國 立 交 通 大 學
生物科技研究所
碩 士 論 文
克雷白氏肺炎桿菌 CG43 中參與莢膜多醣體生合成之核心蛋
白 Wza、Yor5、Yco6 和 Wzx 的功能性研究
Functional analysis of the core elements, Wza, Yor5, Yco6
and Wzx, involved in capsular polysaccharide biosynthesis
in Klebsiella pneumoniae CG43
研 究 生: 李智凱
學 號: 9328507
指導教授:彭慧玲 博士
中文摘要 克雷白氏肺炎桿菌為一株伺機性引起區內感染疾病的格蘭氏陰性菌,其外部 包覆著由多醣體所組成的厚莢膜。此一莢膜可以讓細菌逃避細胞的吞噬作用以及 避免被血清因子所毒殺。克雷白氏肺炎桿菌的莢膜多醣體被認為和大腸桿菌的第 一類莢膜多醣體之生合成與組合途徑相類似,由位於 cps 基因組中的主要因子所 調控,這些主要因子包括了 Wzi、Wza、Yor5、Yco6、Wzx 和 Wzy。本研究的 主要目標為證明其中四個主要因子-Wza、Yor5、Yco6 和 Wzx-的生物功能性。 在實驗室之前的研究中,證明了無論剔除酪胺酸去磷酸酶或是酪胺酸激酶的基 因,yor5 或 yco6 皆會明顯地減少其原本所具有的黏性和莢膜多醣體的量。在此 研究之中,我們亦構築了剔除了 wza 或 wzx 的突變株。其中剔除了位在外膜上的 多醣體輸送蛋白 wza,會使得莢膜多醣體之聚合體無法運送到細菌表面,並且進 而回饋抑制上游的生合成途徑,造成整體生成量明顯地下降。而在 wzx 突變株之 中,雖然莢膜多醣體的總量同樣下降,但是仍然有少量的聚合體存在,這部分的 多醣體合成作用可能不需要經由 Wzx 來調控。此外,剔除 yor5 和剔除 yco6 這 兩個突變株皆完全地喪失了莢膜多醣體的聚合體,而這也證明了由Yco6 和 Yor5 所調控的酪胺酸磷酸化作用對於莢膜多醣體的生成與聚合作用來說相當重要。經 由體外磷酸化試驗,我們證實了克雷白氏肺炎桿菌和大腸桿菌的蛋白質酪胺酸激 酶-Yco6 和 Wzc-都能夠將克雷白氏肺炎桿菌中的 Ugd、Gnd、ManB 和 ManC 這四個酵素磷酸化,而這些酵素和莢膜多醣體醣類核甘酸前驅物的生成相關。而 Ugd 和 Gnd 的磷酸化對於其酵素活性則有明顯的提升效果。另外,以蛋白質酪 胺酸去磷酸酶Yor5 來進行體外去磷酸化試驗則發現,Gnd 和 ManC 可以徹底地 去磷酸化,而KpUgd 則是部分被去磷酸化。相反地,ManB 則是完全不受 Yor5 影響。以上實驗結果明確地證實了這些主要因子對於克雷白氏肺炎桿菌的莢膜多 醣體之生合成與組合作用的重要性和其功能性角色。
Abstract
Klebsiella pneumoniae, an opportunistic gram-negative bacterium causing community-acquired diseases, are mostly encapsulated by a considerable thick polysaccharidic capsule acting to protect the bacteria from phagocytosis and prevent from killing by serum factors. The capsular polysaccharide (CPS) is made from a similar biosynthetic and assembly pathway to that of Escherichia coli group 1 CPS. The core elements encoded in cps gene cluster include Wzi, Wza, Yor5, Yco6, Wzx and Wzy. Our previous study has demonstrated that deletion of either yor5, encoding a protein-tyrosine phosphatase, or yco6, coding for a protein-tyrosine kinase, in K. pneumoniae CG43 reduced significantly the bacterial mucoidy and CPS content. In this study, both wza- and wzx- mutants were generated and the deletion effects were
analyzed and compared. Deletion of wza, encoding an outer membrane polysaccharide export protein, or wzx, coding for a flippase, appeared to affect also the mucoidy and reduce the CPS content. Polymeric CPS were blocked in periplasma in wza- mutants and resulted in feedback inhibition of total CPS synthesis. In contrast,
some polymeric CPS was observed on cell surface in the wzx- mutant suggesting an
alternative translocation system for oligosaccharides through the cytoplasmic membrane. Nonetheless, to demonstrate further a role of tyrosine phosphorylation, several recombinant clones were generated and the proteins, including Yor5, Yco6, Ugd, Gnd, ManB and ManC that have been shown to be required for regulation and synthesis of CPS sugar nucleotide precursors, were purified. Via in vitro phosphorylation assay, the recombinant Yco6 appeared to be able to phosphorylate the enzymes Ugd, Gnd, ManB and ManC, and the phosphorylation increased the enzymatic activities of Ugd and Gnd. Moreover, the phosphorylated Ugd, Gnd and ManC, but not the ManB, could be dephosphorylated by the recombinant Yor5.
致謝 很快地,碩士班生涯的兩年時光就這樣過了。還記得剛進入這個實驗室的 我,仍是個對於「做研究」這門學問絲毫不懂的大學畢業生,經過了在實驗室之 中的訓練、老師的指導,以及學長姊們的幫助之下,才得以完成這個論文的內容。 在這段期間認識了很多人,學到了許多東西,接觸了很多事物,所經歷的一切一 切都要感謝身邊的大家,有你們才會有現在的我和這篇論文。首先要感謝彭老 師,帶我進入這個實驗室和細菌學這個領域,一路上給予我指導和幫助,並且提 供了一個良好的研究環境,以及相當自由的研究空間,才能夠讓我有這樣的研究 成果;感謝鄧文玲老師和邱顯泰老師,百忙之中撥空來擔任我的口試委員,並且 提供了我論文上許多相當寶貴的建議;感謝已經畢業的平輝學長,常常在工作之 餘抽空來關心我的實驗進度,且不厭其煩地回答我提出的實驗問題,讓我能夠在 一開始時可以容易地進入狀況;感謝實驗室的靖婷、盈蓉學姊和新耀、健誠學長, 在實驗上帶我操作許多實驗、提供許多想法、給予很多幫助與建議,讓我得以順 利完成實驗,在生活上陪伴我的喜怒哀樂,讓我研究之外的生活也能過的充實; 感謝和我同屆的育聖和心瑋同學,很開心能夠和你們一起度過碩士班的這兩年; 感謝實驗室的登魁、格維、朝陽這幾個學弟,已畢業的祐俊學長,還有可愛的專 題生們,在各方面的照顧;感謝其他實驗室的阿男、玉淳、琮道、媛婷、程翔等 學長姊,以及所有在這段期間認識的各位,認識你們真好;感謝我的家人,無怨 無悔地在背後默默支持我,讓我順利地完成碩士班學業。最後在這邊要特別感謝 提供了我大腸桿菌Wzc和Ugd蛋白表現載體的Christophe Grangeasse博士,讓我的 實驗能夠有所進展。
Contents Page Abstract in Chinese …………..………..………. i Abstract …………....……….... ii Acknowledgment ……… iii Content ……… iv Table content ………... vi
Figure content ……….... vii
Abbreviation ………... ix
Introduction ……… 1
Materials and methods ………... 6
Results Part 1: Biological roles of the core elements Wza, Yor5, Yco6, and Wzx encoded in the K2 cps gene cluster of Klebsiella pneumoniae CG43 ……… 13
Part 2: Autophosphorylation of protein-tyrosine kinase Yco6 regulates tyrosine phosphorylatoin of the proteins involved in capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43 ……….. 17
References ……….. 25
Table ……….. 34
Figure ………. 39
Table contents
Page Table 1. Bacterial strains used and constructed in this study ………. 34 Table 2. Plasmids used and constructed in this study ………... 35 Table 3. Primers used in this study ………. 37
Figure contents
Page
Fig. 1. The comparison of cps gene clusters ……….. 39
Fig.2. Biosynthesis pathway of lipid-linked K2 repeat units ………. 40
Fig. 3. Identification of the wza- mutation by Southern blot analysis ..…….... 41
Fig. 4. Identification of the wzx- mutation by Southern blot analysis ..……… 42
Fig. 5. Mutants grow faster than wild type strain …...……….... 43
