Cloning and Characterization of Monacolin K
Biosynthetic Gene Cluster from Monascus pilosus
Y
I-P
EIC
HEN,
†,‡C
HING-P
INGT
SENG,
‡L
I-L
INGL
IAW,
†C
HUN-L
INW
ANG,
†I-C
HINGC
HEN,
†W
EN-J
UNGW
U,
†M
ING-D
ERW
U,
† ANDG
WO-F
ANGY
UAN*
,† Bioresource Collection and Research Center, Food Industry Research and Development Institute, and Department of Biological Science and Technology, National Chiao Tung University, HsinChu, TaiwanMonacolin K is a secondary metabolite synthesized by polyketide synthases (PKS) from Monascus, and it has the same structure as lovastatin, which is mainly produced by Aspergillus terreus. In the present study, a bacterial artificial chromosome (BAC) clone, mps01, was screened from the BAC library constructed from Monascus pilosus BCRC38072 genomic DNA. The putative monacolin K biosynthetic gene cluster was found within a 42 kb region in the mps01 clone. The deduced amino acid sequences encoded by the nine genes designated as mokA-mokI, which share over 54% similarity with the lovastatin biosynthetic gene cluster in A. terreus, were assumed to be involved in monacolin K biosynthesis. A gene disruption construct designed to replace the central part of mokA, a polyketide synthase gene, in wild-type M. pilosus BCRC38072 with a hygromycin B resistance gene through homologous recombination, resulted in a mokA-disrupted strain. The disruptant did not produce monacolin K, indicating that mokA encoded the PKS responsible for monacolin K biosynthesis in M. pilosus BCRC38072.
KEYWORDS: Monacolin K; polyketide synthases; Monascus pilosus; bacterial artificial chromosome INTRODUCTION
Monascus spp. are filamentous fungi that have been used in
Chinese fermented foods for thousands of years. They are known
as producers of various secondary metabolites with polyketide
structures, including monacolins, pigments, and citrinin (1–4).
Monacolin K, a cholesterol synthesis inhibitor, was first isolated
from the medium of Monascus ruber (1), and the same substance
was found in Aspergillus terreus as lovastatin (5). It belongs to
polyketide synthesized by the iterative type I PKSs. The
structure of monacolin K shares similarity with HMG-CoA;
therefore, monacolin K competitively inhibits HMG-CoA
re-ductase with HMG-CoA during cholesterol synthesis resulting
in the reduction of cholesterol synthesis (6).
In previous studies, the lovastatin biosynthetic pathway was
proposed in A. terreus. Two polyketide synthases (loVB and
loVF), transesterase (loVD), enoyl reductase (ER) (loVC), and
P450 monooxygenase (loVA) have been proven to be involved
in the structural biosynthesis of lovastatin (7–9). Transformation
of an extra copy of the loVE gene-encoded transcription factor
into the wild-type strain resulted in a 7-10-fold overproduction
of lovastatin (7). Although the lovastatin biosynthetic gene
cluster in A. terreus has been characterized (10), the structural
genes responsible for monacolin K (lovastatin) biosynthesis in
Monascus are still unclear. In the present study, to explore the
monacolin K biosynthetic gene cluster, construction of a
bac-terial artificial chromosome (BAC) library from M. pilosus
BCRC38072 producing monacolin K was carried out. According
to the conserved region of the loVB gene (lovastatin nonaketide
synthase, LNKS), A. terreus was designed as a probe (11), and
the mps01 clone containing the putative monacolin K
biosyn-thetic gene cluster was isolated. Analysis of the disruption of
the polyketide synthase gene (mokA) was conducted to identify
the gene involved in monacolin K biosynthesis.
MATERIALS AND METHODS
Strain Used and Growth Conditions. M. pilosus BCRC38072, which is a monacolin K-producing strain isolated from red rice (anka), was collected from a local traditional market and used in this study. To identify the transcripts from monacolin K biosynthetic genes, the strain was incubated on YM (DIFCO 271120, Detroit, MI) agar for 1 week, and spore suspensions were obtained by washing cultured YM agar plates with distilled water. Mycelia were harvested after incubation for 12 days at 25°C with constant agitation in liquid medium (7% glycerol, 3% glucose, 3% monosodium glutamate, 1.2% polypetone, 0.2% NaNO3, and 0.1% MgSO4· 7H2O).
