing.
2.8. Promoter-reporter activity assay
Cells were plated into 24-well culture plates, transfected with promoter-reporter constructs (1 g/well) and pRL-SV40 in serum-free DMEM for 24 h, washed with 1 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215
PBS, incubated in DMEM supplemented with 2% FBS, and treated with or without LPRP-Et-97543 (10 g/ml) for 2 days. The luciferase assay was performed according to the manufacturer’s instructions (Promega, Madison, WI, USA). The promoter-reporter construct transfection consisted of co-transfecting the pRL-SV40 Renilla luciferase expression plasmid (0.02 g/well) (Promega, Madison, WI, USA) and using it to normalize the basal level luciferase activity.
2.9. Transfection
Huh7 cells were seeded in 60-mm dishes at the same density (as described in the cell viability assay) for 24 h. The cells were then transfected with the pHBV1.2 plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in DMEM which
contained 10% FBS for 2 days which followed the manufacturer’s protocol. The cells were then treated with the LPRP-Et-97543 compound for another 2 days.
2.10.Analysis of intracellular HBV-RNA by Northern blotting
Total RNA from the treated cells was extracted using the Trizolisolation buffer (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Five micrograms of total RNA was denatured, separated on a 1.0% agarose gel using a commercial kit (Amresco, Solon, OH, USA), and transferred to a positively charged Hybond-N+ nylon membrane (Amersham, GE Healthcare, Buckinghamshire, UK).
216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235
After UV cross-linking, the membrane was hybridized with a DIG-labeled full-length HBV genome probe, washed, and exposed to X-ray film. In brief, the full-length HBV probe was produced by the restriction enzymatic digestion of the pHBV2 plasmid with EcoRI (Will et al., 1985), purified and labeled with a DIG high prime DNA labeling kit (Roche, Mannheim, Germany). The total RNA was normalized by hybridizing the membrane with a glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) probe which was produced by the Pst I digestion of the pGAPDH plasmid (provided by Dr. Hsiao-Sheng Liu from the Graduate Institute of Microbiology and Immunology, National Cheng-Kung University, Tainan, Taiwan). The variations in transfection efficiency were controlled by co-transfecting the Huh 7 cells with the pCMV- (Clontech, Palo Alto, CA, USA), which contains the CMV immediate-early promoter driving the -galactosidase (-gal) gene.
2.11.Southern blot and Real-time PCR analysis of intracellular HBV-DNA synthesis
Encapsidated HBV DNA in cells was extracted from the viral core particles and fractionated on 1.0 % agarose gels and transferred onto the Hybond N+ membrane
(GE Healthcare, Buckinghamshire, UK) as described by Pugh et al. (Pugh et al., 1988). HBV DNA was detected by Southern blot analysis using the DIG-labeled full-length HBV probe. In addition, HBV DNA in the virion from conditioned media that 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254
were untreated or treated with LPRP-Et-97543 or Lamivudine (3TC) was isolated and subjected to Southern blot analysis as described above. Real-time PCR analysis of HBV DNA used 5- AGGAGGCTGTAGGCATAAATTGG -3 as the forward primer and 5- CAGCTTGGAGGCTTGAACAGT-3 as the reverse primer (Feng et al., 2009). The PCR reactions were performed using SYBR Green PCR master mix and the primer pair followed the protocol described below: initial denaturation at 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of amplification at 95°C for 15s and annealing/extending at 58°C for 1 min.
2.12.Sodium dodecylsulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blot analyses of intracellular viral antigens and cellular signaling proteins
Whole cell proteins of Huh7 cells were extracted by RIPA buffer with protease inhibitors. For another set of experiments, the nuclear and cytoplasmic proteins of cells were prepared by NE-PER extraction reagents (Thermo Fisher Scientific,
Rockford, IL, USA) according to the manufacturer’s protocol. The BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA) was used to determine the cellular protein concentration and 25 g of protein from each sample was used for the sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis. Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Amersham, GE 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273
Healthcare, Buckinghamshire, UK) by electroblotting and then the proteins were blocked for 1 h with PBST (PBS buffer with 0.05% Tween-20) that contained 4% of nonfat milk. Immunoblots were incubated with the primary antibody followed by a HRP-conjugated secondary antibody. The antibody-bound proteins were detected using an ECL reagent (Amersham
,
GE Healthcare, Buckinghamshire, UK) and then the same membranes were stripped and reprobed using the anti--tubulin antibody (Sigma, St Louis, MO, USA) or anti-PCNA antibody as the loading control.2.13.Nuclear extract preparation and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from Huh7 cells that were transfected with or without pHBV1.2 for 2 days and treated with or without LPRP-Et-97543 (10 g/ml) for another 2 days to detect the DNA/protein-binding activity. For the EMSA, nuclear extracts (8 g) for every reaction was pre-incubated with or without a competitor and concomitantly mixed with 1 g poly[dI-dC] (Sigma) and 1 g of salmon sperm DNA (Sigma) which was placed on ice for 20 min and then incubated with a 3P-labeled
double-stranded DNA probe for 20 min at 30 °C. The mixture was separated on a 5% nondenatured polyacrylamide gel in 0.5 TBE (Tris-borate-EDTA) buffer at 150 V for 90 min and the binding shift was detected from an autoradiograph. The
oligonucleotide-probe for the DNA binding assay was the NF-B CS1- binding site 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292
which corresponded to the HBV adw serotype (nt 207-216 ), 5′-TACAGGCGGGGTTTTTCTTGTTG-3′ (Lin et al.,2009).