Fig. 6. Loss of colony mucoidy-Sedimentation test …….………... 44
Fig. 7. Reduction of the glucuronic acid content ………... 45
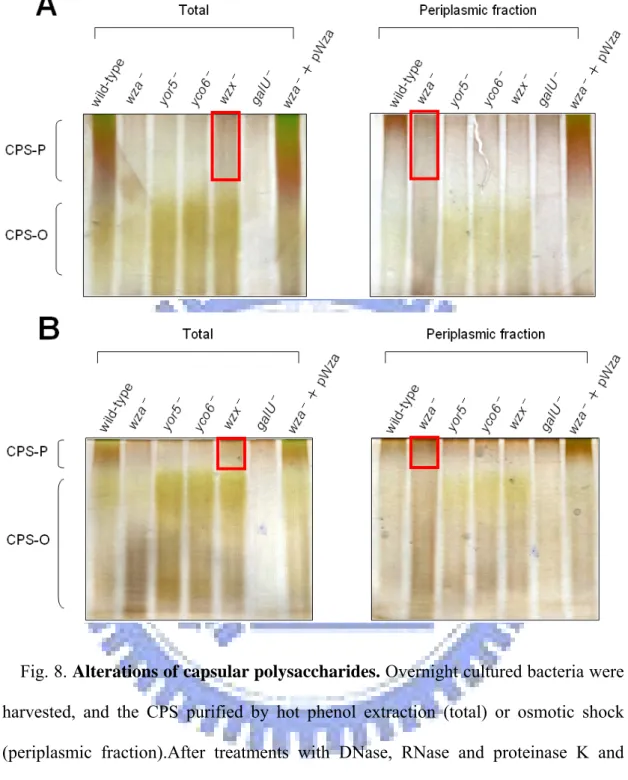
Fig. 8. Alterations of capsular polysaccharides ……….. 46
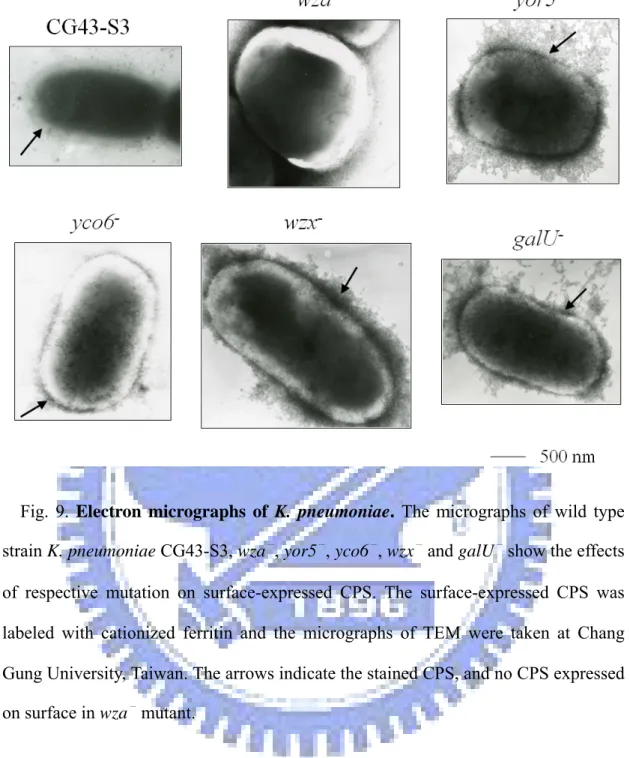
Fig. 9. Electron micrographs of K. pneumoniae ………... 47
Fig. 10. Effects of the susceptibility to polymyxin B ………. 48
Fig. 11. Biofilm formation ability ………..……… 49
Fig. 12. In vitro phosphorylation and dephosphorylation of different proteins ... 50
Fig. 13. Phosphorylation of Ugd enhances its UDP-glucose dehydrogenase activity ……..……….. 51 Fig. 14. Phosphorylation of Gnd enhances its UDP-glucose 6-phosphogluconic
dehydrogenase activity ……… 52 Fig. 15. Schematic model for functional roles of core elements involved in CPS biosynthesis in K. pneumoniae …….……….……….. 53
Appendix 1. Features of the ORFs encoded in Chedid K2 cps region from K. pneumoniae ……… 54
Abbreviation
ABC ATP-binding cassette ATP adenosine triphosphate
BCIP 5- bromo-4-chloro-3-indolyl phosphate bp base pair
CFU colony forming unit(s)
CIAP calf intestine alkaline phosphatase CPS capsular polysaccharide
DOC deoxycholic acid DNA deoxyribonucleic acid
EDTA ethylenediamine-tetraacetic acid EPS exopolysaccharide GDP guanosine 5'-diphosphate IPTG isopropyl-1-thio-β-D-galactopyranoside kb kilobase(s) kDa kiloDalton(s) LB Luria-Bertani mRNA messenger RNA
NAD nicotinamide adenine dinucleotide
NADH nicotinamide adenine dinucleotide (reduced form) NBT nitro blue tetrazolium chloride
ORF open reading frame
PAGE polyacrylamide gel electrophoresis PBS phosphate buffered saline
PCR polymerase chain reaction PPP pentose phosphate pathway PTK protein-tyrosine kinase
PTP protein-tyrosine phosphatase PVDF polyvinylidene difluoride RNA ribonucleic acid
rpm revolutions per minute RT-PCR reverse transcriptase-PCR SDS sodium dodecyl sulfate
TEM transmission electron microscopy UDP uridine 5'-diphosphate
Introduction
Klebsiella pneumoniae, a member of the Enterobacteriaceae, is a rod-shaped and opportunistic gram-negative bacterium. It can cause community-acquired diseases including pneumonia, bacteremia, septicemia, urinary tract and respiratory infections, occurring particularly in immunocompromised patients (Podschun and Ullmann, 1998). Most Klebsiella strains are encapsulated by a polysaccharidic capsule of considerable thickness responsible for the glistening and mucoid colonies on agar plates. The highly virulent clinical isolates are often carry heavy capsules as an important virulence factor to protect the bacteria from phagocytosis and prevent from killing by serum factors (Simoons-Smit et al., 1986).
The capsular serotypes of Klebsiella strains have been classified as 77 K antigens (Ørskov and Ørskov, 1984). On the basis of mouse lethality assay, the strains belonging to serotypes K1 and K2 were found to be the most virulent (Simoons-Smit et al., 1986). K. pneumoniae CG43, showing a strong virulence to Balb/c mice with 50% lethal dose of 10 CFU, is a highly encapsulated clinical isolate of K2 serotype (Chang et al., 1996). The structure of K. pneumoniae K2 capsular polysaccharides (CPS) has been determined as [→)4-Glc-(1→3)-α-Glc-(1→4)-β-Man-(3←1)- α-GlcA)-(1→]n (Wacharotayankun et al., 1992), which is made from a similar
biosynthetic pathway to that of Escherichia coli group 1 CPS (Whitfield and Roberts, 1999).
On the basis of genetic and biosynthetic criteria, CPS in E. coli has been classified into four different groups. These include two fundamentally different polymerization pathways. Group 1 and 4 CPS follow a Wzx/Wzy-dependent polymerization pathway, in which undecaprenol diphosphate (und-PP)-linked repeat units are formed at the
inner leaflet of the inner membrane. These und-PP-linked intermediates are flipped across the membrane by a process involving Wzx protein and then polymerized in a reaction requiring Wzy. Group 2 and 3 CPS are assembled by an ABC (ATP-binding cassette)-transporter-dependent pathway, in which und-PPlinked polymers are formed at the inner leaflet of the membrane by processive glycosyltransfer to the nonreducing terminus of the und-PP-linked intermediate. The nascent polymer is then exported across the inner membrane by an ABC transporter (Whitfield, 2006).
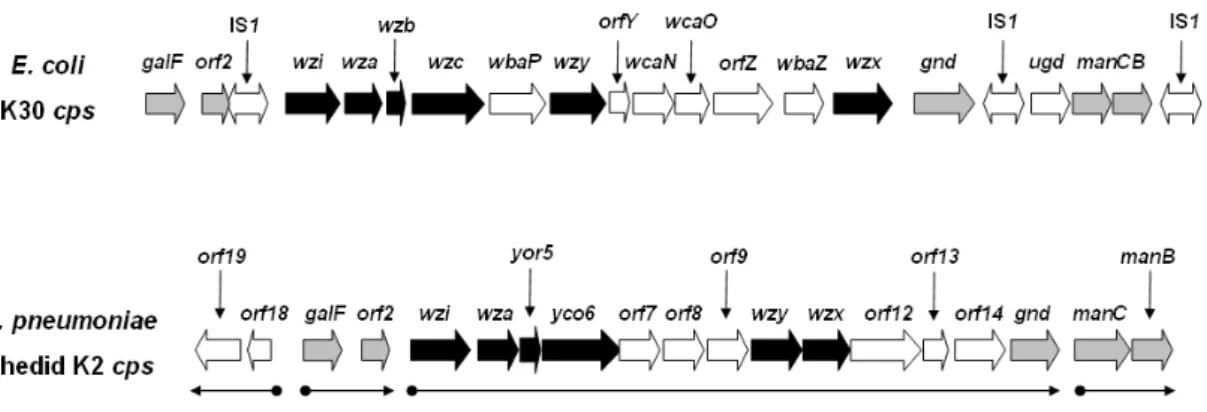
In terms of primary structure and mechanism of synthesis, the Klebsiella capsules resemble those of E. coli group 1 and, in some cases, same polymer structures are found. In E. coli, the genes for group 1 CPS synthesis and translocation were characterized as the core elements including wzi, which is involved in surface assembly of CPS and acts as an anchor of CPS; wza, which encodes a polysaccharide export protein; wzb, encoding a low-molecular-weight phosphatase; wzc, encoding a protein-tyrosine autokinase; wzx, encoding a flippase responsible for oligosaccharides translocation; and wzy, encoding putative polymerase (Drummelsmith and Whitfield, 1999).