BAC Library Construction. The methods of Peterson et al. (12) were used to construct the BAC library. Fragments of genomic DNA ranging in size from 150 to 300 kb were excised from pulse field gel electrophoresis (PFGE) and recovered by electroelution (BioRad, Hercules, CA). The eluted DNA was used for ligation. To perform the ligation, 100-200 ng of electroeluted DNA was mixed with 50 ng of linearized vector DNA (pIndigoBAC-5 HindIII Ready, Epicenter, Madison, WS), after which the ligation products were used to transform * To whom correspondence should be addressed. Tel:
+886-3-5223191ext. 580. Fax: +886-3-5224171. E-mail: address: gfy@ firdi.org.tw.
†
Food Industry Research and Development Institute.
‡National Chiao Tung University.
10.1021/jf800595k CCC: $40.75 2008 American Chemical Society Published on Web 06/26/2008
Escherichia coli strain TransforMax EC 100 electrocompetent
(Epi-center, Madison, WI) by electroporation. Transformed cells were cultured on LB agar plates supplemented with chloramphenicol (12.5
µg mL-1), IPTG (100 µg mL-1), and XGal (50 µg mL-1). The resulting white bacterial colonies were harvested and transferred to 384 well plates for library screening or storage at -80°C in freezing medium (2.5% [w/v] LB, 36 mM K2HPO4, 13.2 mM KH2PO4, 1.7 mM sodium
citrate, 0.4 mM MgSO4, 6.8 mM [NH4]2SO4, and 4.4% v/v
gly-cerol).
Library Screening, Sequencing, and Sequence Analysis. About 12000 clones from the BAC library were cultured on LB agar plates, after which they were transferred to nylon membranes and then subjected to alkali-sodium dodecyl sulfate lysis. The plasmid DNAs extracted from BAC clones were cross-linked to nylon membranes by UV irradiation. According to the conserved region of the ketosynthase domain of the loVB gene in A. terreus (10), the primer set (Mplov1, 5′-TCCACTGCCGTTTATGTTG-3′; Mplov2, 5′-GATGGGGTGAA-GATGACGA-3′) was designed for the probe synthesis using a PCR DIG probe synthesis kit (Roche Diagnostics, Mannheim, Germany). The clones of membranes were screened to find a gene cluster involved in polyketide biosynthesis metabolism. Twenty-five positive BACs were identified in screening by colony hybridization. Two of these, mps01 and mps02, containing the longest inserted DNA, were sequenced. BAC DNA for shotgun sequencing was extracted with a Qiagen Large-Construct kit (Qiagen, Valencia, CA) and sheared by sonication. The DNA fragments were blunted with the Bal31 nuclease and T4 DNA polymerase. Fragments were excised from the gel ranging in size from 1 to 2 kb and inserted into a pUC18/SmaI/CIAP (Amersham Pharmacia Biotech, Piscataway, NJ) vector. The ligation products were transformed into E. coli strain TransforMax EC 100 electrocompent (Epicenter, Madison, WS) by electroporation to construct the subclone library. Transformed cells were cultured on LB agar plates supplemented with ampicillin (50 µg mL-1), IPTG (100 µg mL-1), and XGal (50 µg mL-1). The resulting white bacterial colonies were harvested and transferred to 96 deep well plates for overnight culture in LB broth and ampicillin (50 µg mL-1). The inserts of these subclones were isolated and dissolved in 30 µL of TE. Cycle sequencing reactions were carried out using a BigDye, V3.0 kit with universal primers. DNA sequencing for 10-fold coverage was performed with an ABI Prism 3700 Sequencer (Applied Biosystems, Foster City, CA). The Phred-Phrap-Consed system developed by the Phil Green Laboratory was used to assemble DNA fragments (13, 14). Nucleotide and deduced amino acid sequences were used to interrogate the nonredundant database at GenBank using BLASTN and BLASTX. The BAC mps01 contained the complete monacolin K gene cluster instead of the incomplete mps02.
