Elsevier Editorial System(tm) for Journal of Ethnopharmacology Manuscript Draft
Manuscript Number: JEP-D-10-00094R1
Title: Application of bioactivity database of Chinese herbal medicine on the therapeutic prediction, drug development, and safety evaluation
Article Type: Full Length Article
Corresponding Author: Dr. Tin-Yun Ho,
Corresponding Author's Institution: China Medical University First Author: Hui-Man Cheng
Order of Authors: Hui-Man Cheng; Chia-Cheng Li; Calvin Yu-Chian Chen; Hsin-Yi Lo; Wen-Yu Cheng; Chang-Hsien Lee; Shi-Zhe Yang; Shih-Lu Wu; Chien-Yun Hsiang; Tin-Yun Ho
Dear Dr. Guo,
We have revised the manuscript (JEP-D-10-00094) according to reviewers’ comments. Our point-by-point reply to the reviewers’ comments is described as follows.
Please handle our manuscript at your convenience. Thank you very much.
Sincerely yours, Tin-Yun Ho *Cover Letter
Journal of Ethnopharmacology
AUTHOR CHECKLIST
Dear Author,It frequently happens that on receipt of an article for publication, we find that certain elements of the manuscript, or related information, is missing. This is regrettable of course since it means there must be a delay in processing the article while we obtain the missing details.
In order to avoid such delays in the publication of your article, if accepted, could you please run through the list of items below and check each box. Please enclose a copy of this list with the manuscript
submission.
Overall Manuscript Details
•
Manuscript type– please check one of the following:Research article ■
Review article
□
Ethnopharmacological Communication□
Book Review□
Commentary
□
Other□
•
Do you declare that the abstract is in the requested structured format? ■•
Did you use the right format for the references? ■•
Are the corresponding author’s postal address, telephone and faxnumbers complete on the manuscript? ■
•
Have you provided the corresponding author’s e-mail address? ■•
Do you declare that this manuscript/data, or parts thereof, has not been submitted ■ or published elsewhere for publication?•
Do you declare that all the listed authors have read and approved ■ the submitted manuscript?•
Do you declare that the present study was performed according to ■ international, national and institutional rules considering animal experiments,clinical studies and biodiversity rights?
Revised manuscripts
•
Have you addressed each remark from the referees?□
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Revision notes (JEP-D-10-00094)
Reviewer #1
1. We have requested the phytochemical profile (including HPLC and TLC data) of
each formula from a GMP pharmaceutical company, Sun Ten Pharmaceutical Co, in Taiwan. These profiles have been deposited in Molecular Biology Laboratory, School of Chinese Medicine, China Medical University. We have supplemented
this statement in the “Materials and Methods” section (p. 6, line 13).
2. In this study, we would like to analyze the biological events induced by herbal formulae, predict the therapeutic potentials of formulae, and evaluate the safety of formulae. Therefore, the dose (150 mg/kg) for animal study is equivalent to the dose (9 g/adult) for human usage. We have supplemented this statement in the
“Materials and Methods” section (p. 6, line -7).
3. The organs from three mice were combined into a single sample and the number of microarray replicates was three. We have supplemented this statement in the
“Materials and Methods” section (p. 7, line 4).
4. We have revised the writing according to reviewer’s suggestion.
“large-scaled” “large-scale” (p. 4, lines -7 and -10)
shown “on” Table shown “in” Table (p. 6, line -10; p. 11, line -6) *Revision Notes
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 Reviewer #2
1. Table 1 shows the constituents of each herbal formula and the ration of each constituent in the formula. The herbal constituents are represented by scientific names (genus and species) and the non-herbal constituents are expressed by English names.
2. We have supplemented the ethnopharmacological usage of each formula in Table 1.
3. We have supplemented the detail information about the preparation of each formula in Table 1.
4. We have revised the figure legends to make them more informative (p. 26-27).
We also have supplemented the interpretation of each figure in “Materials and Methods” and “Results” sections.
5. In this study, we would like to analyze the biological events induced by herbal formulae, predict the therapeutic potentials of formulae, and evaluate the safety of formulae. Therefore, the dose (150 mg/kg) for animal study is equivalent to the dose (9 g/adult) for human usage. Additionally, organs from three mice were combined into a single sample and the number of microarray replicates was three. We have supplemented this statement in the “Materials and Methods” section (p. 6, line -7; p. 7, line 4).
6. The aim of this report is to apply transcriptomic tools as a novel platform of translational medicine for Chinese herbal medicine. We illustrate the usefulness of this platform by analyzing the biological events induced by formulae, predicting the therapeutic potential of formulae, and evaluating the safety of formulae. Additionally, the linkage of other literature studies and our microarray analysis also show the reliability of bioactive database. Therefore, this platform will be used not only for the understanding of therapeutic mechanisms involving Chinese herbal medicine and gene interactions, but also for the new theories in drug discovery.
7. We have revised the writing according to reviewer’s suggestion.
Title: Application of bioactivity database of Chinese herbal medicine on the therapeutic prediction, drug development, and safety evaluation (p. 1)
have “be” used have “been” used (p. 4, line 15)
We applied transcriptomic tools as a novel platform of translational medicine for Chinese herbal medicine. The usefulness of this platform was illustrated by analyzing the biological events induced by formulae, predicting the therapeutic potential of formulae, and evaluating the safety of formulae.
Mice were administrated orally with top 15 mostly used Chinese herbal formulae in National Health Insurance
Database in Taiwan for 7 consecutive days
Microarray analysis
Gene expression profile database
Biological events Therapeutic potential Herbal safety Graphical Abstract
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 Abstract
Aim of the study: Chinese herbal medicine has been used for the treatments of various diseases for years. However, it is often difficult to analyze their biological activities and molecule mechanisms because of their complex nature. In this study, we applied DNA microarray to analyze the biological events induced by herbal formulae, predict the therapeutic potentials of formulae, and evaluate the safety of formulae.
Materials and methods: Mice were administrated orally with 15 formulae for 7 consecutive days, and the gene expression profiles in liver or kidney were further analyzed by transcriptomic tools.
Results: Our data showed that most formulae altered the metabolic pathways, such as glutathione metabolism and oxidative phosphorylation, and regulatory pathways, such as antigen processing and presentation and insulin-like growth factor signaling pathway. By comparing the gene expression signatures of formulae with those of disease states or drugs, we found that mice responsive to formula treatments might be related to disease states, especially metabolic and cardiovascular diseases, and drugs, which exhibit anti-cancer, anti-inflammatory, and anti-oxidative effects. Moreover, most formulae altered the expression levels of cytochrome p450, glutathione S-transferase, and UDP glycosyltransferase genes, suggesting that caution should be paid to possible drug interaction of these formulae. Furthermore, the similarities of gene expression profiles between formulae and toxic chemicals were low in kidney, suggesting that these formulae might not induce nephrotoxicities in mice.