2.14.Statistical Analysis and Quantification of data
The data were expressed as means and standard deviations (SD) of the mean for three independent experiments that used GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA). Variance analysis and student t-tests were used for data analysis. Significant differences occurred when P < 0.05. Quantitative data from Northern and Western blot analysis were obtained using the computing densitometer and TotalLab Quant software (Nonlinear Dynamics Ltd.).
3. Results
3.1. Effect of LPRP-Et-97543 on cell viability
To exclude the possibility of HBV production being suppressed by LPRP-Et-97543 due to its cytotoxicity, cell viability of LPRP-Et-97543 on HepG2.2.15 and Huh7 cells were tested using the MTS assay. The results indicated that LPRP-Et-97543 caused cytotoxic effects at doses >10 g/ml in HepG2.2.15 and HBV-transfected Huh7 cells (Fig. 1B). Thus, a non-cytotoxic dose (< 10 g/ml) was used for the antiviral treatment of HepG2.2.15 or HBV-transfected Huh7 cells.
293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312
3.2. Effects of LPRP-Et-97543 on the HBV virions and antigen secretion in HepG2.2.15 cells
LPRP-Et-97543 effects on the expression of viral antigens were examined by measuring the secreted levels of HBsAg and HBeAg by HepG2.2.15 cells using the EIA. Cells were treated with various LPRP-Et-97543 concentrations (2.5, 5 and 10 g/ml) and the HBsAg and HBeAg secretions were significantly suppressed in a dose-dependent manner compared to controls. The HBeAg inhibition rate was higher than the HBsAg inhibition rate (Fig. 2A). LPRP-Et-97543 function on virion secretion was further examined by isolating the viral DNA from the conditioned medium. Real-time PCR was performed to detect the extracellular viral DNA level. The results showed that LPRP-Et-97543 treatment significantly reduced the secreted viral DNA level in a dose-dependent manner (Fig. 2B).
3.3. Effect of LPRP-Et-97543 on the HBV gene expression
The molecular regulatory effect of LPRP-Et-97543 on the HBV gene expression was determined by transiently transfecting the Huh 7 cells with the pHBV1.2 HBV
genome-containing plasmid (Blum et al., 1991) and treating the cells with three doses (2.5, 5 and 10 g/ml) of LPRP-Et-97543 for 2 days. A Northern blot was performed to analyze the HBV viral RNA expression levels. LPRP-Et-97543 treatment
313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331
significantly reduced both precore/pregenomic and major S/preS RNA with the LPRP-Et-97543 inhibition potential higher on surface RNA than on the precore/ pregenomic RNA (Fig. 3A). In addition, immunoblot analysis was used to detect the intracellular levels of HBV large surface (LHBsAg) and core (HBcAg) proteins and the results indicated that LPRP-Et-97543 potently reduced LHBsAg and HBcAg protein levels compared to vehicle controls (Fig. 3B). Furthermore, Southern blotting was used to assess the intracellular HBV DNA level and the results indicated that LPRP-Et-97543 treatment potently inhibits the replication HBV DNA level in HBV transfected Huh7 cells (Fig. 3C).
3.4. Effect of LPRP-Et-97543 on the HBV gene and cellular signaling pathways related to the responsive element that contains the promoter activity
The regulatory effect of LPRP-Et-97543 on the HBV gene expression was examined by isolating and cloning four promoters that corresponded to the viral gene into a pGL4.17 luciferase reporter vector (Huang et al., 2014a) and the LPRP-Et-97543 effect on the promoter’s activity level was tested. In this study, pCore-Luc, pS-Luc (S promoter), pPreS-Luc (preS promoter) or pX-Luc constructs were transfected into Huh7 cells for 24 h and then the cells were treated with the maximum inhibitory LPRP-Et-97543 dose prior to performing the luciferase assay. The results showed that 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350
LPRP-Et-97543 treatment significantly reduced the Core, S and preS promoter activity whereas it had no effects on the X promoter activity (Fig. 4A). Meanwhile, four luciferase reporters that contained the NF-B, AP-1, AP-2 or the ISRE binding elements were co-transfected with pHBV1.2 into Huh7 cells to determine if LPRP-Et-97543 inhibits the HBV gene expression in the host’s mediated cellular signaling pathways. After LPRP-Et-97543 treatment (10 g/ml), cells were harvested and subjected to a promoter activity assay. Figure 4B shows that LPRP-Et-97543
significantly inhibited NF-B-containing promoter activity but did not affect the AP-1, AP-2 and ISRE-containing promoter activity (p < 0.05).