Nucleic Acid Manipulations. Fungal genomic DNA was isolated by liquid nitrogen treatment according to the method developed by Bingle et al. (15). Colony hybridization, Southern hybridization, and Northern hybridization were performed using the DIG system (DIG wash and buffer set) (Roche Diagnostics, Mannheim, Germany). The manipulations of transfer, immobilization, and hybridization of DNA and RNA were carried out as described in Sambrook et al. (16). Total RNAs of M. pilosus BCRC 38072 were isolated with TRIzol reagent (Invitrogen, Carlsbad, CA) for Northern hybridizations and reverse-transcription polymerase chain reaction (PCR) analyses. The probes of monacolin K biosynthetic genes were DIG-labeled by PCR amplification using the PCR DIG probe synthesis kit (Roche Diagnos-tics, Mannheim, Germany). The primer sets of monacolin K biosynthetic genes were as follows: pmkA-f, ATAGCTCCGAGAATGGTCCC, and pmkA-r, CCATCAAGGATGCTCTGTC; pmkB-f, CTAGACTTTGCT-TCCCACGCC, and pmkB-r, CATTGTCGAGCGTTGGAGTC; pmkC-f, TCAGAGATCTTCGTCCCGAC, and pmkC-r, GGCCTGAGCCGA-AGAAGTAC; pmkD-f, TGATGACTTTGCCCTGGCGG, and pmkD-r, TCACCCAATGACTCTAGCCC; pmkE-f, TTCTCTCCCGACAACT-GCCC, and pmkE-r, AATGGTCACCGCCGACTGGA; pmkF-f, GC-CCCGAATCCTACATGAAG, and pmkF-r, GGCCCACCGTAGTTGAT-GTG; pmkG-f, CCTCGCTCTGAATATGACCC, and pmkG-r, TCGGAT-CGGCTTCTCAAACC; pmkH-f, ACCTCATCGCTCCAGACCAT, and pmkH-r, CTGCGAGAGAATGAGAGTGC; and pmkI-f, CCATACAT-TCTACCTTGCGG, and pmkI-r, CTAGACTCGTTCATCGCGGC.
cDNA Analysis. cDNA sequencing of nine genes, designated as
mokA-mokI, was carried out to characterize their structure. First strand
cDNA was synthesized by the ImProm-II Reverse Transcription System (Promega, Madison, WI) and used as the template for PCR. Amplifica-tion of full-length or partial cDNAs was performed with several sets of oligonucleotide primers. Sequences analyses were performed using VectorNTI 9.0 (InforMax, Frederick, MD) software.
Targeted Gene Disruption of mokA. The human cytomegalovirus (CMV) promoter of plasmid pHygEGFP (BD Biosciences Clontech, Palo Alto, CA) was replaced with a 0.5 kb BglII-XhoI fragment of the heat shock protein 90 (hsp 90) promoter from M. pilosus BCRC38072 (GenBank accession no. DQ983312) to obtain the plasmid pMS-hsp. The hph cassette, a hygromycin B resistance gene, was flanked at the 5′-site by 2.0 kb and at the 3′-site by 3.1 kb, respectively, of the mokA gene to obtain the plasmid pMkAko. The transformant that had mokA replaced with pMkAko by homologous recombination was identified by Southern hybridization.
Transformation of M. pilosus BCRC38072. The conidia from a 1 week culture of M. pilosus were incubated in 100 mL of Vogel medium at 30 °C for 16-18 h. The mycelia were harvested on miracloth (Millipore, Bedford, MA) and washed in MA digestion solution (0.1 M maleic acid, pH 5.5, and 1.2 M (NH4)2SO4). The mycelia were
digested for 4-5 h using 100 mg of Yatalase (Takara, Tokyo, Japan), 100 mg of lysing enzyme (Sigma, St. Louis, MO), and 100 µL of
β-glucuronidase (Sigma, St. Louis, MO) in 50 mL of MA digestion
solution. To remove undigested mycelia, protoplasts were harvested by passing through miracloth and by centrifugation at 1000 rpm for 10 min (Sorvall, Wilmington, United States). The protoplasts were maintained in 80% STC (1 M sorbitol, 50 mM Tris, pH 8.0, and 50 mM CaCl2) and 20% PTC (40% PEG 4000, 50 mM Tris, pH 8.0, and
50 mM CaCl2), and dimethylsulfoxide (DMSO) was added to a final
concentration of 1%. For genetic transformation of M. pilosus, the 100
µL of protoplasts was mixed with 5 µg of linearized DNA. The mixtures
were incubated on ice for 30 min. One milliliter of PTC was added and mixed gently. Following incubation at room temperature for 20 min, the protoplast mixtures were added to 15 mL of SYP medium (1 M sorbitol, 0.1% yeast extract, 0.1% peptone, and 2% agar) that contained 60 µg hygromycin B/mL. Plates were incubated at 28°C, and the transformants were detected by PCR and Southern hy-bridization.