Conclusions: This report applied transcriptomic tools as a novel platform of translational medicine for Chinese herbal medicine. This platform will not only for understanding the therapeutic mechanisms involving herbal formulae and gene interactions, but also for the new theories in drug discovery.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Application of bioactivity database of Chinese herbal medicine on the therapeutic prediction, drug development, and safety evaluation
Hui-Man Chenga, Chia-Cheng Lib, Calvin Yu-Chian Chenb, Hsin-Yi Lob, Wen-Yu Chengb, Chang-Hsien Leeb, Shi-Zhe Yangb,c, Shih-Lu Wud, Chien-Yun Hsiange,*,1, and Tin-Yun
Hob,f,*,1
a
Graduate Institute of Integration of Traditional Chinese and Western Medicine, China Medical University, Taichung 40402, Taiwan
b
School of Chinese Medicine, China Medical University, Taichung 40402, Taiwan
c
Department of Radiology, China Medical University Hospital, Taichung 40447, Taiwan
d
Department of Biochemistry, China Medical University, Taichung 40402, Taiwan
e
Department of Microbiology, China Medical University, Taichung 40402, Taiwan
f
Department of Nuclear Medicine, China Medical University Hospital, Taichung 40447, Taiwan
*
Corresponding author at: School of Chinese Medicine, China Medical University, 91 Hsueh-Shih Road, Taichung 40402, Taiwan. Tel: +886 4 22053366 x 3302; fax: +886 4 22032295; E-mail address: cyhsiang@mail.cmu.edu.tw (C.Y. Hsiang).
1
These authors contributed equally.
Abbreviations: SHXXT, San-Huang-Xie-Xin-Tang; PBS, phosphate-buffered saline; MeSH, Medical Subject Headings; GSTs, glutathione S-transferases; UGT, UDP glycosyltransferase; EDGE, Environment, Drugs and Gene Expression; IGF, *Manuscript
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
insulin-like growth factor; CCCTS, Chuan-Chiong-Chaa-Tyau-Saan; MHC-I, major histocompatibility complex class I.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 Abstract
Aim of the study: Chinese herbal medicine has been used for the treatments of various diseases for years. However, it is often difficult to analyze their biological activities and molecule mechanisms because of their complex nature. In this study, we applied DNA microarray to analyze the biological events induced by herbal formulae, predict the therapeutic potentials of formulae, and evaluate the safety of formulae.
Materials and methods: Mice were administrated orally with 15 formulae for 7 consecutive days, and the gene expression profiles in liver or kidney were further analyzed by transcriptomic tools.
Results: Our data showed that most formulae altered the metabolic pathways, such as glutathione metabolism and oxidative phosphorylation, and regulatory pathways, such as antigen processing and presentation and insulin-like growth factor signaling pathway. By comparing the gene expression signatures of formulae with those of disease states or drugs, we found that mice responsive to formula treatments might be related to disease states, especially metabolic and cardiovascular diseases, and drugs, which exhibit anti-cancer, anti-inflammatory, and anti-oxidative effects. Moreover, most formulae altered the expression levels of cytochrome p450, glutathione S-transferase, and UDP glycosyltransferase genes, suggesting that caution should be paid to possible drug interaction of these formulae. Furthermore, the similarities of gene expression profiles between formulae and toxic chemicals were low in kidney, suggesting that these formulae might not induce nephrotoxicities in mice.
Conclusions: This report applied transcriptomic tools as a novel platform of translational medicine for Chinese herbal medicine. This platform will not only for understanding the therapeutic mechanisms involving herbal formulae and gene interactions, but also for the new theories in drug discovery.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 1. Introduction
Chinese herbal medicine has been used for the treatments of various diseases for years. Extracts prepared from medicinal plants and other natural sources contain a variety of molecules with potent biological activities. Unfortunately, it is often difficult to analyze the biological activities of these extracts because of their complex nature and the possible interaction of their components. Therefore, the genome-wide expression monitoring system with high-density microarrays may provide a simple way to test biochemical effects of herbs and thereby gain insights into their potential beneficial effects and negative side effects.
DNA microarray is a popular research and screening tool for differentially expressed genes (Schena et al., 1995). Although certain limitations of the current technology exist and have become more apparent during the past couple of years, their ability to monitor the expression of thousands of genes simultaneously is unsurpassed (Draghici et al., 2006). Microarray-based gene expression patterns have been used as fingerprints of cellular physiology and cancer researches (Altenhein et al., 2006; Bild et al., 2006; Pittman et al., 2004). The large-scale gene expression analysis of toxin-treated cells and animals has yielded a highly accurate capacity to recognize the toxic potentials of novel drug candidates (Ganter et al., 2005). Moreover, the large-scale gene expression profile has been applied to identify the disease target for drug development (Whitfield et al., 2006). Furthermore, the therapeutic efficacy can be predicted on the basis of gene expression signatures in vitro (Gunther et al., 2003).
Chinese herbal medicine has been applied for the treatments of diseases or served as corroborants to improve general physical weakness and fatigue for thousands of years. The clinical trials of herbal formulae have demonstrated their efficacies in
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
patients with illness. However, the molecular mechanisms and molecular effects of most herbal formulae are still unclear. Microarray data have been used to characterize the mechanisms of herbal formulae or herbal compounds. For examples, PC-SPES is used among patients with prostate carcinoma as an alternative to conventional forms of therapy. Biochemical assays and clinical observations suggest that the cytotoxic effects of PC-SPES are mediated at least in part through estrogenic activity (DiPaola et al., 1998). Microarray analysis indicates that alternations in specific genes involved in modulating the cell cycle, cell structure, and androgen response may also be responsible for PC-SPES-mediated cytotoxicity (Bonham et al., 2002). San-Huang-Xie-Xin-Tang (SHXXT) has been used to treat gastritis, gastric bleeding, and peptic ulcers. Microarray data show that SHXXT exhibits an anti-proliferation effect via p53 and DNA damage signaling pathways. Moreover, Rhizoma Coptis shares a similar gene expression profile with SHXXT, suggesting that Rhizoma Coptis is the principle herb that exerts the major effect in SHXXT (Cheng et al., 2008). EGb 761, a well-defined extract from Ginkgo biloba, has been widely used in patients with cerebral disorders. Transcriptomic analysis shows that EGb 761 alters unique pathways and regulates the expressions of some specific neuronal receptor genes exclusively in frontal cortex (Su et al., 2009). Vanillin is one of the most widely used flavor compounds in food and personal products. Microarray data show that vanillin exhibits the anticancer potential by the regulation of cell cycle and apoptosis. Moreover, its regulation may involve the suppression of a central molecule, activator protein 1 (Cheng et al., 2007). Quinoclamine is a chemically synthesized naphthoquinone compound. Comprehensive evaluation of quinoclamine by transcriptomic analysis shows that quinoclamine is a novel nuclear factor-κB inhibitor with anti-cancer potential (Cheng, et al., 2009).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
In this study, we applied DNA microarray to analyze the transcriptomic patterns of top 15 mostly used Chinese herbal formulae in National Health Insurance Database in Taiwan. Our data showed that transcriptomic analysis of formulae can be used as a novel platform of translational medicine to analyze the biological events, predict the therapeutic potentials, and evaluate the safety of herbal formulae.
2. Materials and methods 2.1. Animal experiments
Mouse protocols performed in all experiments have been approved by the China Medical University Animal Ethics Committee. Fifteen herbal formulae were purchased from GMP pharmaceutical company (Sun Ten Pharmaceutical Co., Taipei, Taiwan). The phytochemical profiles of herbal formulae have been deposited in Molecular Biology Laboratory, School of Chinese Medicine, China Medical University. The composition and ethnopharmacological usage of each formula is shown in Table 1. For formula groups, three mice were administered orally with one formula (150 mg/kg), which was resuspended in phosphate-buffered saline (PBS, 137 mM NaCl, 1.4 mM KH2PO4, 4.3 mM Na2HPO4, 2.7 mM KCl, pH 7.2), for 7
consecutive days. The dose for mouse study is equivalent to the dose (9 g/adult) for human usage. For mock group, three mice were administrated orally with the same volume of PBS for 7 consecutive days. Mice were then sacrificed for RNA extraction.