The gene cluster cps (capsular polysaccharide synthesis) region that is responsible for K2 CPS synthesis of K. pneumoniae Chedid has been determined, which contains a total of 19 open reading frames (ORFs) organized into 3 transcriptional units (Arakawa et al., 1995). Among these genes, orf3 to orf6, a highly conserved four-gene-block, are counterparts of E. coli wzi-wza-wzb-wzc (Rahn et al., 1999). Wzi, an orf3 encoding protein, is an outer membrane protein and wzi mutant showed a significant reduction in cell-bound CPS polymer with a corresponding increase in cell-free material. This proposed that Wzi plays a late role in capsule assembly, perhaps in the process that links high-molecular-weight capsule to the cell surface
(Alvarez et al., 2000; Rahn et al., 2003). orf4 encodes for a Wza homolog, orf5 for Yor5, and orf6 for Yco6, respectively. Enzymatic activities of the two proteins, Yor5 and Yco6, which are respectively Wzb and Wzc homolog, have been demonstrated. Similar to the E. coli Wzb and Wzc, Yor5 is a protein-tyrosine phosphatase (PTP) and Yco6 is a protein-tyrosine kinase (PTK) (Preneta et al., 2002). The orf10 encodes a membrane protein containing multi-transmembrane domains, which is likely the homolog of E. coli Wzy, and the orf11 is likely the wzx homolog, which encoding a flippase involved in the export of antigen out to cell surface (Paulsen et al., 1997).
Among these core elements, PTK and PTP are assumed to regulate the tyrosine phosphorylation in the cells. Phosphorylation is a covalent modification that proteins frequently undergo in a post-translational process. Protein phosphorylation on tyrosine is one type of phosphorylation that was long considered to be specific in eukaryotes, which has been shown to be critical in the regulation of a series of fundamental biological functions, including signal transduction, growth control, and malignant transformation (Hunter, 1995). Comparing to the eukaryotic systems, less is known about the physiological roles of protein-tyrosine phosphorylation in bacteria. The first indication of a PTK activity in bacteria has been reported in E. coli (Manai and Cozzone, 1983). Nevertheless, the first bacterial PTK, Ptk, an 81 kDa protein located in the inner-membrane fraction, was identified in Acinetobacter johnsonii, which is able to autophosphorylate on multiple tyrosine residues at the expense of ATP (Duclos et al., 1996; Grangeasse et al., 1997). In recent years, several homologous PTKs were reported in other Gram-negative and Gram-positive bacteria, including AmsA from Erwinia amylovora (Bugert and Geider, 1997), Yco6 from K. pneumoniae (Preneta et al., 2002), EpsB from Pseudomonas solanacearum (Huang and Schell, 1995), ExoP from Sinorhizobium meliloti (Niemeyer and Becker, 2001),
CapB from Staphylococcus aureus (Lin et al., 1994), Waap from Pseudomonas aeruginosa (Zhao and Lam, 2002), and CpsC from Streptococcus pneumoniae (Morona et al., 2000). In addition, bacteria often contain PTPs simultaneously, which antagonize the activity of PTKs. The genes encoding PTKs and PTPs are generally located in the gene clusters involved in biosynthesis and transport of CPS or exopolysaccharides (EPS) (Vincent et al., 1999).
Together, PTKs and PTPs play important roles in the production and processing of CPS and EPS (Nakar and Gutnick, 2003; Stevenson et al., 1996). In E. coli, knock-out of PTK or PTP corresponding genes, wzc or wzb, apparently reduced the production of CPS and colanic acid (Vincent et al., 2000). In S. pneumoniae, autophosphorylation of CpsD kinase decreased its activity which consequently lowered the production of CPS (Morona et al., 2000). The question as to how the regulation affects CPS biosynthesis via tyrosine phosphorylation remains elusive. Recently, a growing number of protein substrates of the PTKs have been reported, which include RNA polymerase heat shock sigma factors in E. coli (Klein et al., 2003), UDP-glucose dehydrogenases in E. coli and Bacillus subtilis (Grangeasse et al., 2003; Mijakovic et al., 2003), and single-stranded DNA-binding proteins in B. subtilis (Mijakovic et al., 2006). Phosphorylation of these proteins appeared to enhance their activities and hence promoted respectively the biological functions.
In the cps gene cluster of K. pneumoniae Chedid, besides the PTK and PTP encoding genes, other cps genes encoding glycosyltransferases or enzymes for producing sugar nucleotide precursors are designated to CPS synthesis include gnd, manC and manB. Gnd is 6-phosphogluconic dehydrogenase involving in pentose phosphate pathway (PPP), which catalyzes the oxidative decarboxylation of 6-phospho-gluconate to ribulose-5-phosphate. In E. coli, PPP is a major route for
intermediate carbohydrate metabolism and plays various roles, including breakdown of carbon sources, generation of reducing power (NADPH) and essential metabolites for biosynthesis, and formation of the components of polysaccharide layer of the cell. Furthermore, Gnd is involved in the metabolism of glucose and gluconate and generally exists in enterobacteria (Sprenger, 1995). GDP-mannose pyrophosphorylase and phosphomannomutase encoded respectively by manC and manB are required for the formation of GDP-mannose, a precursor required for the formation of both CPS and LPS (Jayaratne et al., 1994). As shown in Fig. 2, ManC, ManB, and Gnd proteins are apparently involved in the biosynthesis of CPS repeat units.
In this work, the goal is to demonstrate the biological roles of Wza, Yor5, Yco6, and Wzx by generation of the specific gene deletion mutants and analyses of the mutant phenotypes. In addition, I examined the possibility whether Gnd, ManC, ManB, and Ugd of K. pneumoniae CG43 are the substrates of the PTK Yco6 and the PTP Yor5, and if the phosphorylation and dephosphorylation are able to modulate their enzymatic activities.
Materials and methods
Bacterial strains, plasmids, and growth conditions. The bacterial strains and
plasmids used in this study are listed in Table 1 and Table 2 respectively. The genomic DNA for PCR amplification was prepared from K. pneumoniae CG43, a liver abscess isolate of K2 serotype recovered from Chang Gung Memorial Hospital, Linkou, Taiwan. All bacteria were propagated at 37 ℃ in Luria-Bertani (LB) broth supplemented with appropriate antibiotics when needed including kanamycin (25 µg/ml), ampicillin (100 µg/ml) , tetracycline (20 µg/ml) and streptomycin (500 µg/ml). The plasmids, pQE30-41, pQE30-42 and pQE30-U, are gifts from Institut de Biologie et Chimie des Prote´ines, France (Grangeasse et al., 2003).
DNA manipulation. Plasmids were purified by using the High-Speed Plasmid
Mini kit (Geneaid, Taiwan). All restriction and DNA-modifying enzymes were used as recommended by the manufacturer (Fermentas, USA). PCR amplifications were performed with Taq DNA polymerase (MDBio, Inc, Taiwan). PCR products and DNA fragments were purified using the Gel/PCR DNA Fragments Extraction kit (Geneaid, Taiwan). The primers used in this study were synthesized by MDBio, Inc, Taiwan. Transformation of E. coli cells was performed by following the method of Dower (Dower et al., 1988).
Construction of the specific gene-deletion mutants and complements. K.
pneumoniae CG43 mutants disrupted specifically at yor5, yco6, wza, or wzx genes were constructed by the allelic exchange strategy. The primer sets used for PCR amplification of the DNA fragments flanking sequence are listed in Table 3. The generated DNA fragments were cloned into pKAS46, a suicide vector (a generous gift
from Dr. Skorupski, University of New Hampshire), and the resulting plasmids were then mobilized to K. pneumoniae CG43-S3 through conjugation from E. coli S17-1 λ pir. The trans-conjugants were selected with kanamycin on minimal medium (M9 minimal salts, Sigma). Some of the kanamycin-resistant transconjugants was picked and then spread onto a LB plate supplemented with streptomycin. When the occurrence of a double cross-over, the streptomycin-resistant and kanamycin-sensitive colonies were isolated, and the deletions of wza, yor5, yco6, and wzx were verified by PCR and by Southern blot analysis with a gene specific probe. To obtain the complement of wza, yor5, yco6 and wzx, the genes were amplified from K. pneumoniae CG43-S3 with the primers listed in Table 3 and the DNA fragment was ligated into pRK415, a broad host range plasmid (Keen et al., 1988).
Quantification of the glucuronic acid content. The CPS was extracted by using
the method described previously (Domenico et al., 1989). Briefly, bacteria were collected from 500 µl of overnight culture and mixed with 100 μl of 1% Zwittergent 3-14 detergent (Sigma-Aldrich, Milwaukee, WI) in 100 mM citric acid (pH 2.0). The mixture was incubated at 50℃ for 20 min. After centrifugation, 250 μl of the supernatant was transferred to a new tube and CPS was precipitated with 1 ml of absolute ethanol. The pellet was then dissolved in 200 μl distilled water and a 1200 μl of 12.5 mM borax (Sigma-Aldrich, Milwaukee, WI) in H2SO4 was added. The
mixture was vigorously vortexed and boiled for 5 min, subsequently cooled on ice. Then 20 μl of 0.15% 3-hydroxydiphenol (Sigma-Aldrich, Milwaukee, WI) was added and the absorbance at 520 nm was measured. The uronic acid content was determined from a standard curve of glucuronic acid (Sigma-Aldrich, Milwaukee, WI) and expressed as μg per 109 CFU.
Bacterial fractionation. Cell pellet was collected from 5 ml of overnight culture,
and washed three times with STE buffer (10mM Tris-HCl, pH 7.5, 10mM EDTA, 25% sucrose). After centrifugation, the pellet was resuspended in 0.4 ml ice-cold water and proceeded with osmotic shock (put on ice for 10 min and subsequently stand at 37℃ for 10 min, repeat three times). After centrifugation at 12000 ×g for 5 min, the supernatant was assigned as the periplasmic fraction, and the pellet resuspended in 0.4 ml T buffer (50mM Tris-HCl, pH 8.0, 30mM magnesium acetate, 2mM dithiothreitol). After disruption by sonication and centrifugation at 180000 ×g for 60 min, the supernatant and pellet were collected as the cytoplasmic fraction and membrane fraction, respectively.