Measurement of Monacolin K. The aliquots of M. pilosus culture were cleared of cells and filtered through a 0.2 mm filter. The supernatants were analyzed by high-performance liquid chromatography (HPLC) performed on a Waters system (Waters, Milford, MA) fitted with a reverse-phase C18column (LichroCART 250-4, Rp-18e, 5 µm).
The HPLC parameters were as follows: solvent A, 0.1% phosphorus acid in water; solvent B, acetonitrile; 35% A and 65% B in 30 min; flow rate, 1.5 mL min-1; and detection by UV spectroscopy (Waters 600 pump and 996 pgotodiode array detector). The monacolin K purified from the cultivation of M. pilosus BCRC38072 was verified by mass and 1H NMR spectroscopic analyses. A standard monacolin K
compound (Sigma, St. Louis, MO) was used to confirm the analysis by HPLC.
Nucleotide Sequence Accession Number. The nucleotide sequence of the monacolin K biosynthetic gene cluster was submitted to GenBank under the accession number DQ176595.
RESULTS
Cloning of Monacolin K Biosynthetic Gene Cluster.
Studies on fungal polyketide biosynthetic genes indicate that
metabolites are largely synthesized by iterative multifunctional
polyketide synthase systems (17). Each PKS minimally carries
keto-synthase (KS), acyltransferase (AT), and acyl carrier protein
(ACP) domains to catalyze different modifications. To search
for the genes related to monacolin K biosynthesis, degenerate
primers designed according to the conserved region of the KS
domain of the loVB gene in A. terreus (11), were used to amplify
genomic DNA from M. pilosus BCRC38072. The candidate
PCR products were sequenced and analyzed. The result of the
amplified DNA showed that the PCR product shared a 75%
similarity with the KS domain of the loVB gene in A. terreus
(data not shown). This DNA fragment was further used to design
a specific probe for cloning of the PKS gene.
A BAC library consisting of 12000 clones was constructed
from the total DNA of M. pilosus BCRC38072. By screening
the library with the specific PKS probe, 25 positive BACs were
identified. To evaluate the sizes of the BACs, PFEG and
Southern hybridization were carried out. The BAC designated
as mps01 was selected for shotgun sequencing. It yielded a
contig of approximately 160 kb. Database searches and open
reading frame (ORF) prediction further provided information
on the putative gene loci. The whole sequences of mps01 were
annotated by BLASTN and BLASTX, and 30 ORFs were
predicted.
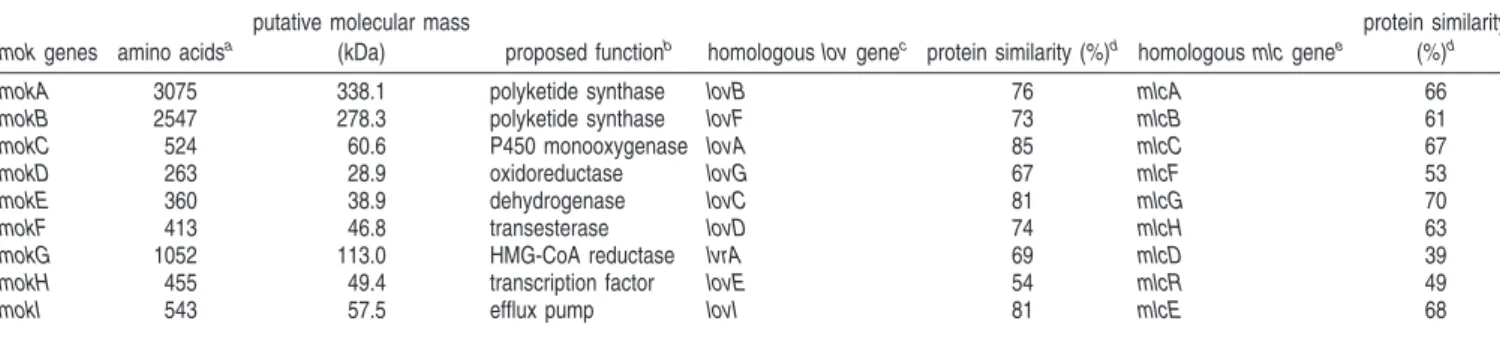
Identification of Genes Involved in Monacolin K
Biosyn-thesis. Within the 30 putative genes, nine genes were found to
have strong homology to the genes involved in the lovastatin
biosynthetic gene cluster (10), and they were presumed to
encode proteins required for monacolin K biosynthesis (Table
1). The putative gene cluster of monacolin K covered 42 kb
(Figure 1A) (GenBank accession no. DQ176595). Moreover,
they also shared high similarity with the genes involved in the
compactin biosynthetic gene cluster of Penicillium citrinum (18).