2.2. Total RNA isolation
Total RNA was extracted from liver or kidney using a RNeasy Mini kit (Qiagen, Valencia, CA, USA). Total RNA was quantified using the Beckman DU800
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
spectrophotometer (Beckman Coulter, Fullerton, CA, USA). Samples with A260/A280 ratios greater than 1.8 were further evaluated using Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The RNA sample with a RNA integrity number greater than 7.0 was accepted and the RNA samples from three mice were combined into a single sample for microarray analysis.
2.3. Microarray and pathway analysis
Microarray analysis was performed as described previously (Hsiang et al., 2009). Briefly, fluorescent amplified RNA targets were hybridized to the Mouse Whole Genome OneArrayTM (Phalanx Biotech Group, Hsinchu, Taiwan), and the fluorescent signals on the array were scanned by an Axon 4000 scanner (Molecular Devices, Sunnyvale, CA, USA). Number of replicates was three. The fluorescent intensity of each spot was analyzed by genepix 4.1 software (Molecular Devices, Sunnyvale, CA, USA). The signal intensity of each spot was corrected by subtracting background signals in surroundings. We filtered out spots that signal-to-noise ratio was less than 1 or control probes. Spots that passed these criteria were normalized by the limma package of the R program (Smyth, 2005). Normalized data were analyzed using the “geneSetTest” function implemented in the “limma” package of R program to detect groups of up-regulated genes in biological pathways. This function computes a p-value to test the hypothesis that the selected genes tend to be up-regulated. Then, we defined the score of each pathway in formula treatments as follows: score = -log (p) if p-value ≦ 0.5 or score = log (2(1-p)) if p-value > 0.5. Three-hundred and fifty-two
pathways, including Kyoto Encyclopedia of Genes and Genomes pathways (http://www.genome.jp/kegg/pathway.html) and PathArtTM pathways (http://www.jubilantbiosys.com/pathart.html), were extracted from ArrayTrack
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
(http://www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/) and used in this analysis. Finally, the scores of pathways were displayed using TIGR Multiexperiment Viewer (http://www.tm4.org/index.html) (Eisen et al., 1998).
2.4. Connection analysis of formulae-altered genes and diseases-altered genes
To connect the gene expression profiles of herbal formulae with those of diseases, we built the diseases-genes gene sets from the genetic association database (Becker et al., 2004) according to Medical Subject Headings (MeSH) terms (http://www.nlm.nih.gov/mesh/meshhome.html). There are 735 MeSH disease terms used in this analysis. We performed the “geneSetTest” to detect groups of differentially-expressed genes in MeSH disease terms. This function computes a p-value to test the hypothesis that the selected genes tend to be differentially expressed. Then, we calculated the score of each MeSH disease term in formula treatments as the negative logarithm of its p-value computed by “geneSetTest” function and ranked the MeSH disease terms in descending of MeSH disease term scores in each formula treatment. We calculated the average rank for each MeSH disease term in 15 formula treatments and the weighted MeSH disease term scores in each formula treatment were calculated by multiplying the original MeSH disease term scores with a ratio of the rank of MeSH disease terms over the average rank of those MeSH disease terms.
2.5. Connectivity Map analysis of formulae-altered genes and small molecules-altered genes
To connect the expression signatures of formulae-regulated genes with those of drugs-regulated genes, we analyzed gene expression profiles by the Connectivity Map
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
(Lamb et al., 2006; Lamb, 2007). We first converted the symbols of top 500 up-regulated and down-regulated genes in each formula group into Affymetrix ID according to U133A probe set. Then, we uploaded and analyzed these gene lists on the Connectivity Map (http://www.broad.mit.edu/cmap/). Using Connectivity Map, an imported query was compared with predefined signatures of therapeutic compounds and ranked according to a connectivity score, representing relative similarity to the imported gene list.
2.6. Analysis of expression levels of genes associated drug metabolism and toxicity The expression levels of phase I drug metabolism genes, including alcohol dehydrogenases, aldehyde dehydrogenases and cytochrome P450 families genes, and phase II drug metabolism genes, including glutathione S-transferases (GSTs), sulfotransferase families and UDP glycosyltransferase (UGT) families genes, in formula-treated kidney were analyzed. Furthermore, we compared the alterations in gene expression resulting from formula treatments and chemical exposures. The transcriptional profiles of 22 chemicals and 223 treatment conditions built in Environment, Drugs and Gene Expression (EDGE) website (http://edge.oncology.wisc.edu/edge.php) were used in this analysis (Hayes et al., 2005b). The comparison were performed by hierarchical clustering analysis and visualized by TIGR Multiexperiment Viewer.
3. Results
3.1. Pathway analysis of formulae-regulated gene expression profiles in livers
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
treatments by transcriptomic analysis. The “geneSetTest” function was used to test which biological pathways were transcriptionally regulated by formulae in livers. Scores of pathways calculated using aforementioned method were then visualized by TIGR Multiexperiment Viewer. As shown in Fig. 1, >90% biological pathways were regulated by formulae. The scores of metabolism-associated pathways were positive in most formula treatments, indicating that most formulae could upregulate the metabolism processes. Additionally, these formulae shared several pathways in common in livers. For examples, low-density lipoprotein signaling pathway involved in atherosclerosis, 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) degradation involved in biosynthesis of secondary metabolites, and serum response factor-mediated pathway involved in cell adhesion were negatively regulated by all formulae. Regulatory pathways, including antigen processing and presentation, complement and coagulation cascades, limonene and pinene degradation, metabolism of xenobiotics by cytochrome p450 and type I diabetes mellitus, were positively regulated by all formulae. Insulin-like growth factor (IGF) signaling pathway, Gas6 signaling pathway, and prothymosin signaling pathway, which are involved in cell cycle, and retinoic acid signaling pathway and Wnt signaling pathway, which are involved in neurogenesis, were upregulated by all formulae. Moreover, metabolic pathways, including glutathione metabolism, oxidative phosphorylation and tyrosine metabolism, were also upregulated by all formulae. These findings suggested that these formulae might alter similar biological pathways in livers.
3.2. Gene expression connection between formula treatments and disease states
We next tested groups of regulated genes in MeSH disease terms by the “geneSetTest”, and the weighted MeSH disease term scores of each formula treatment
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
were calculated to interpret whether the mice treated with these formulae were related to disease states. Among 735 tested MeSH disease terms, top 30 disease terms were selected, and their weighted MeSH disease term scores were visualized by TIGR Multiexperiment Viewer. As shown in Fig. 2, half of diseases belonged to metabolic and cardiovascular diseases, such as diabetes, myocardial infarction, hyperlipoproteinemia, thrombosis, hypolipoproteinemia, diabetic angiopathies, hypertriglyceridemia, hyperlipidemia, coronary restenosis, arteriosclerosis, and carotid artery diseases. Additionally, gene expression signatures of neuroskeletal diseases, such as Parkinson disease, apnea, dyskinesias, eclampsia, and seizures, were related to formula treatments. These findings suggested that mice responsive to formula treatments might be related to disease states, especially metabolic and cardiovascular diseases.