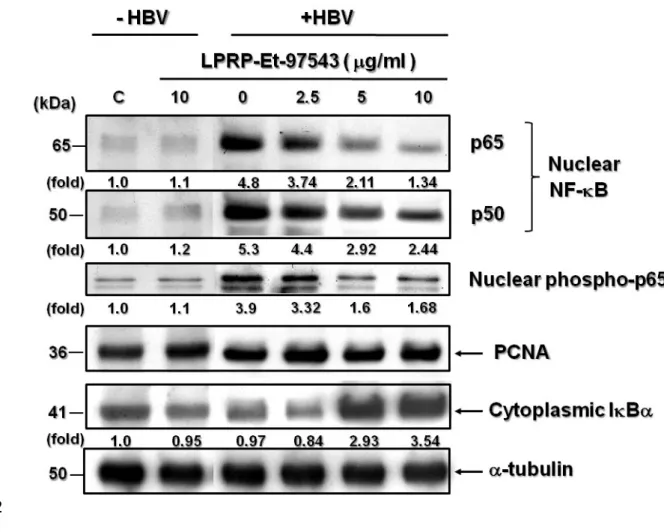
3.5. Effects of LPRP-Et-97543 on NF-B and IB protein expressions in Huh7 cells During LPRP-Et-97543 treatment, NF-B had a regulatory role on the HBV gene expression; thus Huh7 cells were transfected with pHBV1.2 and then treated with LPRP-Et-97543. Western blot was conducted to determine the effect of this
compound on the NF-B signaling pathway relative protein expression. The results showed that LPRP-Et-97543 treatment reduced nuclear p65/p50 NF-B protein expression and reduced phosphorylated NF-B p65 (Fig. 5), whereas cytoplasmic IB expression was significantly enhanced in HBV-expressed cells. To our surprise, when the LPRP-Et-97543 was used to treat Huh7cells that without HBV expression, 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369
the expression levels of nuclear NF-B p65/p50, phosphor-p65 was not impaired during treatment but significantly increased by HBV expression (Fig. 5). However, IB protein slightly decreased by LPRP-Et-97543 in HBV-nontransfected cells.
3.6. NF-B is essential for the inhibitory effects of LPRP-Et-97543 on HBV
Several putative NF-B binding sites were identified in the HBV genome (Lin et al., 2009). Among them, a specific NF-B-binding site was proposed that have a positive regulatory role in HBV gene expression (Huang et al., 2014b). This site was located in the HBV genome at nt207 to 216 (CS1) which corresponds to HBV surface gene coding region. To determine whether LPRP-Et-97543 regulated viral gene expression by NF-B as mediated through the CS1 site, nuclear extracts were prepared from Huh7 cells with or without HBV transfection and tested for the NF-B binding activity to CS1 site by EMSA. The results showed that LPRP-Et-97543 significantly reduced the binding activity of NF-B to the CS1 DNA sequence in HBV-expressing or HBV non-transfected nuclear extracts (Fig. 6A). Moreover, this binding is
mediated through NF-B p65 protein because of occurring supershift band when p65 antibody was added in EMSA binding reaction. The results also showed that
transfection of HBV genome significantly enhanced the binding activity of NF-B to the CS1 site (Fig. 6A). To determine if LPRP-Et-97543 could affect CS1 containing 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388
promoter activity, p5CS1-Luc reporter was constructed and co-transfected into Huh7 cells with or without HBV expression. The transfections were then treated with or without LPRP-Et-97543 (10 g/ml) for 2 days and tested for luciferase assay. Results revealed that CS1- containing promoter activity increased in HBV-expressing cells, but not in HBV non-transfected cells. Furthermore, treatment with LPRP-Et-97543 significantly decreased CS1-containing promoter activity in HBV-expressed cells, but not in HBV-nontransfected cells (Fig. 6B). To further examine whether the regulatory effect of LPRP-Et-97543 on the HBV is mediated through the inhibition of the NF-B signaling pathway, the p65 expression plasmid was co-transfected with pHBV1.2 in Huh7 cells and treated with LPRP-Et-97543 (10 g/ml). The HBV DNA from the cultural medium was extracted for Real-time PCR analysis and the results showed that an increase in p65 expression dose (0.5 and 2.5 g) significantly reversed the
inhibitory effect of LPRP-Et-97543 on the HBV DNA level as compared to the empty vector transfection (Fig. 6C).