The structure of monacolin K differs from that of compactin,
in which a methyl group derived from S-adenosyl-
L-methionine
(SAM) is introduced at the C-6 position of the
nonaketide-derived backbone. An extensive comparison analysis of these
nine genes indicated the presence of two polyketide synthase
genes by BLAST. One was predicted to be responsible for the
synthesis of the nonaketide skeleton (mokA), while the other
was for the synthesis of the diketide skeleton (mokB). Also
included were a P450 monooxygenase gene (mokC), an
oxi-doreductase gene (mokD), a dehydrogenase gene (mokE), a
tran-sesterase gene (mokF), an HMG-CoA reductase gene (mokG),
a transcription factor gene (mokH), and an efflux pump gene
(mokI).
To assess the transcription of the predicted monacolin K
biosynthetic genes, Northern hybridizations were performed with
DIG-labeled probes. Transcripts of the putative monacolin K
biosynthetic genes could be detected on the eighth day (Figure
1B). The cDNA sequences were used to annotate the nine gene
sequences. For this, several sets of oligonucleotide primers were
designed to amplify cDNAs by reverse-transcription PCR. Only
the mokD gene revealed no intron, and others contained at least
one intron with sizes ranging from 52 to 109 bp. The deduced
amino acid sequences of the putative monacolin K biosynthetic
gene cluster were confirmed, and the sequence similarity among
corresponding genes of A. terreus and P. citrinum is shown in
Table 1. Several conserved domains were recognized in MokA
and MokB by comparing their amino acid sequences with those
of known PKSs. The domains of KS, AT, DH, MeT, KR, and
ACP are included in both MokA and MokB. Additionally,
MokB comprised an additional ER domain similar to the
corresponding gene loVF of A. terreus, as shown in Figure 1C.
The mokH gene-encoded transcription factor was suggested to
be a positive regulatory protein for the production of monacolin
K just like loVE, which is involved in lovastatin biosynthesis
in A. terreus (Figure 2) (10). The arrangement of a
cysteine-rich nucleotide-binding domain indicated that the consensus
sequence CX
2CX
6CX
11CX
2CX
6C represented a Zn
2Cys
6type
zinc finger (7, 10).
Polyketides are frequently synthesized from CoA
thioesteri-fied carboxylic acids, and the extent of keto-group processing
varies from one condensation cycle to another (7). In this study,
the phylogeny was further constructed according to the
con-served domain of KS (Figure 3A). The result showed that the
PKSs were divided into three clades. The clade of mokA and
mokB was subdivided into two subclades belonging to the
structural type of highly reduced polyketide. Because the MeT
domain in mlcA of the compactin biosynthetic gene cluster is
assumed to be inactive (18), the domain was compared among
corresponding PKSs from M. pilosus, P. citrinum, and A.
terreus, and our results showed that the amino acid residues of
mlcA in consensus motifs were different from those of the other
PKS, as boxed (Figure 3B).