3.3. Gene expression connection between formula treatments and drugs
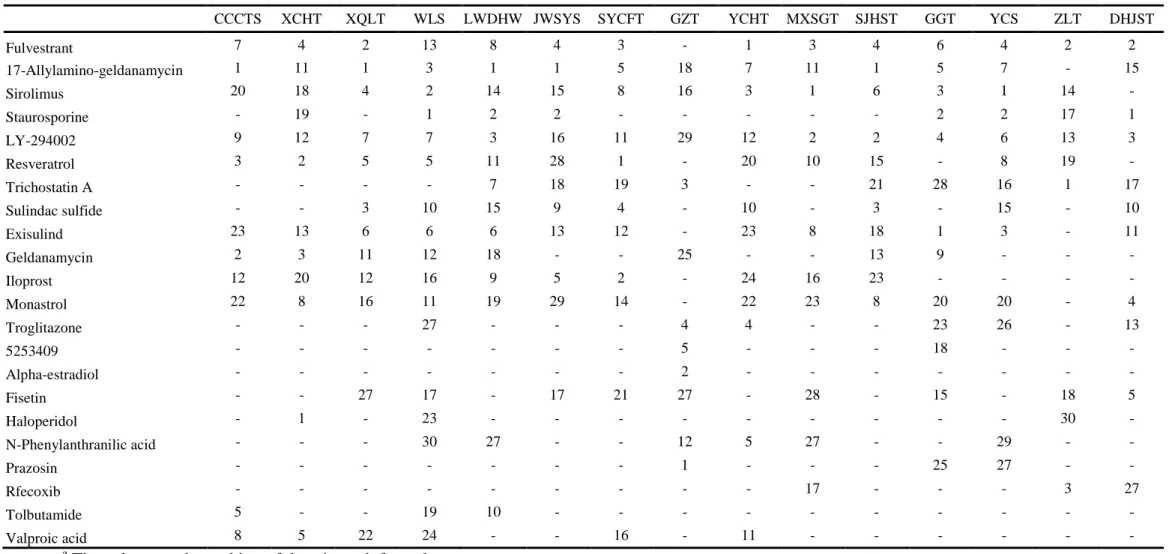
To further interpret whether the formula treatments in livers were similar to those induced by drugs, we connected the gene expression signatures by Connectivity Map. The top 500 up- and down-regulated genes were selected and analyzed to obtain the connectivity score. Top five drugs that exhibited highly connectivity scores in each formula were selected and further ranked in the order of the gene expression similarities with 15 formulae. As shown in Table 2, six compounds, including fulvestrant, 17-allylamino-geldanamycin, sirolimus, staurosporine, LY-294002 and resveratrol, were ranked top five drugs in at least five formulae. Fulvestrant, 17-allylamino-geldanamycin, and staurosporine have been known to exhibit the anti-cancer effects (Chia and Gradishar, 2008; Sharp and Workman, 2006; Stepczynska et al., 2001). Sirolimus is an immunosuppressant drug and resveratrol
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
exhibits a number of beneficial health effects, such as anti-aging, anti-inflammatory, and antioxdiative effects (Baur and Sinclair, 2006; Morelon et al., 2001). These findings suggested that mice responsive to formula treatments might be similar to drugs, which exhibit anti-cancer, anti-inflammatory, and antioxidative effects.
3.4. Formulae affected the expression levels of several genes involved in drug metabolism
To evaluate whether formula treatments affected drug metabolism in liver, we analyzed the expression levels of genes encoding phase I and II drug metabolism enzymes. Most of genes encoding cytochrome p450 family 4, GSTs, and UGTs, were significantly altered by formulae (Fig. 3). ALDH6A1, CYP24A1, CYP2B13, CYP2C38, SFT1B1, and UGT2A1 genes were down-regulated by most formulae, while UGT1A1 gene was up-regulated by most formulae. Among 15 formulae, Chuan-Chiong-Chaa-Tyau-Saan (CCCTS) upregulated the expression levels of almost all genes involved in drug metabolism. These findings suggested that most formulae altered the expression levels of CYP4, GSTs, and UGT genes. Moreover, CCCTS differed from other formulae in upregulating most genes involved in drug metabolism.
3.5. Analysis of nephrotoxicities of formulae
To analyze the nephrotoxicities of 15 formulae in kidneys, we compared the gene expression profiles of 223 chemical treatments with those of 15 formula treatments. One thousand five hundred and eighteen genes were selected from EDGE websites, and the comparison was performed by hierarchical clustering analysis. As shown in Fig. 4, the distances among formula treatments, except CCCTS, were close to each other and very far from most chemical treatments. Furthermore, the gene expression
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
patterns between chemical and formula treatments were different. Therefore, these findings suggested that these formulae might not induce nephrotoxicities in mice.
4. Discussion
In this study, we analyzed the gene expression profiles of 15 Chinese herbal formulae in mice by transcriptomic tools. The gene expression signatures were further applied to analyze the biological events induced by formulae, to predict the therapeutic potentials of formulae, and to evaluate the safety of formulae. By pathway analysis, we found that more than 90% of biological pathways were regulated by formulae. In addition to the metabolic pathways, formulae upregulated several regulatory pathways, such as antigen processing and presentation and IGF signaling pathways. Major histocompatibility complex class I (MHC-I) genes, such as H2-Q1, H2-Q2, H2-Q5, H2-Q6, H2-Q7, H2-Q8, H2-K1 and H2-T23 genes, in the antigen processing and presentation pathway were upregulated by all formulae in the livers. MHC-I antigen presentation pathway is active in most cell types to present peptides which are synthesized in the cells on the cell surface. MHC-I antigens play critical roles in the interaction between tumors and immune system by presenting tumor-associated peptides to cytotoxic T cells and regulating the cytotoxic activity of natural killer cells. Additionally, MHC-I antigen presents viral peptides to cytotoxic T cells, evoking the protective immunity to viral infection (Purcell and Elliott, 2008). It has been shown that Xiao-Qing-Long-Tang inhibits influenza virus and ganciclovir-resistant human cytomegalovirus through stimulation of mucosal immune system and induction of interferon-β, respectively (Murayama et al., 2006; Nagai and Yamada, 1998). Moreover, Ge-Gen-Tang suppresses the replication of influenza
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
viruses by increasing body temperature, enhancing the phagocytic activity of macrophages, and increasing interleukin-12 level (Kurokawa et al., 2002; Muraoka et al., 2004). All formulae upregulated the MHC-I antigen presentation pathway, suggesting that these formulae might exhibit anti-viral and anti-cancer effects through MHC-I pathway.