Analysis of the capsular polysaccharide. Capsular polysaccharides were purified
as described (Whitfield et al., 1992). Briefly, cells were harvested from overnight culture and pelleted at 13200 rpm for 10 min. The pellet was resuspended in 500 µl PBS and extracted with hot-phenol, and then dialyzed against water overnight. The crude extract was treated with DNase (4 mg/ml), RNase (0.2 mg/ml) and proteinase K (4 mg/ml) and dialyzed against water. The extracted CPS was then analyzed by 5% or 10% DOC-PAGE. After electrophoresis, the gel was immediately immersed in 100 ml of alcian blue solution (0.005% alcian blue, 40% ethanol, 5% acidic acid) and gently rocked for 30 min as described (Reuhs et al., 1998). This was followed by a change to 100 ml of fresh solution and overnight rocking. The gel was then rinsed for 5 min in dH2O and oxidized in 100 ml of 0.7% sodium metaperiodate for 10 min, and washed
five times in 200 ml of dH2O for 5 min each time. After the gel was stained in 100 ml
of silver solution (10% Bio-Rad silver concentrate) for 10 min and rinsed in dH2O for
5 min, 100 ml of developer (Bio-Rad) was added with agitation until dark precipitate formed and immediately drained to remove all precipitate. Finally, the color was
developed with developer again for 5 min. The development was stopped by 100 ml of 5% acetic acid for 10 min followed by a rinse of 200 ml dH2O.
Visualization of the CPS by electron microscopy. As described by Reid (Reid
and Whitfield, 2005), the cells were grown to mid-exponential phase in LB broth containing the appropriate antibiotics. The cultures with an optical density at 600 nm of about 0.5 were collected by centrifugation at 5,600 ×g for 5 min. The cells were washed twice with 0.5 ml of phosphate buffered saline (PBS) and the pellets were incubated with 20 μl cationized ferritin solution (Sigma F-7879) for 30 min at room temperature with gentle agitation. Binding of the cationized ferritin binds to negatively charged capsule prevents it from dehydration for analysis by electron microscopy. Finally, the cells were collected by centrifugation and washed twice with 0.5 ml of PBS to remove the unbound ferritin and then processed for visualization by transmission electron microscopy (JEOL JEM 2000EXII).
Polymyxin B sensitivity assay. The Polymyxin B sensitivity assay was performed
by using the method described (Campos et al., 2004). Cells were grown at 37°C in LB medium and harvested at the exponential phase of growth, and a suspension containing approximately 105 CFU/ml was prepared in 1% (wt/vol) tryptone-PBS (pH 7.4). Then, 100 μl of the cell suspension was mixed with various concentrations of polymyxin B in a volume of 1 ml, followed by incubation at 37°C for 30 min. Finally, 100 μl of the suspension was plated on LB agar plates and the plates were incubated overnight at 37°C for colony formation. The colony counts were determined and expressed in percentage to the colony counts of bacteria not exposed to polymyxin B.
Biofilm formation assay. The measurement of biofilm formation was performed
bacteria were diluted (1:100) in culture medium, and 100 μl diluted bacteria were inoculated into each well of a 96-well microtiter dish (TPP industries, France) and incubated at 37°C for 48 h to allow the formation of biofilm. After incubation, each well was washed with water and 150 μl of 1% crystal violet was added for incubation at room temperature for 30 min. Subsequently, after washing with water, 150 μl of 1% SDS was added to each well and the microtiter dish was shaken to dissolve the dye. The capability of biofilm formation was quantitated by determining absorbance at 595 nm and each biofilm result represented the mean of three separate experiments.
Western immunoblotting. The purified proteins were analyzed by SDS-PAGE
and the resolved proteins were transferred to a polyvinylidene difluoride (PVDF) membrane electrophoretically in the transfer buffer containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 and 20% methanol. The His6-tagged
proteins were detected by anti-His-tag monoclonal antibody (Novagen) and phosphotyrosine proteins were detected by anti-phosphotyrosine monoclonal antibody (Sigma P4110). The secondary antibody, an anti-mouse IgG conjugate alkaline phosphatase antibody (Sigma), was then applied and the bound complex was detected by using nitro blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) as the substrates.
Constructions of the recombinant His6-tagged proteins. For construction of the
kinase domain expression plasmid pET30-Yco6E23, 0.85-kb DNA fragment of yco6 (from nucleotide 1351 to 2166), which encoding the C-terminal PTK domain of Yco6, was PCR-amplified from K. pneumoniae CG43-S3 using the primer pair Yco6E2 and Yco6E3 (Table 3). The DNA fragment was restricted by EcoRI/SalI and then ligated into pET-30b vector, which resulting of the plasmid pET30-Yco6E23. To generate other recombinant plasmids including pET30-Yor5, pET30-Ugd, pET30-Gnd,
pQE31-ManC and pQE31-ManB, the respective coding region of the genes were PCR amplified using the specific primer pair (Table 3), and each of the DNA cloned into pET or pQE expression vector.
Overexpression and purification of the His6-tagged proteins. The bacterial cells
were incubated in 100 ml of LB medium at 37℃ with shaking until OD600 reached
0.6. Isopropyl-1-thio-β-D-galactopyranoside (IPTG) was then added to a final concentration of 0.5 mM and the growth was continued for 4 h at 37℃. Subsequently, the cells were harvested by centrifugation at 5000 rpm for 10 min, resuspended in buffer A (20 mM Tris-HCl, 500 mM NaCl, 5 mM imidazole, pH 7.9), and the cell suspension disrupted by sonication and then the cell debris removed by centrifugation at 13000 rpm for 10 min. Finally, the His6-tagged proteins were purified from the
supernatant via affinity chromatography using His-Bind resin (Novagen), and the elution was carried out with buffer B (20 mM Tris-HCl, 500 mM NaCl, 1 M imidazole, pH 7.9). Aliquots of the collected fractions were analyzed by SDS-PAGE and the fractions containing most of the purified His6-tagged kinase were dialyzed
against the buffer containing 50 mM sodium phosphate (pH 7.4), 150 mM NaCl, 5 mM MgCl2, and 10% glycerol. The fractions containing His6-Yor5, His6-Ugd,
His6-Gnd, His6-ManC or His6-ManB were dialyzed against the buffer containing 50
mM Tris-HCl (pH7.5), 100 mM NaCl, 1 mM EDTA, and 10% glycerol.
In vitro phosphorylation assay. The phosphorylation reaction was carried out as
described (Grangeasse et al., 2003) by incubating the mixture (20 μl) containing about 2 μg of the purified kinase and/or substrate proteins and 4 µCi (10 μM) [γ-32P]ATP in 25 mM Tris-HCl (pH 7.0), 1 mM DTT, 5 mM MgCl2, 1 mM EDTA at
37℃ for 10 min. The reaction was stopped by addition of the sample buffer and heating at 100℃ for 10 min. After electrophoresis (10% SDS-PAGE), the gel was
visualized by autoradiography with InstantImagerTM (Packard Instrument Company) and stained with Coomassie Blue.
Protein dephosphorylation assay. This assay was performed as described
(Grangeasse et al., 2003). The purified His6-tagged proteins were phosphorylated in
the presence of [γ-32P] ATP as described above, and then desalted by Microcon YM-10 columns (Millipore) to remove [γ-32P] ATP before dephosphorylation. The dephosphorylation reaction was carried out by adding 1 μg of the purified Yor5 phosphatase with the buffer containing 100 mM sodium citrate (pH6.5) and 1 mM EDTA, and the mixtures were incubated at 37℃ for 30 min. The reaction mixture was stopped and analyzed as described above.
UDP-glucose dehydrogenase assay. The activity of the purified Ugd was
determined according to the described spectrophotometric assay (Pagni et al., 1999). The assay was carried out essentially for the purified Ugd, the phosphorylated Ugd, and the dephosphorylated Ugd in the buffer containing 100 mM Tris-HCl (pH 9.0), 100 mM NaCl, 2 mM DTT, 5 mM NAD+ and 5 mM UDP-glucose and the absorbance at 340 nm was measured by spectrophotometer (Ultrospec 3100 pro UV/Visible Spectrophotometer, Amersham Biosciences Ltd).
6-phosphogluconic dehydrogenase assay. The 6-phosphogluconic dehydrogenase
activity was determined according to the described assay (Corpas et al., 1998) for the purified Gnd, the phosphorylated Gnd, and the dephosphorylated Gnd in the buffer containing 50 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 2 mM NAD+ and 2 mM
Results
Part 1: Biological roles of the core elements Wza, Yor5, Yco6, and Wzx encoded in the K2 cps gene cluster of Klebsiella pneumoniae CG43
Comparative analysis of the K. pneumoniae cps gene clusters. In a previous
study, K. pneumoniae CPS has been shown to be made from a similar biosynthetic pathway to that of E. coli group 1 CPS (Whitfield and Roberts, 1999). The sequence of K2 cps gene cluster of K. pneumoniae Chedid has been determined, which is composed of 19 open reading frames organized into 3 transcriptional units identified (Arakawa et al., 1995). K. pneumoniae CG43, as well as K. pneumoniae Chedid, produce CPS of K2 serotype (Chang et al., 1996). Comparative analysis of the cps gene cluster of K. pneumoniae Chedid with that of E. coli K30 revealed several conserved genes involved in regulation of high-level polymerization and translocation of CPS, and hence they are called as core elements. These proteins, Wzi, Wza, Yor5, Yco6, Wzx and Wzy, of K. pneumoniae Chedid exhibit striking sequence homologies with the proteins, Wzi, Wza, Wzb, Wzc, Wzx and Wzy, of E. coli K30 (identity and similarity: 97.6% and 98.4%, 91.0% and 95.5%, 53.1% and 64.6%, 66.5% and 78.3%, 14.1% and 31.4%, 15.9% and 30.3%, respectively). Besides, the genes, galF, orf2, gnd and manCB, are also conserved in the two genomes (Fig. 1).