4. Discussion
In this study, the LPRP-Et-97543 compound was used to evaluate the potential antiviral effects against HBV. The results demonstrated that LPRP-Et-97543 inhibited viral HBsAg and HBeAg secretion and supernatant viral DNA levels in HepG2.2.15 cells (Fig. 2A and 2B). To elucidate the inhibitory mode of LPRP-Et-97543 on HBV, 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407
the HBV-transfected Huh7 cells were used as a platform to determine how this compound regulates viral gene expression and viral propagation, which was facilitated by the low sensitivity of determination for the viral gene products in HepG2.2.15cells (Huang et al., 2014b). LPRP-Et-97543 significantly down-regulated viral S/preS relative to precore/pregenomic RNA levels (Fig. 3A). LPRP-Et-97543 treatment significantly inhibited the synthesized viral protein expression level of a large form of viral surface protein (LHBsAg) and core protein (HBcAg) (Fig. 3B). Southern blot analysis was used to detect the isolated viral DNA in cells and the results revealed that intracellular HBV DNA was decreased by LPRP-Et-97543 (Fig. 3C). In view of our results demonstrated that LPRP-Et-97543 could regulate the HBV gene at the transcription level, therefore, viral promoter activity was further examined and the results showed that HBV core, S, and preS promoters were attenuated by LPRP-Et-97543 but the X promoter activity in the HBV-expressed Huh7 cells were not affected (Fig. 4A). These results suggest that LPRP-Et-97543 could potentially inhibit the HBV gene expression by regulating its promoter activity. In addition, It is important to figure out if viral covalently closed circular DNA (cccDNA) was affected during treatment in HBV transfected cells to distinguish the exact transcription potential of produced viral genome than the transfected plasmid. However, there is no detectable level of cccDNA was observed that when we 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426
conducted the experiments for cccDNA isolation and for Southern blot analysis (data not shown). This may due to the very low level of cccDNA during treatment or poor detecting efficiency in Southern blot.
To examine the possible mechanism by which LPRP-Et-97543 affects HBV function is mediated through host’s cellular signaling pathways; transcriptional activities of four major signaling pathways, AP-1, AP-2, NF-B and ISRE were analyzed using a promoter-reporter assay. After transfection and treatment, LPRP-Et-97543 specifically decreased NF-B promoter activity, while there were no effects on AP-1, AP-2 and ISRE promoters in the HBV-transfected Huh7cells. This result indicates that LPRP-Et-97543 may regulate the NF-B signaling pathway in HBV transfected host cells (Fig. 4B). Moreover, immunoblots revealed that the nuclear expression of NF-B p65/p50 proteins and the phosphorylated p65 protein level were decreased by LPRP-Et-97543 while the cytoplasmic IB protein level was
augmented by the same treatment in a dose-dependent manner (Fig. 5). These results suggest that LPRP-Et-97543 could potentially enhance IB expression in cytoplasm and inhibits the p65/p50 NF-B nuclear translocation and then attenuates the p65 phosphorylation in subsequent activation. Surprisingly, the immunoblot also reveals that without compound treatment, HBV transfection could enhance NF-B nuclear translocation of NF-B protein and subsequent activation of p65 in Huh7 cells, while 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445
the expression of NF-B member proteins was not changed in HBV non-transfected cells, suggests that HBV infection might be required for the actions of NF-B factors on gene promoter activity and LPRP-Et-97543 could interfere this regulation.
Previous reports have shown that several binding elements for NF-B are essential for HBV viral gene regulation. Among them, a study by Kwon and Rho (2002) clearly demonstrated that the HBV core protein (HBcAg) upregulates the HBV enhancer II/ pregenomic promoter through the NF-B binding site located upstream of the promoter. In addition, our recent studies also indicated that several natural occurring compounds could inhibit HBV gene expression and genome replication base on the interfering NF-B factor on viral surface promoter activity (Huang et al., 2014b). Therefore, the importance of NF-B in HBV gene regulation is the main issue we further addressed. On the other hand, to determine the modulatory role of LPRP-Et-97543 on HBV gene regulation probably raised the possible mechanisms that HBV genes could be regulated by HBV viral protein. It makes sense since previous studies showed that transcription activators like HBx (Doria et al., 1995), HBcAg (Kwon and Rho, 2002), a C-terminally truncated form of HBV middle surface antigen (MHBst)
(Meyer et al., 1992), and HB large surface antigens (LHBs) (Hildt et al., 1996) are capable of NF-B activators. Thus, the effects of LPRP-Et-97543 on HBV gene regulation may suggest that link to cellular NF-B signaling pathway.