By comparing the similarities of gene expression signatures between formulae and disease states, we found that mice responsive to formula treatments might be related to metabolic, cardiovascular, neuroskeletal, and hepatic diseases, such as diabetes, myocardial infarction, arteriosclerosis, Parkinson disease, liver cirrhosis, and chronic hepatitis C virus infection. Indeed, Liow-Wey-Diyh-Huang-Wan has been used for the treatment of diabetes and anti-aging for a long time (ChPC, 2000). Xiao-Chia-Hu-Tang and Yin-Chen-Hao-Tang have been used to treat liver diseases, such as hepatitis and liver cirrhosis. Moreover, administration of Jia-Wey-Shiau-Yau-Saan is effective against the tremor associated with Parkinsonism (Ishikawa et al., 2000). Previous studies indicate that IGF-1 is associated with diabetes-associated, cardiovascular, and hepatic diseases. It has been shown that levels of IGF-1 in the blood are related to the risks of arteriosclerosis, type II diabetes, and myocardial infarction (Sievers et al., 2008). Additionally, increasing the level of IGF has been shown to suppress liver cirrhosis and to exhibit hepatoprotective effects (Tutau et al., 2008). Our findings showed that all formulae positively regulated the IGF signaling pathways and upregulated the expression of IGF-1 gene. These data suggested that these formulae might be used for the metabolic, cardiovascular, neuroskeletal, and hepatic diseases through the regulation of IGF signaling pathway.
We further linked the formulae-altered genes associated with drugs or compounds by Connectivity Map. Connectivity Map is a method that compares lists of
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
differential expressed genes to a library of experiments assessing the effects of small molecules and genetic events on gene expression (Lamb et al., 2006; Lamb, 2007). Connectivity Map finds connections among molecules sharing similar mechanisms of action. By connecting the gene expression signatures of formulae with those of drugs, we found that gene expression profiles induced by formulae were similar to those induced by anti-cancer, anti-inflammatory, anti-oxidative, and anti-diabetic drugs. For examples, fulvestrant is used for the treatment of hormone receptor-positive metastatic breast cancer in postmenopausal women (Chia and Gradishar, 2008). 17-Allylamino-geldanamycin that binds heat shock protein 90 has shown a promising anti-tumor activity in preclinical studies (Sharp and Workman, 2006). Staurosporine is a potent protein kinase C inhibitor and has the potential for anti-cancer (Stepczynska et al., 2001). Sirolimus is an immunosuppressant drug that blocks the signal transduction pathway required for cytokine-stimulated T cells replication and has been used to prevent rejection in organ transplantation and certain autoimmune disorders (Morelon et al., 2001). Resveratrol exhibits a number of beneficial effects, such as anti-cancer, anti-aging, neuroprotective, antioxidant, and anti-angiogenic effects (Baur and Sinclair, 2006). Troglitazone is an insulin sensitizer and a peroxisome proliferator-activated receptor γ agonist and has been used for diabetes (Ghanim et al., 2001). Based on the similarity of gene expression profile, we proposed that 15 formulae might display the anti-cancer, anti-inflammatory, antioxidative, and anti-diabetic effects in mice.
The expression levels of genes encoding phase I and phase II drug metabolism enzymes in formulae-treated mice were analyzed. Most of genes encoding cytochrome p450, GSTs, and UGTs were significantly altered by formulae. Cytochromes p450 are external monooxygenases that catalyze the incorporation of a
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
single atom of molecular oxygen into a substrate with the concomitant reduction of the other atom to water. Cytochrome p450 is involved in the biotransformation of drugs, the bioconversion of xenobiotics, the metabolism of chemical carcinogens, and the degradation of herbicides and insecticides (Bernhardt, 2006). Many chemical compounds which can induce or inhibit the cytochrome p450 enzyme function can also alter the metabolism of themselves or other compounds (Elbarbry et al., 2008). GSTs are considered to contribute to the phase II biotransformation of xenobiotics. GSTs conjugate xenobiotics with reduced glutathione to facilitate the solubility of xenobiotics in the aqueous cellular and extracellular environments and therefore enhance the excretion of xenobiotics. This activity is useful for the detoxification of endogenous compounds, such as peroxidised lipids (Hayes et al., 2005a; Margis et al., 2008). UGT superfamily is comprised of 2 families (UGT1 and UGT2) and 3 subfamilies (UGT1A, UGT2A, and UGT2B). A glucuronidation reaction catalyzed by UGT enzymes makes the drug molecule more hydrophilic and thus the drug molecule tends to be excreted easily (Ouzzine et al., 2003). Glucuronidation usually abolishes the pharmacological activity in most cases. The upregulation of UGTs by formulae might increase UGT-mediated glucuronidation and improve the drug excretion. Among 15 formulae, CCCTS differed from others in upregulating almost all genes involved in drug metabolism. CCCTS has been used for the treatment of common cold in traditional Chinese medicine. CCCTS consists of eight herbs, including Glycyrrhiza uralensis, Ligusticum chuanxiong, Notopterygium incisium, Angelica dahurica, Mentha haplocalyx, Asarum sieboldii, Schizonepeta multifida and Saposhnikovia divaricata, and five out of eight herbs have been shown to alter drug metabolism. For examples, falcarindiol from Notopterygium incisium induces phase II drug-metabolizing enzymes (Ohnuma et al., 2009). Glycyrrhiza uralensis,
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Ligusticum chuanxiong, Notopterygium incisium, Angelica dahurica, and Saposhnikovia divaricata modulate cytochrome p450 activity and participate in the interaction with conventional drugs (Ioannides et al., 2002; Tang et al., 2006). Therefore, our findings suggested that caution should be paid to possible drug interactions of these formulae, especially CCCTS.
The nephrotoxicities of formulae were further evaluated by comparing the gene expression profiles between formulae and other chemicals established in EDGE. The pattern of transcriptional activity is a highly sensitive indicator of chemical exposure and can be used as diagnostic fingerprint to predict toxicity and/or carcinogenicity (Thomas et al., 2001). EDGE is a prototype resource for sharing toxicogenomics information and used to develop algorithms for efficient chemical classification and hazard prediction (Hayes et al., 2005b). By comparing the gene expression signatures, we found that the distances of most formulae were close to each other and far from chemicals treatments, such 2,3,7,8-tetrachlorodibenzo-p-dioxin, phenylhydrazine, lipopolysaccharide, and phenobarbital . These data suggested that the nephrotoxicities of these formulae might be ignored. However, among 15 formulae, the distance of CCCTS was far from other formulae. The aristolochic acid I, which is a known nephrotoxin, is found in Asarum sieboldii, might explain why CCCTS displayed the different transcriptomic pattern with others (Jong et al., 2003).
5. Conclusion
In conclusion, this report applied transcriptomic tools as a novel platform of translational medicine for Chinese herbal medicine. The usefulness of this platform was illustrated by analyzing the biological events induced by formulae, predicting the
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
therapeutic potential of formulae, and evaluating the safety of formulae. This platform will be used not only for the understanding of therapeutic mechanisms involving Chinese herbal medicine and gene interactions, but also for the new theories in drug discovery.
Acknowledgements
This work was supported by grants from National Research Program for Genomic Medicine, National Science Council, and Committee on Chinese Medicine and Pharmacy at Department of Health (CCMP 96-RD-201 and CCMP 97-RD-201), Taiwan.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 References
Altenhein, B., Becker, A., Busold, C., Beckmann, B., Hoheisel, J.D., Technau, G.M., 2006. Expression profiling of glial genes during Drosophila embryogenesis. Developmental Biology 296, 545-560.
Baur, J.A., Sinclair, D.A., 2006. Therapeutic potential of resveratrol: the in vivo evidence. Nature Reviews. Drug Discovery 5, 493-506.
Becker, K.G., Barnes, K.C., Bright, T.J., Wang, S.A., 2004. The genetic association database. Nature Genetics 36, 431-432.