Characterizations of the deletion mutants. In addition to yor5- and yoc6-
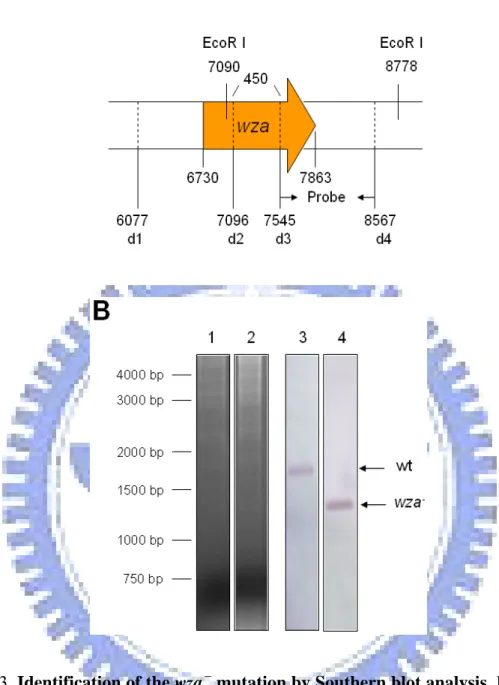
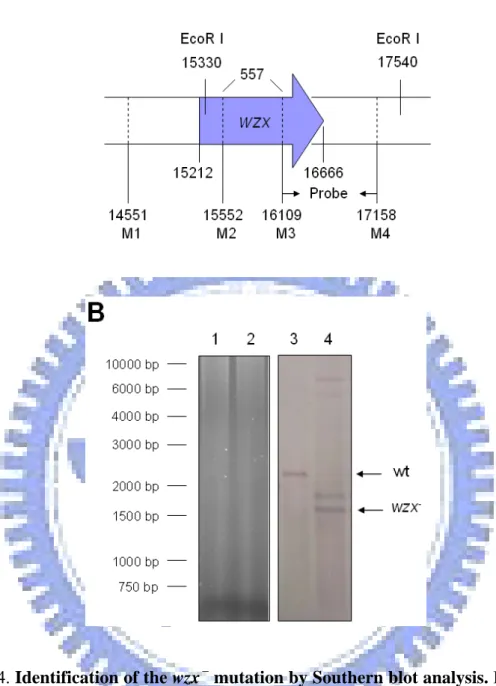
mutants, which have been constructed previously (白平輝, 2004), deletion mutations of wza and wzx were also constructed by using allelic exchange strategy. As shown in Fig. 3 and Fig. 4, Southern blotting analysis using the probes amplified by specific
primer pairs, wzad3 and wzad4 for wza-
mutation and wzxM3 and wzxM4 for wzx- mutation, demonstrated that the specific deletions have been introduced. As shown in Fig. 3, 1688-bp signal was detected in the wild type strain and 1238-bp signal was detected in the wza- mutant. Although 2210-bp and 1653-bp signal were detected respectively in wild type strain and wzx-
mutant, additional unexpected band was also observed (Fig. 4). The additional unexpected band was about 1800-bp and might be de to incomplete digestion of chromosome or additional deletion of wzx homolog. In addition, analysis of Western blotting and RT-PCR indicated that Yco6 protein was not detected in the yor5-
mutant and yco6 mRNA was apparently less than wild type strain (data not shown), suggesting a polar effect of the yor5 deletion.
Phenotypes of the mutants. Colony morphology of all four mutants appeared to
be smaller compared to that of the wild type strain CG43-S3 and the mucoidy was dramatically reduced as determined by string formation (data not shown). As shown in Fig. 5, all the mutants showed higher growth rates than wild type strain in LB broth. In the sedimentation test using a galU deletion mutant (U9451) as a negative control, the mutants appeared to precipitate readily, particularly the wza- mutant (Fig. 6). Mucoidy of the wza complementing strain appeared to be recovered and the defect in the sedimentation test was also partially restored.
Capsular analysis of the deletion mutants. Since the core elements were assumed
to play critical roles in CPS biosynthesis, the defects of these mutants ought to result from modification of CPS biosynthesis. As shown in Fig. 7, the glucuronic acid contents, which serve as indicator of K2 CPS amount (Vodonik and Gray, 1988), of all the mutants were significantly reduced. Furthermore, the CPS purified and analyzed in Fig. 8 showed that all, except the wza- mutant, lost their polymeric CPS, and in turn increased the corresponding oligomeric CPS. In the periplasmic fraction
of the wza-
mutant, some polymeric CPS were detected indicating that Wza is involved in translocation of polymeric CPS but not in CPS polymerization. Block of the polymeric CPS translocation could feedback-inhibit the upstream CPS biosynthesis and hence resulted in the reduction of total CPS amount. In addition, some polymeric CPS was found in wzx-
mutant suggesting the existence of an alternative system with low efficiency for the translocation of the CPS oligomer. In the yor5-
and yco6-
mutants, disappearance of the high-molecular-weight CPS demonstrated a regulatory role of the tyrosine phosphorylation executed by Yco6 and Yor5 in polymerization of CPS.
Visualization of K. pneumoniae CPS by transmission electron microscopy (TEM). The surface-expressed CPS of each of the mutants was labeled with
cationized ferritin (Sigma F-7879) as shown in the electron micrograph (Fig. 9), K. pneumoniae wild-type strain CG43-S3 appeared to carry an intact thick capsule but the mutants exhibited different levels of unorganized capsules. The disorderly electron-dense materials seen on the surfaces of mutants in micrographs were labeled CPS which might result form dehydration of cells, the treatment for TEM. Notably, the wza-
mutant had the most smooth surface, and the wzx-
mutant appeared to carry more of the disrupted CPS on the surface than the other mutants.
Reduction of CPS affected the bacterial susceptibility to polymyxin B and ability of biofilm formation. According to the above mentioned results, deletions of
the genes encoding the core elements indeed affected normal CPS expression, but the influences of these defects on bacterial physiology are still not clear. It has been demonstrated in K. pneumoniae that increasing amount of CPS and upregulated cps transcription were induced by polymyxin B thereby led to the phenotype of polymyxin B resistance (Campos et al., 2004). As shown in Fig. 10, yor5-
mutants appeared to be more sensitive to polymyxin B. Capsular expression has been shown to exert significant effect on K. pneumoniae biofilm formation (Schembri et al., 2005). Increasing capability of biofilm formation was also observed in either the yor5 - or the wzx- mutant (Fig. 11).
Part 2: Autophosphorylation of protein-tyrosine kinase Yco6 regulates tyrosine phosphorylatoin of the proteins involved in capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43
Tyrosine phosphorylation of Ugd is catalyzed by the cytoplasmic PTK domain of Yco6. Although the tyrosine phosphorylation regulated by PTK and PTP has been
shown to modulate biosynthesis and assembly of CPS (Vincent et al., 2000), the mechanism is still unknown. Since the recent reports suggested that E. coli Ugd and B. subtilits Ugd are the substrates for PTK (Grangeasse et al., 2003; Mijakovic et al., 2003), we herein intend to demonstrate in K. pneumoniae CG43 that the Ugd could also be phosphorylated by the PTK, Yco6. In comparing the amino acid sequences of K. pneumoniae Ugd (KpUgd) (NTUH-K2044 strain, NCBI accession No. BAD03946) with E. coli Ugd (K12 strain, NCBI accession No. NP_416532) and B. subtilis YwqF (strain 168, NCBI accession No. CAB15640), approximately 83% and 26% sequence identities were observed respectively. Interestingly, 99.2% amino acid sequence identity was found between KpUgd and the Ugd of E. coli K30 strain (NCBI accession No. AAC45348) indicating they are more related. To demonstrate if a similar control of Ugd by PTK Yco6 is present in K. pneumoniae, the purified His6-KpUgd was incubated with the phosphorylated Yco6, His6-Yco6 (Arg451-Lys722),
and [γ-32P] ATP. As shown in Fig. 12A, Yco6 could auto-phosphorylate itself in the presence of [γ-32P] ATP (lane 5) and the phosphorylation of KpUgd could be detected (lane 6) by comparing to the non-radioisotope-labeled His6-KpUgd while the protein
incubated with only [γ-32P] ATP (lane 1). While incubating with the phosphorylated His6-Wzc (Ser447-Ala704), the PTK of E. coli and deleted from its tyrosine cluster, in
12B, lane 2), suggesting that the two PTK, Yco6 and Wzc, exert a similar mechanism via tyrosine phosphorylation to modulate the activity of Ugd.