446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464
In view of the potential inhibitory effect of LPRP-Et-97543 on HBV surface transcripts from Northern blot analysis (Fig. 3A) and a putative CS1 site was identified available for NF-B on HBV surface gene upregulation (Lin et al., 2009) (Huang et al., 2014b). We further examine the possible role of LPRP-ET-97543 on the binding activity of NF-B to DNA elements and CS1 NF-B binding sequence was synthesized and used for an EMSA analysis. As seen in result Fig. 6A, LPRP-Et-97543 treatment significantly decreased the binding activity of NF-B to the CS1 site and this binding was specific because the protein/DNA shift band can be supershift when p65 NF-B antibody was used in the binding reaction and the similar result has been reported previously (Huang et al., 2014b). In addition, Lin et al. (2009) have proposed that CS1 site of HB surface gene is capable for p65 homodimer binding. In view of important role of CS1 NF-B binding sequence for the action of LPRP-Et-97543 on HBV surface gene expression, we constructed CS1 site containing
promoter-reporter (luciferase) and examine whether LPRP-Et-97543 could regulate CS1 containing gene expression. The result indicates that LPRP-Et-97543 compound could inhibit CS1-containing promoter activity and can be enhanced by HBV co-transfection (Fig. 6B). These results clearly indicate that HBV viral proteins might be involved in LPRP-Et-97543 regulated viral gene expression and showed that host NF-B signaling pathways inhibited by LPRP-Et-97543 were investigated in an HBV-465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483
dependent manner.
To determine if LPRP-Et-97543 effects HBV gene expression and viral propagation is indeed correlated with NF-B regulation, the pCMV-p65 NF-B expression plasmid was co-transfected with pHBV1.2 to Huh7cells. The secreted HBV viral particles were isolated and Southern blot analysis detected the viral DNA level by viral DNA real-time PCR. As seen in Fig.6C, when cells were treated with a maximum dose (10 g/ml) of LPRP-Et-97543, the secretion of the HBV viral particle was inhibited about 41% compared to the vehicle treated controls. However, the inhibition effect of LPRP-Et-97543 on the HBV viral particle secretion was
significantly reversed when cells were co-transfected with an increasing dose (0.5 and 2.5 g) of pCMV-p65 plasmid using the same treatment. There was no difference in the secreted levels of HBV DNA in a maximum transfection dose of pCMV-p65 plasmid combined with LPRP-Et-97543 treatment compared to non-treated controls. This result suggests the LPRP-Et-97543 effects on HBV gene expression and viral DNA replication might be mediated through the NF-B inhibition.
For medicinal applications, controlling the HBV well enough to ameliorate the disease is the most crucial issue in affecting the antiviral therapeutic strategies, as well as preventing the viral resistance commonly caused by viral reverse transcriptase inhibitors. In this study, LPRP-Et-97543 regulated the HBV gene expression and viral 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502
DNA replication by interfering with the host cell’s NF-B signaling pathway. In addition, previous studies have proposed that NF-B activation in cells that were infected with HBV (Doria et al., 1995; Hildt et al., 1996; Kwon and Rho, 2002; Meyer et al., 1992) might be a possible treatment for the HBV and that activation of the NF-B pathway is mediated by viral proteins (Huang et al., 2014b). However, the discrepancy was raised that the inhibitory effect of tumor necrosis factor alpha (TNF-) or interleukin-1 (IL-1) on HBV replication was mediated through of NF-B activation in previous studies (Lin et al., 2009). In contrast, elevated expression of NF-B in HBV-infected hepatocytes seemed to be critical to HBV gene upregulation and viral replication in our study. A possible explanation is that the inhibition of HBV by NF-B activation is mediated through indirect protein interaction between NF-B and other factor such as SP-1 (Lin et al., 2009) and do not bind directly to putative NF-B site in HBV genome. However, our study clearly indicated that the possible upregulatory role of viral protein on HBV gene expression and via through direct binding of NF-B to its element and this regulation can be interfered by drug during treatment. Thus, the activation of NF-B by which to inhibit or activate HBV gene expression and viral replication seems to have distinct pathways. On the other hand, several issues remain to be elucidated in the future, including how viral factor (i.e., Core and X proteins) involved in LPRP-Et-97543 action on HBV gene regulation; 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521
how NF-B is involved in regulating viral gene promoter activity other than the surface gene and how HBV switches the regulatory machinery and takes over cellular signaling pathways in the chronic-infected phase of HBV in response to drug
treatment. In conclusion, the mechanism underlying the anti-HBV effects has been elucidated and a novel drug, LPRP-Et-97543, for treating HBV infected liver disease was discovered.