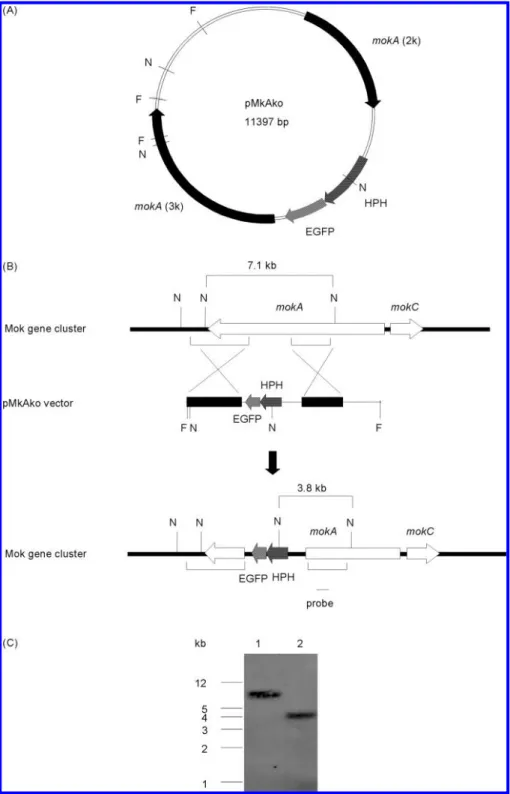
Disruption of mokA Gene in M. pilosus BCRC38072. The
mokA gene-encoded polyketide synthase was suggested to
synthesize the nonaketide of monacolin K. In this study, the
mokA gene was disrupted in M. pilosus BCRC38072 by
homologous recombination to identify the gene involved in
monacolin K biosynthesis. Plasmid pMkAko was linearized at
the FspI sites and was used to transform strain BCRC38072
(Figure 4A). Forty-four transformants were isolated, one of
which had a MokA
sgenotype by Southern hybridization.
Southern hybridization analysis of NdeI-digested DNA
in-dicated that instead of a 7.1 kb fragment corresponding to the
mokA gene in wild-type BCRC38072, a 3.8 kb NdeI fragment
was present in the disruptant BCRC38135 (Figure 4B,C). Thus,
our result revealed that disruption of mokA gene had occurred.
A precise gene replacement, in which the nonfunctional mokA
gene construct (pMkAko) replaced the functional chromosomal
mokA gene, yielded NdeI fragments of 3.8 kb. In addition, the
amount and structure of monacolin K produced from M. pilosus
BCRC38072 and BCRC38135 on the eighth day of cultivation
were further determined by HPLC, mass, and
1H NMR
spectroscopic analyses. Monacolin K was detectable in
wild-type M. pilosus BCRC38072, as confirmed by UV light
Table 1. Summary of Genes Identified in BAC mps01 Obtained from M. pilosus BCRC38072
mok genes amino acidsa
putative molecular mass
(kDa) proposed functionb homologous lov genec protein similarity (%)d homologous mlc genee
protein similarity (%)d
mokA 3075 338.1 polyketide synthase lovB 76 mlcA 66
mokB 2547 278.3 polyketide synthase lovF 73 mlcB 61
mokC 524 60.6 P450 monooxygenase lovA 85 mlcC 67
mokD 263 28.9 oxidoreductase lovG 67 mlcF 53
mokE 360 38.9 dehydrogenase lovC 81 mlcG 70
mokF 413 46.8 transesterase lovD 74 mlcH 63
mokG 1052 113.0 HMG-CoA reductase lvrA 69 mlcD 39
mokH 455 49.4 transcription factor lovE 54 mlcR 49
mokI 543 57.5 efflux pump lovI 81 mlcE 68
aThe deduced amino acid sequences were determined from cDNA sequences.bThe proposed gene functions were based on their homology to proteins in the GenBank database.cThe lovastatin biosynthetic gene cluster.dSimilarity was obtained by alignment using VectorNTI 9.0 (InforMax) software.eThe compactin biosynthetic gene cluster.
absorption, mass, and
1H NMR spectra (Figure 5). The peak
of monacolin K from M. pilosus BCRC38072 was identified
by comparison to the monacolin K standard, which showed three
maximum absorptions at (λ
max) 230, 237, and 246 nm (Figure
Figure 1. Identification of the monacolin K biosynthetic gene cluster of M. pilosus BCRC38072. (A) The genes involved in the biosynthesis of monacolin K. For proposed functions of assigned ORFs, see Table 1. The small black bars indicated the probes for Northern hybridization analysis. The lovastatin biosynthetic gene cluster in A. terreus was obtained from the GenBank database using the following accession numbers: AF141924, AF151722, and AF141925. The compactin biosynthetic gene cluster in P. citrinum was obtained from the GenBank database using the following accession number: AB072893. (B) Northern hybridization analyses. Total RNA isolated after 12 days of cultivation was blotted. Total RNA (6 µg per lane) was separated on 0.8% agarose gels by electrophoresis. The size of each transcript was estimated by comparison with markers of known size. (C) Arrangement of functional domains of genes encoding PKSs in the biosynthesis of monacolin K. DH, dehydratase; MeT, methyltransferase; KR, keto-reductase; and ACP, ACP.