Phosphorylation enhances the Ugd activity. Ugd is UDP-glucose dehydrogenase
catalyzing a NAD+-dependent transformation of UDP-glucose into UDP-glucuronic acid (Pagni et al., 1999). Phosphorylation of the UDP-glucose dehydrogenases in E. coli and B. subtilis enhancing their enzymatic activities have recently been reported (Grangeasse et al., 2003; Mijakovic et al., 2003). Consistent with the findings, the enzymatic activity could be enhanced when the recombinant KpUgd was incubated with His6-Yco6(Arg451-Lys722) and ATP (Fig. 13). When KpUgd was incubated with
His6-Wzc(Ser447-Ala704) and ATP, its activity was also enhanced. In addition,
incubation of the phosphorylated His6-KpUgd with calf intestine alkaline phosphatase
(CIAP) (Fermentas) dramatically reduced the Ugd activity, which indicating that phosphorylation of Ugd indeed enhanced its activity, and the purified His6-KpUgd
contained a small fraction of the phosphorylated form.
Tyrosine phosphorylation of either of Gnd, ManC and ManB could be catalyzed by Yco6 and Wzc. In the biosynthesis of E. coli group 1 CPS, there are
several glycosyltransferases and enzymes, including ManBC and Ugd, involved in the synthesis of sugar nucleotide precursors (Whitfield and Paiment, 2003). Besides, Gnd, a 6-phosphogluconic dehydrogenase, is also required for the formation of the cellular polysaccharide layer (Sprenger, 1995). As shown in Fig. 1, ManBC and Gnd are encoded by the genes in the cps gene cluster of E. coli and K. pneumoniae. The possibility of ManBC and Gnd are also regulated by tyrosine phosphorylation was investigated. When the purified His6-Gnd, His6-ManC and His6-ManB were incubated
respectively with [γ-32P] ATP and the phosphorylated His6-Yco6(Arg451-Lys722),
comparing to those when the proteins incubated only with [γ-32P] ATP (Fig. 12A, lane 2, 3 and 4). When the purified His6-Gnd, His6-ManC and His6-ManB were incubated
respectively with [γ-32P] ATP and the phosphorylated His6-Wzc(Ser447-Ala704), the
phosphorylation signals of these proteins were also observed (Fig. 12B, lane 3, 4 and 5). This indicated that phosphorylations of these proteins did not require the tyrosine cluster of PTK.
Phosphorylation of Gnd enhances its 6-phosphogluconic dehydrogenase activity. Gnd is a 6-phosphogluconic dehydrogenase, which catalyzes the oxidative
decarboxylation of 6-phospho-gluconate to ribulose-5-phosphate (Sprenger, 1995). When the purified His6-Gnd was incubated with ATP and either
His6-Yco6(Arg451-Lys722) or His6-Wzc(Ser447- Ala704), increasing enzymatic activity
was observed (Fig. 14). Incubation of the His6-Gnd with CIAP (Fermentas) also
appeared to reduce its activity indicating that the purified His6-Gnd contained some of
the phosphorylated form.
The phosphorylated form of Ugd, Gnd, and ManC could be modified by Yor5.
In general, bacteria most often contain antagonistic PTKs and PTPs, and the genes encoding PTKs and PTPs are located next to each other in the same gene clusters. In bacteria, PTK and PTP have been reported to play important roles in regulation of the CPS and EPS production (Nakar and Gutnick, 2003; Stevenson et al., 1996). Many of the PTPs could specifically dephosphorylate the cognate PTKs and also the substrates of PTK (Grangeasse et al., 1998; Mijakovic et al., 2005; Morona et al., 2002; Musumeci et al., 2005; Vincent et al., 1999). To demonstrate if the phosphorylations demonstrated above could be modified specifically by Yor5, aliquots of the purified His6-Yor5 were added respectively into the above phosphorylation mixtures and
(lane 9) and ManC (lane 11) were removed. On the contrary, no effect on the phosphorylated ManB (lane 13) was found. Interestingly, the phsophorylated KpUgd appeared to be a poor substrate for the recombinant Yor5 (Fig. 12A, lane 7). Moreover, significant amount of Yco6 and Wzc remained to be phosphorylated (Fig. 12A, lane 7, 9, 11, 13, and Fig. 12B, lane 6). I speculate that Yor5 could efficiently remove the phosphorylated tyrosine residues at the C-terminal tyrosine cluster of Yco6. However, the modification at Tyr570 which is related to the kinase activity of Yco6 could be low. Therefore, Yor5 would regulate the activity of KpUgd, Gnd, ManC and Yco6 by desphosphorylation. On the other hand, ManB is likely regulated by the other phosphatase.
Discussion
The presence of homologues of Wzi, Wza, Yor5, Yco6, Wzx, and Wzy in K. pneumoniae raises the question of whether these proteins are functional conserved. The gene, wzi, is only found in the cps loci of K. pneumoniae and E. coli group 1 CPS serotype. Its role in linking high-molecular-weight CPS to cell surface has been demonstrated recently (Alvarez et al., 2000; Rahn et al., 2003). Although the precise way of the linkage is not clear, it is noticeable that wzi is confined to the bacteria wherein the capsule is a major virulence factor and their CPS polymers are tightly linked to the cell surface. Wza is assumed to be a multimeric outer membrane protein complex required for surface expression of CPS (Nesper et al., 2003), and wza
-mutation in K. pneumoniae also showed apparent loss of surface CPS in this study. The block of translocation of polymeric CPS also resulted in reduction of total CPS biosynthesis. The block of translocation of CPS polymer has been suggested to feedback inhibit the upstream CPS biosynthesis, but the exact mechanism remains to be determined (Nesper et al., 2003). As shown in Fig. 6, expression of Wza in wza
-mutant could only partially restore the phenotype might result from an inappropriate amount of the expressed protein. Nevertheless, three-dimensional structure of Wza has recently been resolved, which indicated that Wza is an octameric complex with a tetrameric (C4) rotational symmetry and is organized as a tetramer of dimeric subunits (Beis et al., 2004).
Compared with Wza, the studies of Wzx and Wzy are less and confined in O-antigen synthesis, due to the difficulties of purification of the membrane protein and verification of their activities. Some polymeric CPS found in wzx- mutant (Fig. 8)
Wzx might not be the only translocator required in the process. The amino sequence of Wzy is variable among strains (identities are less than 16% among K. pneumoniae Chedid and NTUH K2044, E. coli K12 and K30), and this implies that it is specific to each serotype. So far, no Wzy homolog has been purified and studied at biochemical level to confirm its polymerase activity. In E. coli serotype K30 (Drummelsmith and Whitfield, 1999) and K40 (Amor and Whitfield, 1997), wzy mutants lack capsules and add only a single K antigen repeat unit to the LPS lipid A core, namely KLPS, implying
the role of Wzy for CPS polymerization. Both yco6 and yor5 mutants lost high-molecular-weight CPS, suggesting that Yco6 and Yor5 are involved in polymerization of CPS. Overall, mutation in either of the core elements exerted capsule-lacking phenotype. Using the assays of polymyxin B sensitivity and biofilm formation, yor5 and wzx mutants were found to be more susceptible to polymyxin B and increase capability of biofilm formation. Nevertheless, the mechanism is still a question to be investigated.
After thorough analyses, it is believed that PTKs are involved in the regulation of CPS or EPS production (Vincent et al., 2000). The finding that UDP-glucose dehydrogenases are phosphorylated by PTKs strongly supports this viewpoint, although PTKs might play other roles via other mechanisms (Klein et al., 2003; Mijakovic et al., 2006). Herein, we have demonstrated that, in addition to Ugd, Gnd and ManBC are also regulated by tyrosine phosphorylation via Yco6. The phosphorylation appeared also to increase the enzymatic activity, respectively. Although the effect of phosphorylation on ManBC activity has not yet been verified, enhancement of the activity is predictable. Through dephosphorylation by Yor5, the phosphorylated residues is at tyrosine could be inferred. To identify the phosphorylated residue, the amino acid sequences of the enzymes, KpUgd, Gnd and
ManBC, and known substrates of PTK, Ugd and Ssb, were comparatively analyzed. There are 14 to 25 tyrosine residues existing in each protein, however, no conserved motif was found. The phosphorylated site may be determined according to the three-dimensional structure of protein substrate but not specific primary amino acid sequence. In recently, mass spectrometry allowed to pinpoint the phosphorylated tyrosine residue in B. subtilis Ssb (Mijakovic et al., 2006), and the phosphorylated residues of KpUgd, Gnd and ManBC might be analyzed by the same method.
Besides the enzymes for CPS precursor synthesis, it is known that the PTKs also autophosphorylate themselves and the phosphorylations at C-terminal tyrosine residues seem to be unrelated to activations of their substrates (Grangeasse et al., 2003). Their kinase activity appeared to be relative to the upstream tyrosine residue and Walker A and B motifs. Moreover, the phosphorylation of the upstream tyrosine and the substrates could not or slowly be dephosphorylated by their cognate PTPs. Only the tyrosine residues in the C-terminal tyrosine cluster of PTKs could be entirely modified by the cognate PTPs implying that different PTP existed to exert the activity to dephosphorylate the upstream tyrosine residue and the tyrosine of certain substrates. The three-dimensional structure of Wzc (Collins et al., 2006) and the finding of the Wza-Wzc interaction (Nesper et al., 2003) indicated that Wzc, as a tetrameric complex with C4 rotational symmetry, forms multienzyme complex with Wza and other membrane proteins which is involved in polymerization and translocation of CPS. The overall level of phosphorylation in the tyrosine cluster is important for capsule assembly, but not the upstream tyrosine (Paiment et al., 2002). Briefly, the PTP-modifiable phosphorylations on the C-terminal tyrosine clusters of PTKs are probably related to conformational changes of PTKs which result in modulating conformation of the multienzyme complex to open or close the translocation channel
of CPS.