Acknowledgements
This work was funded by grants from the Committee on Chinese Medicine and Pharmacy, Department of Health, Executive Yuan, Taiwan (CCMP102-RD-116), China Medical University Hospital (DMR-103-042). The authors express heartfelt thanks to Dr. Cheng-Chan Lu for the pHBV1.2 and pHBV2 plasmids, to Dr. Hsiao-Sheng Liu for the pGAPDH plasmid, and to Dr. Cheng-Wen Lin for pAP1-Luc, pAP2-Luc, pNF-kB-Luc, and pISRE-Luc promoter–reporter constructs.
References
Bai, X., Chen, X., Liu, Y., Tian, L., Zhou, Q., Liu, S., Fang, J., Chen, J., 2009a. Effects of water extract and crude polysaccharides from Liriope spicata var. prolifera on InsR/IRS-1/PI3K pathway and glucose metabolism in mice. J. Ethnopharmacol. 125, 482-486. 522 523 524 525 526 527 528 529 530 531 532 533 534 535 536 537 538 539 540 541
Bai, X., Chen, X., Liu, Y., Tian, L., Zhou, Q., Liu, S., Fang, J., Chen, J., 2009b. Effects of water extract and crude polysaccharides from Liriope spicata var. prolifera on InsR/IRS-1/PI3K pathway and glucose metabolism in mice. J. Ethnopharmacol. 125, 482-486.
Bent, S., 2008. Herbal medicine in the United States: review of efficacy, safety, and regulation: grand rounds at University of California, San Francisco Medical Center. J. Gen. Intern. Med. 23, 854-859.
Bent, S., Ko, R., 2004. Commonly used herbal medicines in the United States: a review. Am. J. Med. 116, 478-485.
Blum, H.E., Galun, E., Liang, T.J., von Weizsacker, F., Wands, J.R., 1991. Naturally occurring missense mutation in the polymerase gene terminating hepatitis B virus replication. J. Virol. 65, 1836-1842.
Chen, X., Bai, X., Liu, Y., Tian, L., Zhou, J., Zhou, Q., Fang, J., Chen, J., 2009. Anti-diabetic effects of water extract and crude polysaccharides from tuberous root of Liriope spicata var. prolifera in mice. J. Ethnopharmacol. 122, 205-209.
Choi, S.I., Lee, H.R., Goo, J.S., Kim, J.E., Nam, S.H., Hwang, I.S., Lee, Y.J., Prak, S.H., Lee, H.S., Lee, J.S., Jang, I.S., Son, H.J., Hwang, D.Y., 2011. Effects of Steaming Time and Frequency for Manufactured Red Liriope platyphylla on the Insulin Secretion Ability and Insulin Receptor Signaling Pathway. Lab. Animal 542 543 544 545 546 547 548 549 550 551 552 553 554 555 556 557 558 559 560
Doria, M., Klein, N., Lucito, R., Schneider, R.J., 1995. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J 14, 4747-4757.
Feng, Y., He, F., Zhang, P., Wu, Q., Huang, N., Tang, H., Kong, X., Li, Y., Lu, J., Chen, Q., Wang, B., 2009. Inhibitory effect of HMGN2 protein on human
hepatitis B virus expression and replication in the HepG2.2.15 cell line. Antiviral Res. 81, 277-282.
Gish, R.G., 2005. Current treatment and future directions in the management of chronic hepatitis B viral infection. Clin. Liver Dis. 9, 541_565.
Gitlin, N., 1997. Hepatitis B: diagnosis, prevention, and treatment. Clin. Chem. 43, 1500-1506.
Hildt, E., Saher, G., Bruss, V., Hofschneider, P.H., 1996. The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology 225, 235-239. Hu, Z.F., Chen, L.L., Qi, J., Wang, Y.H., Zhang, H., Yu, B.Y., 2011. Two new
benzofuran derivatives with anti-inflammatory activity from Liriope spicata var. prolifera. Fitoterapia 82, 190-192.
Huang, T.J., Liu, S.H., Kuo, Y.C., Chen, C.W., Chou, S.C., 2014a. Antiviral activity of chemical compound isolated from Artemisia morrisonensis against hepatitis B virus in vitro. Antiviral Res. 101, 97-104.
Huang, T.J., Chou, B.H., Lin, C.W., Weng, J.H., Chou, C.H., Yang, L.M., Lin, S.J., 562 563 564 565 566 567 568 569 570 571 572 573 574 575 576 577 578 579 580 581
2014b. Synthesis and antiviral effects of isosteviol-derived analogues against the hepatitis B virus. Phytochemistry 99, 107-114.
Hur, J., Lee, P., Kim, J., Kim, A.J., Kim, H., Kim, S.Y., 2004. Induction of Nerve Growth Factor by Butanol Fraction of Liriope platyphylla in C6 and Primary Astrocyte Cells. Biol. Pharm. Bull. 27, 1257-1260.
Kim, S.-W., Chang, I.-M., Oh, K.-B., 2002. Inhibition of the Bacterial Surface Protein Anchoring Transpeptidase Sortase by Medicinal Plants. Biosci., Biotechnol., Biochem. 66, 2751-2754.