Bernhardt, R., 2006. Cytochromes P450 as versatile biocatalysts. Journal of Biotechnology 124, 128-145.
Bild, A.H., Yao, G., Chang, J.T., Wang, Q., Potti, A., Chasse, D., Joshi, M.B., Harpole, D., Lancaster, J.M., Berchuck, A., Olson, J.A. Jr, Marks, J.R., Dressman, H.K., West, M., Nevins, J.R., 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353-357.
Bonham, M., Arnold, H., Montgomery, B., Nelson, P.S., 2002. Molecular effects of the herbal compound PC-SPES: identification of activity pathways in prostate carcinoma. Cancer Research 62, 3920-3924.
Cheng, W.Y., Lien, J.C., Hsiang, C.Y., Wu, S.L., Li, C.C., Lo, H.Y., Chen, J.C., Chiang, S.Y., Liang, J.A., Ho, T.Y., 2009. Comprehensive evaluation of a novel nuclear factor-κB inhibitor, quinoclamine, by transcriptomic analysis. British Journal of Pharmacology 157, 746-756.
Cheng, W.Y., Hsiang, C.Y., Bau, D.T., Chen, J.C., Shen, W.S., Li, C.C., Lo, H.Y., Wu, S.L., Chiang, S.Y., Ho, T.Y., 2007. Microarray analysis of vanillin-regulated gene expression profile in human hepatocarcinoma cells. Pharmacological Research 56, 474-482.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Cheng, W.Y., Wu, S.L., Hsiang, C.Y., Li, C.C., Lai, T.Y., Lo, H.Y., Shen, W.S., Lee, C.H., Chen, J.C., Wu, H.C., Ho, T.Y., 2008. Relationship between San-Huang-Xie-Xin-Tang and its herbal components on the gene expression profiles in HepG2 cells. American Journal of Chinese Medicine 36, 783-797. Chia S, Gradishar W., 2008. Fulvestrant: expanding the endocrine treatment options
for patients with hormone receptor-positive advanced breast cancer. Breast 17, S16-S21.
DiPaola, R.S., Zhang, H., Lambert, G.H., Meeker, R., Licitra, E., Rafi, M.M., Zhu, B.T., Spaulding, H., Goodin, S., Toledano, M.B., Hait, W.N., Gallo, M.A., 1998. Clinical and biological activity of an estrogenic herbal combination (PC-SPES) in prostate cancer. New England Journal of Medicine 339, 785-791.
Draghici, S., Khatri, P., Eklund, A.C., Szallasi, Z., 2006. Reliability and reproducibility issues in DNA microarray measurements. Trends in Genetics 22, 101-109.
Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D., 1998. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America 95, 14863-14868.
Elbarbry, F.A., Marfleet, T., Shoker, A.S., 2008. Drug-drug interactions with immunosuppressive agents: review of the in vitro functional assays and role of cytochrome P450 enzymes. Transplantation 85, 1222-1229.
Ganter, B., Tugendreich, S., Pearson, C.I., Ayanoglu, E., Baumhueter, S., Bostian, K.A., Brady, L., Browne, L.J., Calvin, J.T., Day, G.J., Breckenridge, N., Dunlea, S., Eynon, B.P., Furness, L.M., Ferng, J., Fielden, M.R., Fujimoto, S.Y., Gong, L., Hu, C., Idury, R., Judo, M.S.B., Kolaja, K.L., Lee, M.D., McSorley, C., Minor, J.M., Nair, R.V., Natsoulis, G., Nguyen, P., Nicholson, S.M., Pham, H., Roter,
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
A.H., Sun, D., Tan, S., Thode, S., Tolley, A.M., Vladimirova, A., Yang, J., Zhou, Z., Jarnagin, K., 2005. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. Journal of Biotechnology 119, 219-244.
Ghanim, H., Garg, R., Aljada, A., Mohanty, P., Kumbkarni, Y., Assian, E., Hamouda, W., Dandona, P., 2001. Suppression of nuclear factor-κB and stimulation of inhibitor κB by troglitazone: evidence for an anti-inflammatory effect and a potential antiatherosclerotic effect in the obese. Journal of Clinical Endocrinology and Metabolism 86, 1306-1312.
Gunther, E.C., Stone, D.J., Gerwien, R.W., Bento, P., Heyes, M.P., 2003. Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro. Proceedings of the National Academy of Sciences of the United States of America 100, 9608-9613.
Hayes, J.D., Flanagan, J.U., Jowsey, I.R., 2005a. Glutathione transferases. Annual Review of Pharmacology and Toxicology 45, 51-88.
Hayes, K.R., Vollrath, A.L., Zastrow, G.M., McMillan, B.J., Craven, M., Jovanovich, S., Rank, D.R., Penn, S., Walisser, J.A., Reddy, J.K., Thomas, R.S., Bradfield, C.A., 2005b. EDGE: a centralized resource for the comparison, analysis, and distribution of toxicogenomic information. Molecular Pharmacology 67: 1360-1368.
Hsiang, C.Y., Chen, Y.S., Ho, T.Y., 2009. Nuclear factor-κB bioluminescence imaging-guided transcriptomic analysis for the assessment of hoist-biomaterial interaction in vivo. Biomaterials 30, 3042-3049.
Ioannides, C., 2002. Pharmacokinetic interactions between herbal remedies and medicinal drugs. Xenobiotica 32, 451-478.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Ishikawa, T., Funahashi, T., Kudo, J., 2000. Effectiveness of the Kampo kami-shoyo-san (TJ-24) for tremor of antipsychotic-induced parkinsonism. Psychiatry and Clinical Neurosciences 54, 579-582.
Jong, T.T., Lee, M.R., Hsiao, S.S., Hsai, J.L., Wu, T.S., Chiang, S.T., Cai, S.Q., 2003. Analysis of aristolochic acid in nine sources of Xixin, a traditional Chinese medicine, by liquid chromatography/atmospheric pressure chemical ionization/tandem mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis 33, 831-837.
Kurokawa, M., Tsurita, M., Brown, J., Fukuda, Y., Shiraki, K., 2002. Effect of interleukin-12 level augmented by Kakkon-to, a herbal medicine, on the early stage of influenza infection in mice. Antiviral Research 56, 183-188.
Lamb, J., Crawford, E.D., Peck, D., Modell, J.W., Blat, I.C., Wrobel, M.J., Lerner, J., Brunet, J.P., Subramanian, A., Ross, K.N., Reich, M., Hieronymus, H., Wei, G., Armstrong, S.A., Haggarty, S.J., Clemons, P.A., Wei, R., Carr, S.A., Lander, E.S., Golub, T.R., 2006. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929-1935.
Lamb, J., 2007. The Connectivity Map: a new tool for biomedical research. Nature Review 7, 54-60.
Margis, R., Dunand, C., Teixeira, F.K., Margis-Pinheiro, M., 2008. Glutathione peroxidase family - an evolutionary overview. FEBS Journal 275, 3959-3970. Morelon, E., Mamzer-Bruneel, M.F., Peraldi, M.N., Kreis, H., 2001. Sirolimus: a new
promising immunosuppressive drug. Towards a rationale for its use in renal transplantation. Nephrology, Dialysis, Transplantation 16, 18-20.