As shown in Fig. 15, we propose a model which predicts that Yco6 would regulate both CPS biosynthesis and translocation by phosphorylation. The PTK activity of Yco6 is first enhanced by autophosphorylation at Tyr and the resulting activation of Yco6 stimulates interphosphorylation at C-terminal tyrosine cluster and also the phosphorylations of its substrates. The phosphorylations of these substrates increase their activities resulting in elevated synthesis of sugar nucleotide precursors. CPS lipid-linked repeat unit composed of sugar nucleotide precursors then is flipped across the inner membrane in a process requiring Wzx and polymerized through Wzy-dependent reaction. Finally, CPS polymer is exported to cell surface via the channel formed by Wza and associated on the surface per the outer membrane protein, Wzi. Although Yco6 also plays roles in CPS polymerization and translocation, it is not yet clear whether these activities require the phosphorylation of Yco6 at specific tyrosine in the C-terminal tyrosine cluster.
References
Alvarez, D., Merino, S., Tomas, J.M., Benedi, V.J., and Alberti, S. (2000) Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect Immun
68: 953-955.
Amor, P.A., and Whitfield, C. (1997) Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his-region containing gene clusters for multiple cell-surface polysaccharides. Mol Microbiol 26: 145-161.
Arakawa, Y., Wacharotayankun, R., Nagatsuka, T., Ito, H., Kato, N., and Ohta, M. (1995) Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol 177: 1788-1796.
Beis, K., Collins, R.F., Ford, R.C., Kamis, A.B., Whitfield, C., and Naismith, J.H. (2004) Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J Biol Chem 279: 28227-28232.
Bugert, P., and Geider, K. (1997) Characterization of the amsI gene product as a low molecular weight acid phosphatase controlling exopolysaccharide synthesis of Erwinia amylovora. FEBS Lett 400: 252-256.
Campos, M.A., Vargas, M.A., Regueiro, V., Llompart, C.M., Alberti, S., and Bengoechea, J.A. (2004) Capsule polysaccharide mediates bacterial resistance
to antimicrobial peptides. Infect Immun 72: 7107-7114.
Chang, H.Y., Lee, J.H., Deng, W.L., Fu, T.F., and Peng, H.L. (1996) Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb Pathog 20: 255-261.
Collins, R.F., Beis, K., Clarke, B.R., Ford, R.C., Hulley, M., Naismith, J.H., and Whitfield, C. (2006) Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J Biol Chem 281: 2144-2150.
Corpas, F.J., Barroso, J.B., Sandalio, L.M., Distefano, S., Palma, J.M., Lupianez, J.A., and Del Rio, L.A. (1998) A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J 330 (Pt 2): 777-784.
Domenico, P., Schwartz, S., and Cunha, B.A. (1989) Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect Immun 57: 3778-3782.
Dower, W.J., Miller, J.F., and Ragsdale, C.W. (1988) High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16: 6127-6145. Drummelsmith, J., and Whitfield, C. (1999) Gene products required for surface
expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol Microbiol 31: 1321-1332.
Duclos, B., Grangeasse, C., Vaganay, E., Riberty, M., and Cozzone, A.J. (1996) Autophosphorylation of a bacterial protein at tyrosine. J Mol Biol 259: 891-895.
Grangeasse, C., Doublet, P., Vaganay, E., Vincent, C., Deleage, G., Duclos, B., and Cozzone, A.J. (1997) Characterization of a bacterial gene encoding an autophosphorylating protein tyrosine kinase. Gene 204: 259-265.
Grangeasse, C., Doublet, P., Vincent, C., Vaganay, E., Riberty, M., Duclos, B., and Cozzone, A.J. (1998) Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J Mol Biol
278: 339-347.
Grangeasse, C., Obadia, B., Mijakovic, I., Deutscher, J., Cozzone, A.J., and Doublet, P. (2003) Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J Biol Chem 278: 39323-39329.
Huang, J., and Schell, M. (1995) Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol 16: 977-989.
Hunter, T. (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80: 225-236.
Jayaratne, P., Bronner, D., MacLachlan, P.R., Dodgson, C., Kido, N., and Whitfield, C. (1994) Cloning and analysis of duplicated rfbM and rfbK genes involved in the formation of GDP-mannose in Escherichia coli O9:K30 and participation of rfb genes in the synthesis of the group I K30 capsular polysaccharide. J Bacteriol 176: 3126-3139.
broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene
70: 191-197.
Klein, G., Dartigalongue, C., and Raina, S. (2003) Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol Microbiol 48: 269-285.
Lin, W.S., Cunneen, T., and Lee, C.Y. (1994) Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol 176: 7005-7016.
Manai, M., and Cozzone, A. (1983) Characterization of the amino acids phosphorylated in E. coli proteins. FEMS Microbiol Lett 17: 87-91.
Mijakovic, I., Poncet, S., Boel, G., Maze, A., Gillet, S., Jamet, E., Decottignies, P., Grangeasse, C., Doublet, P., Le Marechal, P., and Deutscher, J. (2003) Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. Embo J 22: 4709-4718.
Mijakovic, I., Musumeci, L., Tautz, L., Petranovic, D., Edwards, R.A., Jensen, P.R., Mustelin, T., Deutscher, J., and Bottini, N. (2005) In vitro characterization of the Bacillus subtilis protein tyrosine phosphatase YwqE. J Bacteriol 187: 3384-3390.
Mijakovic, I., Petranovic, D., Macek, B., Cepo, T., Mann, M., Davies, J., Jensen, P.R., and Vujaklija, D. (2006) Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res 34: 1588-1596.
phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol Microbiol 35: 1431-1442. Morona, J.K., Morona, R., Miller, D.C., and Paton, J.C. (2002) Streptococcus
pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J Bacteriol 184: 577-583.
Musumeci, L., Bongiorni, C., Tautz, L., Edwards, R.A., Osterman, A., Perego, M., Mustelin, T., and Bottini, N. (2005) Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis. J Bacteriol 187: 4945-4956.
Nakar, D., and Gutnick, D.L. (2003) Involvement of a protein tyrosine kinase in production of the polymeric bioemulsifier emulsan from the oil-degrading strain Acinetobacter lwoffii RAG-1. J Bacteriol 185: 1001-1009.
Nesper, J., Hill, C.M., Paiment, A., Harauz, G., Beis, K., Naismith, J.H., and Whitfield, C. (2003) Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30. Structural and functional analysis of the outer membrane lipoprotein Wza. J Biol Chem 278: 49763-49772.
Niemeyer, D., and Becker, A. (2001) The molecular weight distribution of succinoglycan produced by Sinorhizobium meliloti is influenced by specific tyrosine phosphorylation and ATPase activity of the cytoplasmic domain of the ExoP protein. J Bacteriol 183: 5163-5170.
O'Toole, G.A., and Kolter, R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a
genetic analysis. Mol Microbiol 28: 449-461.
Ørskov, I., and Ørskov, F. (1984) Serotyping of Klebsiella. Methods Microbiol 14: 143-164.
Pagni, M., Lazarevic, V., Soldo, B., and Karamata, D. (1999) Assay for UDPglucose 6-dehydrogenase in phosphate-starved cells: gene tuaD of Bacillus subtilis 168 encodes the UDPglucose 6-dehydrogenase involved in teichuronic acid synthesis. Microbiology 145 (Pt 5): 1049-1053.
Paiment, A., Hocking, J., and Whitfield, C. (2002) Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J Bacteriol 184: 6437-6447.
Paulsen, I.T., Beness, A.M., and Saier, M.H., Jr. (1997) Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 143 (Pt 8): 2685-2699.
Podschun, R., and Ullmann, U. (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11: 589-603.
Preneta, R., Jarraud, S., Vincent, C., Doublet, P., Duclos, B., Etienne, J., and Cozzone, A.J. (2002) Isolation and characterization of a protein-tyrosine kinase and a phosphotyrosine-protein phosphatase from Klebsiella pneumoniae. Comp Biochem Physiol B Biochem Mol Biol 131: 103-112.
Rahn, A., Drummelsmith, J., and Whitfield, C. (1999) Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens:
relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J Bacteriol 181: 2307-2313.
Rahn, A., Beis, K., Naismith, J.H., and Whitfield, C. (2003) A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J Bacteriol 185: 5882-5890.
Reid, A.N., and Whitfield, C. (2005) Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J Bacteriol 187: 5470-5481.
Reuhs, B.L., Geller, D.P., Kim, J.S., Fox, J.E., Kolli, V.S., and Pueppke, S.G. (1998) Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl Environ Microbiol 64: 4930-4938.
Schembri, M.A., Blom, J., Krogfelt, K.A., and Klemm, P. (2005) Capsule and fimbria interaction in Klebsiella pneumoniae. Infect Immun 73: 4626-4633.
Simoons-Smit, A.M., Verweij-van Vught, A.M., and MacLaren, D.M. (1986) The role of K antigens as virulence factors in Klebsiella. J Med Microbiol 21: 133-137. Sprenger, G.A. (1995) Genetics of pentose-phosphate pathway enzymes of
Escherichia coli K-12. Arch Microbiol 164: 324-330.