Kwon, J.A., Rho, H.M., 2002. Hepatitis B viral core protein activates the hepatitis B viral enhancer II/pregenomic promoter through the nuclear factor kappaB binding site. Biochem Cell Biol 80, 445-455.
Lee, Y.C., Lee, J.C., Seo, Y.B., Kook, Y.B., 2005. Liriopis tuber inhibit OVA-induced airway inflammation and bronchial hyperresponsiveness in murine model of asthma. J. Ethnopharmacol.101, 144-152.
Lin, Y.C., Hsu, E. C., Ting, L. P., 2009. Repression of hepatitis B viral gene
expression by transcription factor nuclear factor-kappaB. Cell Microbiol. 11, 645-660.
Li, T., Peng, T., 2013. Traditional Chinese herbal medicine as a source of molecules with antiviral activity. Antiviral Res. 97, 1-9.
582 583 584 585 586 587 588 589 590 591 592 593 594 595 596 597 598 599 600
Baeuerle, P.A., 1992. Hepatitis B virus transactivator MHBst: activation of NF-kappa B, selective inhibition by antioxidants and integral membrane localization. EMBO J 11, 2991-3001.
Nguyen, A.T., Fontaine, J., Malonne, H., Duez, P., 2006. Homoisoflavanones from Disporopsis aspera. Phytochemistry 67, 2159-2163.
Park, N.H., Song, I.H., Chung, Y.H., 2006. Chronic hepatitis B in hepatocarcinogenesis. Postgrad. Med. J. 82, 507-515.
Pugh, J.C., Yaginuma, K., Koike, K., Summers, J., 1988. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J. Virol.62, 3513-3516.
Sambrook, J., Fritsch, E.F., Maniatis, T., 1989. Molecular Cloning: A Laboratory Manual, Second ed. Cold Spring Harbor Laboratory Press.
Sells, M.A., Chen, M.L., Acs, G., 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. U. S. A. 84, 1005-1009.
Sun, L., Lin, S., Zhao, R., Yu, B., Yuan, S., Zhang, L., 2010. The Saponin Monomer of Dwarf Lilyturf Tuber, DT-13, Reduces Human Breast Cancer Cell Adhesion and Migration during Hypoxia via Regulation of Tissue Factor. Biol. Pharm. Bull. 33, 1192-1198.
Tsai, Y.C., Chiang, S.Y., El-Shazly, M., Wu, C.C., Beerhues, L., Lai, W.C., Wu, S.F., 602 603 604 605 606 607 608 609 610 611 612 613 614 615 616 617 618 619 620 621
Yen, M.H., Wu, Y.C., Chang, F.R., 2013. The oestrogenic and anti-platelet activities of dihydrobenzofuroisocoumarins and homoisoflavonoids from Liriope platyphylla roots. Food Chem. 140, 305-314.
Wang, H.C., Wu, C.C., Cheng, S., Kuo, C.Y., Tsai, Y.C., Chiang, S.Y., Wong, T.-S., Wu, Y.-C., Chang, F.-R., 2013. Active Constituents from Liriope platyphylla Root against Cancer Growth In Vitro. Evid. Based Complementary Altern. Med. http://dx.doi.org/10.1155/2013/857929
Wang, K.W., Zhang, H., Shen, L.Q., Wang, W., 2011. Novel steroidal saponins from Liriope graminifolia (Linn.) Baker with anti-tumor activities. Carbohydr Res 346, 253-258.
Will, H., Cattaneo, R., Darai, G., Deinhardt, F., Schellekens, H., Schaller, H., 1985. Infectious hepatitis B virus from cloned DNA of known nucleotide sequence. Proc. Natl. Acad. Sci. U. S. A. 82, 891-895.
Wu, F., Cao, J., Jiang, J., Yu, B., Xu, Q., 2001. Ruscogenin glycoside (Lm-3) isolated from Liriope muscari improves liver injury by dysfunctioning liver-infiltrating lymphocytes. J. Pharm. Pharmacol. 53, 681-688.