Muraoka, K., Yoshida, S., Hasegawa, K., Nakanishi, N., Fukuzawa, I., Tomita, A., Cyong, J.C., 2004. A pharmacologic study on the mechanism of action of
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Kakkon-to: body temperature elevation and phagocytic activation of macrophages in dogs. Journal of Alternative and Complementary Medicine 10, 841-849.
Murayama, T., Yamaguchi, N., Iwamoto, K., Eizuru, Y., 2006. Inhibition of ganciclovir-resistant human cytomegalovirus replication by Kampo (Japanese herbal medicine). Antiviral Chemistry and Chemotherapy 17, 11-16.
Nagai, T., Yamada, H., 1998. In vivo anti-influenza virus activity of Kampo (Japanese herbal) medicine "sho-seiryu-to"-stimulation of mucosal immune system and effect on allergic pulmonary inflammation model mice. Immunopharmacology and Immunotoxicology 20, 267-281.
Ohnuma, T., Komatsu, T., Nakayama, S., Nishiyama, T., Ogura, K., Hiratsuka, A., 2009. Induction of antioxidant and phase 2 drug-metabolizing enzymes by falcarindiol isolated from Notopterygium incisum extract, which activates the Nrf2/ARE pathway, leads to cytoprotection against oxidative and electrophilic stress. Archives of Biochemistry and Biophysics 488, 34-41.
Ouzzine, M., Barré, L., Netter, P., Magdalou, J., Fournel-Gigleux, S., 2003. The human UDP-glucuronosyltransferases: structural aspects and drug glucuronidation. Drug Metabolism Review 35, 287-303.
Pharmacopoeia Commission of People’s Republic of China (ChPC), 2000. Pharmacopoeia of People’s Republic of China (ChP), vol. 1. Chemical Industry Press, Beijing.
Pittman, J., Huang, E., Dressman, H., Horng, C.F., Cheng, S.H., Tsou, M.H., Chen, C.M., Bild, A., Iversen, E.S., Huang, A.T., Nevins, J.R., West, M., 2004. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proceedings of the National Academy of Sciences of the United States of America 101, 8431-8436.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Purcell, A.W., Elliott, T., 2008. Molecular machinations of the MHC-I peptide loading complex. Current Opinion in Immunology 20, 75-81.
Schena, M., Shalon, D., Davis, R.W., Brown, P.O., 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467-470.
Sharp, S., Workman, P., 2006. Inhibitors of the HSP90 molecular chaperone: current status. Advances in Cancer Research 95, 323-348.
Sievers, C., Schneider, H.J., Stalla, G.K., 2008. Insulin-like growth factor-1 in plasma and brain: regulation in health and disease. Frontiers in Bioscience 13, 85-99. Smyth, G.K., 2005. Limma: linear models for microarray data. In: Gentleman, R.,
Carey, V., Dudoit, S., Irizarry, R., Huber, W. (Eds.), Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, pp. 397-420.
Stepczynska, A., Lauber, K., Engels, I.H., Janssen, O., Kabelitz, D., Wesselborg, S., Schulze-Osthoff, K., 2001. Staurosporine and conventional anticancer drugs induce overlapping, yet distinct pathways of apoptosis and caspase activation. Oncogene 20, 1193-1202.
Su, S.Y., Hsieh, C.L., Wu, S.L., Cheng, W.Y., Li, C.C., Lo, H.Y., Ho, T.Y., Hsiang, C.Y., 2009. Transcriptomic analysis of EGb 761-regulated neuroactive receptor pathway in vivo. Journal of Ethnopharmacology 123, 68-73.
Tang, J.C., Zhang, J.N., Wu, Y.T., Li, Z.X., 2006. Effect of the water extract and ethanol extract from traditional Chinese medicines Angelica sinensis (Oliv.) Diels, Ligusticum chuanxiong Hort. and Rheum palmatum L. on rat liver cytochrome P450 activity. Phytotherapy Research 20, 1046-1051.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
Thomas, R.S., Rank, D.R., Penn, S.G., Zastrow, G.M., Hayes, K.R., Pande, K., Glover, E., Silander, T., Craven, M.W., Reddy, J.K., Jovanovich, S.B., Bradfield, C.A., 2001. Identification of toxicologically predictive gene sets using cDNA microarrays. Molecular Pharmacology 60, 1189-1194.
Tutau, F., Rodríguez-Ortigosa, C., Puche, J.E., Juanarena, N., Monreal, I., García Fernández, M., Clavijo, E., Castilla, A., Castilla-Cortázar, I., 2009. Enhanced actions of insulin-like growth factor-I and interferon-alpha co-administration in experimental cirrhosis. Liver International 29: 37-46.
Whitfield, M.L., George, L.K., Grant, G.D., Perou, C.M., 2006. Common markers of proliferation. Nature Review of Cancer 6, 99-106.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 Figure captions
Fig. 1. Pathway analysis of gene expression profiles in livers responsive to formula treatments. Normalized data were analyzed using the “geneSetTest” to compute a p-value and to detect groups of up-regulated genes in biological pathways. The score of each pathway in formula treatments was defined as follows: score = -log (p) if p-value ≦ 0.5 or score = log (2(1-p)) if p-value > 0.5. The scores of pathways were
then displayed by TIGR Multiexperiment Viewer. Scores are color-coded according to the legend on the top.
Fig. 2. Gene expression connection between formulae and disease states. Normalized data were analyzed using the “geneSetTest” to compute a p-value and to detect groups of differentially-expressed genes in MeSH disease terms. The score of each MeSH disease term in formula treatments was calculated as the negative logarithm of its p-value computed by “geneSetTest” function. The weighted MeSH disease term scores in each formula treatment were calculated by multiplying the original MeSH disease term scores with a ratio of the rank of MeSH disease terms over the average rank of those MeSH disease terms. The weighted MeSH disease term scores were displayed by TIGR Multiexperiment Viewer. Scores are color-coded according to the legend on the top.
Fig. 3. Expression levels of genes involved in drug metabolism. The expression levels of alcohol dehydrogenase genes, aldehyde dehydrogenase genes, cytochrome P450 family genes, GST genes, sulfotransferase genes, and UGT family genes in formula-treated kidney were selected. Normalized log2 expression values of these
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59
according to the legend on the top.
Fig. 4. Hierarchical clustering analysis of gene expression profiles altered by chemicals and formulae. The transcriptional profiles of 22 chemicals and 223 treatment conditions built in EDGE website were used in this analysis. Normalized log2 expression values are color-coded according to the legend on the top. DEHP,
Di-(2-ethylhexyl)phthalate; LPS, lipopolysaccharide; IL1B, interleukin-1β; PHB, phenobarbital; PHNZ, phenylhydrazine; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43
Table 1 Herbal formulae, their constituents, and ethnopharmacological usage.