Figure 2. Deduced amino acid sequences alignment of transcription factors from the mokH, lovE, and mlcR genes. The cysteine-rich nucleotide-binding domain represented a Zn2Cys6type zinc finger with the consensus sequence CX2CX6CX11CX2CX6C shown boxed.
5A). The mass spectrum of monacolin K revealed that the
molecular weight was 404, which also agreed with the standard
(C
24H
36O
5) (Figure 5B). Moreover, the structure of monacolin
K was further verified by
1H NMR spectrum [
1H NMR (400
Hz, CDCl
3): δ 5.98 (1H, d, J ) 10.0 Hz, H-5), 5.76 (1H, dd, J
) 6.4, 6.0 Hz, H-6), 5.50 (1H, d, J ) 2.8 Hz, H-4), 5.36 (1H,
dt, J ) 4.8, 3.2 Hz, H-1), 4.60 (1H, m, H-5
′), 4.33 (1H, m,
H-3
′), 2.72 (1H, dd, J ) 5.2, 4.8 Hz, H
ax-2′), 2.62 (1H, ddd,
J ) 3.6, 3.2, 2.4, H
eq-2′), 2.43 (2H, m, H-3), 2.38 (1H, m,
H-7), 2.36 (1H, m, H-2
′′), 2.27 (1H, dd, J ) 2.8, 2.8 Hz, H-8a),
1.98 (1H, m, H
eq-4′), 1.95 (2H, m, H-2), 1.89 (1H, m, H-6′),
1.72 (1H, m, H-8), 1.65 (1H, m, H
ex-4′), 1.63 (1H, m, H-3′′),
1.48 (1H, m, H-7
′), 1.42 (1H, m, H-3′′), 1.38 (1H, m, H-7′),
1.29 (1H, m, H-6
′), 1.11 (3H, d, J ) 7.2 Hz, H-2′′-CH
3), 1.06
(3H, d, J ) 7.2 Hz, H-3-CH
3), 0.88 (3H, d, J ) 7.2 Hz,
H-7-CH
3), 0.86 (3H, t, J ) 7.6 Hz, H-4
′′)]. However, the disruptant
M. pilosus BCRC38135 did not produce monacolin K, indicating
that the mokA gene is responsible for monacolin K biosynthesis
in M. pilosus BCRC38072 (Figure 5C).
DISCUSSION
Monacolin K, also known as lovastatin, is a polyketide used
to reduce serum cholesterol levels in humans. Over the past
years, it has become clear that polyketides are assembled in a
variety of mechanistically complex ways (7). Studies on the
lovastatin biosynthetic gene cluster of A. terreus have shown
18 putative ORFs based on the sequence alignment and
characterization of genetically related fungal strains (10).
Surprisingly, only nine genes in the BAC of M. pilosus
BCRC38072 have revealed high homology to the genes involved
in lovastatin biosynthetic gene cluster of A. terreus (Table 1).
Moreover, the genomic arrangement of monacolin K
biosyn-thetic genes in M. pilosus BCRC38072 has corresponded to the
lovastatin biosynthetic genes in A. terreus (Figure 1A). The
high homology between gene clusters of mok and loV implies
that the mok gene cluster was responsible for monacolin K
biosynthesis. To prove this, we disrupted the mokA
gene-encoded polyketide synthase from wild-type M. pilosus
BCRC38072. The phenotype of lost monacolin K productivity
in the disruptant BCRC 38135 indicates that the mokA gene
was essential for monacolin K production (Figure 5C).
In particular, these genes also showed significant homology
to genes identified in the compactin biosynthetic gene cluster
of P. citrinum (18). However, the genomic arrangement of
compactin biosynthetic genes was different from that of the
monacolin K or lovastatin biosynthetic gene clusters (Figure
1A). The order and direction of P450 monooxygenase (mokC),
polyketide synthase (mokA), oxidoreductase (mokD),
dehydro-genase (mokE), and transesterase (mokF) was the same in M.