Figure legends
Fig. 1. (A) Schematic illustration of the LPRP-Et-97543 structure. (B) Cytotoxicity of
622 623 624 625 626 627 628 629 630 631 632 633 634 635 636 637 638 639 640
were plated in 96-well plates for 24 h and treated with serial dilutions (0.625~40 g/ml) of LPRP-Et-97543 for 3 days. After treatment, the cells were subjected to a cytotoxic assay. The data are expressed as mean and the standard deviation of the mean. (n = 4) (*P<0.05 vs untreated cells)
Fig. 2. The effects of LPRP-Et-97543 on (A) the secretion of HBV viral antigens and
(B) replicated viral DNA levels in HepG2.2.15 cells. Cells were treated with various concentrations (2.5, 5 and 10 g/ml) of LPRP-Et-97543 for 3 days and the
conditioned media were collected for the EIA analysis of HBsAg and HBeAg. In addition, the HBV DNA was isolated from virion particles and subjected to Real time-PCR analysis. The data are expressed as the mean and the standard deviation of the mean. (n = 3) (*P<0.05 vs untreated cells)
Fig. 3A. The effects of LPRP-Et-97543 on HBV gene expression and viral DNA
replication in Huh7 cells. Huh7 cells were transfected with pHBV1.2 plasmid for 2 days and treated with three concentrations (2.5, 5 and 10 g/ml) of LPRP-Et-97543 for another 2 days. Treated cells were harvested and subjected to total RNA, protein and DNA isolation. (A) The total RNA from the transfected and treated Huh7cells were subjected to Northern blot analysis using the HBV whole genome DNA as the 642 643 644 645 646 647 648 649 650 651 652 653 654 655 656 657 658 659 660
probe, as described in the Materials and Methods section. GAPDH was used as an RNA loading control and -gal was used to detect the efficiency of each transfection. (B) Total cellular proteins were extracted and immunoblotting was used to detect the LHBsAg, HBcAg and -tubulin for comparison. (C) Intracellular HBV DNA were isolated and subjected to Southern blot analysis. RC, relaxed circular; SS, single stranded; RI, replicated intermediates; HBV standard, 2 ng of 3.2 kb linear HBV genome marker. The intensity of each protein band was quantitated with a
densitometer and the relative amount was normalized with an internal control. Data shown in (A), (B) and (C) are representative of three sets of experiments.
Fig. 4. The effects of LPRP-Et-97543 on the HBV gene promoter or the cellular
signaling pathway responsive to the promoter activity. Huh7 cells were seeded in 24-well plates for 24 h and co-transfected with pHBV1.2 and with either (A) pCore-Luc, pS-Luc, pPreS-Luc, and pX-Luc viral gene promoter reporter constructs or (B) pAP1-Luc, pAP2-pAP1-Luc, pNF-B-Luc and pISRE-Luc cellular signaling pathway responsive to the promoter reporter constructs together with pRL-SV40 for 24 h and then treated with LPRP-Et-97543 (10 g/ml) for another 24 h. The cellular lysates were prepared for the luciferase assay as described in the Materials and Methods Section. The data are expressed as the mean and the standard deviation of the mean. (n=3) (*p0.05 vs 661 662 663 664 665 666 667 668 669 670 671 672 673 674 675 676 677 678 679
untreated cells)
Fig. 5. The effects of LPRP-Et-97543 on nuclear NF-B p65/p50, phospho- p65 and cytoplasmic IB protein expressions in Huh7 cells that with or without HBV genome expression. Huh7 cells were transfected with or without pHBV1.2 for 2 days and treated with three concentrations (2.5, 5 and 10 g/ml) of LPRP-Et-97543 for another 2 days. Nuclear fraction of cellular proteins was extracted and
immunoblotting was used to detect p65, p50, and phosphorylated p65 of NF-B protein. The cytoplasmic fraction of cellular proteins was used to detect IB for comparison. PCNA protein was detected in immunoblot as control for nuclear protein expression. The data shown are representative of the three sets of experiments.
Fig. 6. Effect of the NF-B signaling pathway on LPRP-Et-97543 mediated HBV gene inhibition. (A) For the DNA binding activity assay, Huh7 cells were transfected with pHBV1.2 for 2 days, treated with or without LPRP-Et-97543 (10 g/ml) and then the cellular nuclear proteins were extracted and subjected to EMSA analysis. (B) For the CS1 site-containing promoter activity, Huh7 cells were transfected with either the p5CS1-Luc promoter-reporter constructs together with or without pHBV1.2 plasmid for 24 h and treated with or without LPRP-Et-97543 for 2 days. (C) In the 680 681 682 683 684 685 686 687 688 689 690 691 692 693 694 695 696 697 698
other set of experiments, Huh7 cells were co-transfected with pHBV1.2 and with two doses (0.5, and 2.5 g) of pCMV-p65 plasmid for 2 days and then treated with LPRP-Et-97543 (10 g/ml) for another 2 days. The secreted viral particles from the cells were isolated by the kit and subjected to Real-time PCR analysis of HBV DNA. For comparison, total protein of transfected and treated cells were extracted for NF-B p65 protein immunoblot. (n=3) (*p0.05 vs untreated cells)
Figure 699 700 701 702 703 704 705 706 707 708 709 710 711 712
Fig. 1. 713 714 715 716 717 718
Fig. 2.
719
720 721
722
Fig. 3. 724 725 726 727 728
729
Fig. 4. Fig. 5. 731 732 733 734 735 736
737
Fig. 6.
739 740 741