Herbal formulae Constituentsa Ethnopharmacological usage
Gui-Zhi-Tang (GZT) Cinnamomum cassiab (6), Paeonia lactiflorac (6), Zingiber officinaled (6), Zizphus jujubee (5), Glycyrrhiza uralensisc (4)
Respiratory diseases
Ma-Xing-Shi-Gan-Tang
(MXSGT) Ephedra sinica
f
(4), gypsum (CaSO4·2H2O) (8), Prunus armeniacag (3), Glycyrrhiza
uralensisc (2)
Respiratory diseases and asthma
Xiao-Qing-Long-Tang (XQLT) Ephedra sinicaf (4), Cinnamomon cassiab (4), Glycyrrhiza uralensisc (4), Paeonia lactiflorac (4), Schisandra chinensise (1.5), Asarum sieboldiih (1.5), Pinellia ternated (4), Zingiber officinaled (4)
Respiratory diseases and asthma
Xiao-Chai-Hu-Tang (XCHT) Bupleurum chinensec (8), Scutellaria baicalensisc (3), Pinellia ternated (5), Zingiber officinaled (3), Panax ginsengc (3), Ziziphus jujubee (2), Glycyrrhiza uralensisc (3)
Liver diseases
Wu-Ling-Saan (WLS) Alisma orientalisi (7.5), Poria cocosj (4.5), Polyporus umbellatusj (4.5), Cinnamomum cassiab (3), Atractylodes macrocephalad (4.5)
Cardiovascular diseases Zhu-Ling-Tang (ZLT) Polyporus umbellatusj (1), Poria cocosj (1), Alisma orientalisi (1), talc
(Mg3Si4O10(OH)2) (1), donkey-hide gelatin (1)
Cardiovascular diseases
Ge-Gen-Tang (GGT) Pueraria lobatac (6), Ephedra sinicaf (4.5), Ramulus cinnamomib (3), Paeonia
lactiflorac (3), Zingiber officinaled (4.5), Zizphus jujubee (4), Glycyrrhiza uralensisc (3)
Fever
Yin-Chen-Hao-Tang (YCHT) Artemisia scopariaf,l (3), Gardenia jasminoidese (1), Rheum officinaled (1) Liver diseases Jia-Wey-Shiau-Yau-Saan
(JWSYS)
Angelica sinensisc (4), Atractylodes macrocephalad (4), Gardenia jasminoidese (2.5), Paeonia suffruricosac (2.5), Poria cocosj (4), Mentha haplocalyxl (2), Bupleurum
chinensec (4), Paeonia lactiflorac (4), Zingiber officinaled (4), Glycyrrhiza uralensisc (2)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 Shu-Jing-Hwo-Shiee-Tang (SJHST)
Angelica sinensisc (2), Glycyrrhiza uralensisc (1), Ligusticum chuanxiongd (1), Prunus persicag (2), Achyranthes bidentatac (2), Citrus reticulatak (2), Rehmannia glutinosad (2), Atractylodes japonicad (2), Clematis chinensisd (2), Notopterygium incisiumc,d (1), Angelica dahuricac (1), Poria cocosj (1), Saposhnikovia divaricatac (1), Zingiber
officinaled (3), Gentiana scabrac.d (1), Stephania tetrandrac (1), Paeonia lactiflorac (2.5)
Pain in muscle, joint, and lumbar
Shin-Yi-Ching-Fey-Tang (SYCFT)
Ophiopogon japonicusc (3), Glycyrrhiza uralensisc (1.5), Eriobotya japonical (3), Scutellaria baicalensisc (3), Bupleurum chinensec (3), Anemarrhena asphodeloidesd (3), Lilium brownief,l (3), gypsum (CaSO4·2H2O) (3), Magnolia liliforam (2), Cimicifuga
foetidad (1)
Respiratory diseases
Chuan-Chiong-Chaa-Tyau-Saan (CCCTS)
Glycyrrhiza uralensisc (2), Ligusticum chuanxiongd (4), Notopterygium incisiumc,d (2), Angelica dahuricac (2), Mentha haplocalyxl (8), Asarum sieboldiih (1), Schizonepeta multifidah (4), Saposhnikovia divaricatac (1.5)
Pain
Yn-Chyau-Saan (YCS) Glycine maxg (2.5), Platycodon grandiflorumc (3), Lophatherum gracilel (2), Forsythia suspensee (5), Lancer japonicam (5), Schizonepeta tenuifoliah (2), Arctium lappae (3), Glycyrrhiza uralensisc (2.5), Mentha haplocalyxl (3)
Respiratory diseases
Dwu-Hwo-Jih-Sheng-Tang (DHJST)
Angelica sinensisc (1), Eucommia ulmoidesn (1), Glycyrrhiza uralensisc (1), Ligusticum chuanxiongd (1), Panax ginsengc (1), Paeonia suffruticosac (1), Achyranthes bidentatac (1), Rehmannia glutinosad (1), Angelica pubescensc (1.5), Gentiana macrophylla c (1), Poria cocosj (1), Asarum sieboldiih (1), Saposhnikovia divaricatac (1), Cinnamomum cassian (1), Taxillus chinensisf,l (1)
Respiratory diseases and pain
Liow-Wey-Diyh-Huang-Wan (LWDHW)
Dioscorea oppositei (4), Cornus officinalise (4), Alisma plantagoi (3), Poria cocosj (3), Rehmannia glutinosad (8), Paeonia suffruticosac (3)
Diabetes and anti-aging
a
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 b
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43
Table 2 Gene expression similarities between formulae and drugs.
CCCTS XCHT XQLT WLS LWDHW JWSYS SYCFT GZT YCHT MXSGT SJHST GGT YCS ZLT DHJST
Fulvestrant 7 4 2 13 8 4 3 - 1 3 4 6 4 2 2 17-Allylamino-geldanamycin 1 11 1 3 1 1 5 18 7 11 1 5 7 - 15 Sirolimus 20 18 4 2 14 15 8 16 3 1 6 3 1 14 - Staurosporine - 19 - 1 2 2 - - - 2 2 17 1 LY-294002 9 12 7 7 3 16 11 29 12 2 2 4 6 13 3 Resveratrol 3 2 5 5 11 28 1 - 20 10 15 - 8 19 - Trichostatin A - - - - 7 18 19 3 - - 21 28 16 1 17 Sulindac sulfide - - 3 10 15 9 4 - 10 - 3 - 15 - 10 Exisulind 23 13 6 6 6 13 12 - 23 8 18 1 3 - 11 Geldanamycin 2 3 11 12 18 - - 25 - - 13 9 - - - Iloprost 12 20 12 16 9 5 2 - 24 16 23 - - - - Monastrol 22 8 16 11 19 29 14 - 22 23 8 20 20 - 4 Troglitazone - - - 27 - - - 4 4 - - 23 26 - 13 5253409 - - - 5 - - - 18 - - - Alpha-estradiol - - - 2 - - - - Fisetin - - 27 17 - 17 21 27 - 28 - 15 - 18 5 Haloperidol - 1 - 23 - - - 30 - N-Phenylanthranilic acid - - - 30 27 - - 12 5 27 - - 29 - - Prazosin - - - 1 - - - 25 27 - - Rfecoxib - - - 17 - - - 3 27 Tolbutamide 5 - - 19 10 - - - - Valproic acid 8 5 22 24 - - 16 - 11 - - - - a
3.00 -3.00
Glutathione metabolism IGF signaling pathway
LDL signaling Pathway(Atherosclerosis) 1,1,1-Trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) degradation Oxidative phosphorylation SRF-mediated pathway Gas6 signaling
Pathway(Cell-cycle) Prothymosin signaling pathway
Retinoic acid signaling Pathway(Neurogenesis) Wnt signaling pathway Tyrosine metabolism
PRL signaling pathway
Antigen processing and presentation Complement and coagulation cascades
Limonene and pinene degradation Metabolism of xenobiotics by cytochrome p450
Type I diabetes mellitus Figure 1