Stevenson, G., Andrianopoulos, K., Hobbs, M., and Reeves, P.R. (1996) Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol 178: 4885-4893.
Vincent, C., Doublet, P., Grangeasse, C., Vaganay, E., Cozzone, A.J., and Duclos, B. (1999) Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol 181: 3472-3477. Vincent, C., Duclos, B., Grangeasse, C., Vaganay, E., Riberty, M., Cozzone, A.J., and
Doublet, P. (2000) Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J Mol Biol 304: 311-321.
Vodonik, S.A., and Gray, G.R. (1988) Analysis of linkage positions in a polysaccharide containing nonreducing, terminal alpha-D- glucopyranosyluronic groups by the reductive-cleavage method. Carbohydr Res 172: 255-266.
Wacharotayankun, R., Arakawa, Y., Ohta, M., Hasegawa, T., Mori, M., Horii, T., and Kato, N. (1992) Involvement of rcsB in Klebsiella K2 capsule synthesis in Escherichia coli K-12. J Bacteriol 174: 1063-1067.
Whitfield, C., Perry, M.B., MacLean, L.L., and Yu, S.H. (1992) Structural analysis of the O-antigen side chain polysaccharides in the lipopolysaccharides of Klebsiella serotypes O2(2a), O2(2a,2b), and O2(2a,2c). J Bacteriol 174: 4913-4919.
Whitfield, C., and Roberts, I.S. (1999) Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol 31: 1307-1319. Whitfield, C., and Paiment, A. (2003) Biosynthesis and assembly of Group 1 capsular
in other bacteria. Carbohydr Res 338: 2491-2502.
Whitfield, C. (2006) Biosynthesis and Assembly of Capsular Polysaccharides in Escherichia coli. Annu Rev Biochem.
Zhao, X., and Lam, J.S. (2002) WaaP of Pseudomonas aeruginosa is a novel eukaryotic type protein-tyrosine kinase as well as a sugar kinase essential for the biosynthesis of core lipopolysaccharide. J Biol Chem 277: 4722-4730. 白平輝 (2004) 克雷白氏肺炎桿菌 CG43 莢膜多醣體的產生和酪胺酸磷酸化作用
Table 1. Bacterial strains used and constructed in this study
Strain Genotype or relevant property Reference or source
E. coli:
JM109 RecA1 supE44 endA1 hsdR17 gyrA96 rolA1 thi
△ (lac-proAB)
Laboratory stock NovaBlue(DE3) endA1 hsdR17(rk12-mk12+) supE44 thi-1 recA1
gyrA96 relA1 lac[F,pro AB lacqZ△M15:: Tn10](DE3);Tetr Novagen BL21-RIL F- ompT hsdSB(rB B - mB -
)gal dcm(DE3) Laboratory stock
S17-1 λpir TpR SmR recA, thi, pro, hsdR- M+ [RP4-2-Tc::Mu:KmrTn7] (pir)
De Lorenzo et al., 1994 M15(pREP4) NalS, StrS, RifS, Thi-, Lac-, Ara+, Gal+, Mtl-, F-,
RecA+, Uvr+, Lon+
Qiagen
K. pneumoniae:
CG43 Clinical isolate of K2 serotype Laboratory stock CG43-S3 rspl mutant, StrepR Laboratory stock CG43-S3-U9451 galU deletion mutant Laboratory stock CG43-S3-wza
-wza deletion mutant in CG43-S3 This study
CG43-S3-yor5
-yor5 deletion mutant in CG43-S3 白平輝, 2004
CG43-S3-yco6
-yco6 deletion mutant in CG43-S3 白平輝, 2004
CG43-S3-wzx
-wzx deletion mutant in CG43-S3 This study
P. aeruginosa:
Table 2. Plasmids used and constructed in this study
Plasmid Description Reference or
source
pKAS46 Suicide vector, AmpR, KmR Laboratory stock pRK415 Shuttle vector, mob+, TcR Laboratory stock pKAS46-wza- A 2.0-kb fragment containing a 500-bp deletion
in wza locus cloned into pKAS46, AmpR, KmR
This study pKAS46-yor5- A 2.0-kb fragment containing a 150-bp deletion
in yor5 locus cloned into pKAS46, AmpR, KmR
白平輝, 2004 pKAS46-yco6- A 2.0-kb fragment containing a 1800-bp deletion
in yoc6 locus cloned into pKAS46, AmpR, KmR
白平輝, 2004 pKAS46-wzx- A 2.0-kb fragment containing a 600-bp deletion
in wzx locus cloned into pKAS46, AmpR, KmR
This study pKAS46-wzy- A 2.0-kb fragment containing a 1200-bp deletion
in wzy locus cloned into pKAS46, AmpR, KmR
This study pRK415-Wza pRK415 derivative carrying wza on an KpnⅠ
/BamHⅠ fragment, TcR
This study pRK415-Yor5 pRK415 derivative carrying yor5 on an EcoRⅠ/
EcoRⅠ fragment, TcR
This study pRK415-Yco6 pRK415 derivative carrying yco6 on an BamHⅠ/
BamHⅠ fragment, TcR
This study pRK415-Wzx pRK415 derivative carrying wzx on an BamHⅠ/
BamHⅠ fragment, TcR
This study pRK415-Wzy pRK415 derivative carrying wzy on an HindⅢ/
BamHⅠ fragment, TcR
This study pET-30a-c Overexpression of His6 fusion proteins, KmR Novagen
pQE30-32 Overexpression of His6 fusion proteins, AmpR Qiagen
pET30-Yco6E23 Encoding Yco6 from Arg451 to Lys722,
His6–Yco6(Arg451-Lys722), cloned in EcoRⅠ/Sal
Ⅰ sites, KmR
This study
pET30-Yor5 Encoding Yor5 from Met1 to Ala144, His
6–Yor5,
cloned in EcoRⅤ/SacⅠ sites, KmR
白平輝, 2004 pET30-Ugd Encoding KpUgd from Met1 to Asp388,
His6–KpUgd, cloned in EcoRⅠ/SalⅠ sites,
KmR
白平輝, 2004
pET30-Gnd Encoding Gnd from Met1 to Glu468, His
6–Gnd,
cloned in BamHⅠ/SacⅠ sites, KmR
白平輝, 2004
His6–ManC, cloned in BamHⅠ/HindⅢ sites,
AmpR
pQE31-ManB Encoding ManB from Met1 to Lys468,
His6–ManB, cloned in BamHⅠ/HindⅢ sites,
AmpR
This study
pQE30-41 Overexpression of His6–Wzc(Ser447-Lys720),
AmpR
Grangeasse et al., 2003 pQE30-42 Overexpression of His6–Wzc(Ser447-Ala704),
AmpR
Grangeasse et al., 2003 pQE30-U Overexpression of His6–Ugd(Lys2-Asp388), AmpR Grangeasse et al.,
Table 3. Primers used in this study Primer Sequence (5’ to 3’) WzaD1 TTTCTATGGGCAGATGGTTG WzaD2 CAGAATTCACCCAGTTACCG WzaD3 CCGCAGTGGTATGACAATTG WzaD4 GCTGACTATCGGGAAGCATC Yor5D1 GGTTGGCGATATCTTAATGG Yor5D2 GCTGATTGGTGAGCTCCATG Yor5D3 CGAAGCACGAGCTCAGACA Yor5D4 AATCGCTCCGGACCATTGGC Yco6D1 GCTGACTGGCGATATCTTGTGT Yco6D2 CACCAACCAATCGATCAAAGTC Yco6D3 GCATAATCGATTGGCGGAATTA Yco6D4 CTCCTTCTGGAGCTCGTTTTCA WzxD1 TCGATATCGCTATATTGACAACC WzxD2 AGAAAGCTTAATCTATCCAGCC WzxD3 CTGCAAAGCTTGGTTTATGCA WzxD4 CTGATAGATGAGCTCTGGAATG WzyD1 CAACCATGGAGCTCGATTAGATAT WzyD2 GCAGTCGACTTATATTATGCATGTC WzyD3 CGAAAATGACGTCGACTAAAGTTA WzyD4 GTTTGCTACCATACGCATTGCT
WzaE1 CACC AAGAAAAAAATTGTTAGATTTTCG WzaE2 CTAAACATATTATGGCCAATCC Yor5E1 CTCCATTGGTTCGTTGGAAT Yor5E2 GCATTCGCTTGTTTCTGTTC Yco6N1 CTGGATCCCAGGAAATAATGCATGACTTC Yco6N2 CACTGCAGGTCCCCTTCTTGAGTAACCT Yco6E2 CGTAAAGGAATTGAAACTCCAGA Yco6E3 AAGGGGATTCTTCGTCCCCT WzxN1 CTGGATCCGCGAAGGTATTACTAGATGAGT WzxN2 CACTGCAGATGATTTGGCAGCATATTTA WzyE1 CACCATTCGAAGAAAGTTTTCTCG WzyE2 CTTTAGTTGATGTCATTTTCG UgdE1 CGAATGAAAATTACTATTTCCGG UgdE2 CCAGTGTCAGACAGGCAGAA GndE1 GACCACACCAGACAGGAGCAAGT GndE2 CTCGGGCGGCATATAAAGA
ManCE1 CACCTTGCTTCCTGTGATTATGGC ManCE2 TCTGCGATTTGTCCCGAA
ManBE1 ATGGTCGTTGCTAATTTTTTCG ManBE2 GGCTTCCATTATGGATAATGC