pilosus, A. terreus, and P. citrinum, whereas the organization
of other genes of the compactin biosynthetic gene cluster was
different (10, 18). Furthermore, polyketide synthase (mokB),
monooxygenase (mokC), oxidoreductase (mokD), dehydrogenase
(mokE), and an efflux pump (mokI) appeared to have the same
number of introns and similar intron positions among M. pilosus,
A. terreus, and P. citrinum.
A. terreus and P. citrinum both belong to the family
Trichocomaceae, but they are different from M. pilosus, which
Figure 3. (A) Phylogenetic tree of PKSs from M. pilosus BCRC38072 and various organisms. The phylogeny of PKSs based on the conserved KS domains described by Kroken et al. (21) was constructed and rooted using KS domains of Saccharopolyspora erythraea DEBS (X56107 and X62569). Accession numbers for the polyketide synthase genes were used as follows: A. terreus lovF (AF141925), A. terreus lovB (AF151722), P. citrinum mlcA, mlcB (AB072893), Phoma sp. SQTKS (AY217789), Cochliobolus heterostrophus pks1 (U68040), Gibberella moniliformis FUM1 (AF155773), Monascus purpureus pksCT (AB167465), Emericella nidulans wA (X65866), Emericella nidulans stcA (AAC49191), Aspergillus parasiticus aflC (AY371490), Penicillium patulum 6-MSAS (X55776), Aspergillus parasiticus pksL2 (U52151), and A. terreus pksM (U31329). Bootstrap values were shown in the nodes according to 1000 replications. Only bootstrap values >50% are shown. The tree was constructed by the neighbor-joining method (23). (B) Comparison of the MeT domain. The three MeT consensus motifs were shown boxed. The conserved residues of MeT are described by Kagan and Clarke (24).
belongs to the family Monascaceae (19). Interestingly, lovastatin
biosynthetic genes from A. terreus revealed a higher homology
with monacolin K biosynthetic genes from M. pilosus than with
compactin biosynthetic genes from P. citrinum. Because
polyketides play an ecological role in the environment regarding
microbial competition, genetic differences might reflect extreme
environmental stress and subsequent genetic changes in these
species (20). In addition, many of the predicted PKSs in the
PKS clade that produce highly reduced polyketides have
di-vergent and presumably nonfunctional MeT domains (21). The
structure of monacolin K differs from that of compactin, in which
a methyl group derived from SAM is introduced at the C-6 position
of the nonaketide-derived backbone (18). The MeT domain in
mokA and loVB genes is assumed to be active instead of the mlcA
gene. The consensus motif of the MeT domain of mlcA was found
to be different from the relative PKSs (mokA and loVB) in some
amino acid residues, AfL, GfI, QMfHL, and IfT (Figure 3B).
Furthermore, there were two more introns located at the MeT
Figure 4. (A) Plasmid map of pMkAko for targeted gene disruption of mokA. (B) Disruption of the mokA gene in M. pilosus BCRC38072. The strategy for disrupting mokA gene was done by homologous recombination. A pMkAko vector containing the fusion protein of the hygromycin B resistance gene (HPH) with enhanced green fluorescent protein (EGFP) was flanked at the 5′-site by 2.0 kb (mokA2k) and at the 3′-site by 3.1 kb (mokA3k). pMkAko was linearized at the FspI sites and transformed into M. pilosus BCRC38072. The homologous recombination event between the Monascus genome and the FspI-digested pMkAko fragment resulted in a truncated ORF for mokA gene. (C) Southern hybridization analysis of disruption of mokA gene in the genomes of M. pilosus BCRC38072 (lane 1) and disruptant BCRC38135 (lane 2) hybridized with the probe indicated by a small black bar. The abbreviation F indicates an FspI restriction enzyme site, and N indicates an NdeI restriction enzyme site.
domain of mlcA, whereas mokA and loVB contained the same
number of introns and similar intron positions. Therefore, the
differences among amino acid residues could be the reason for
the lack of MeT activity of mlcA in P. citrinum. These results could
form the basis for the study of site-directed mutagenesis to
understand the MeT activity of PKSs (18). Among these genes
shown in Table 1, the transcription factor (mokH, loVE, and mlcR)
and HMG-CoA reductase (mokG, lVrA, and mlcD) were found to
have fewer similarities to each other. The number and positions
of introns were also different from one another. Nevertheless, the
transcription factor and HMG-CoA reductase genes were assumed
to be regulators responsible for up-regulation and down-regulation,
respectively (7, 22). HMG-CoA reductase (mokG) could play a
role in conferring resistance to monacolin K (22), and theoretically,
there was no effect upon the structure of the polyketides (7).
The data for the monacolin K biosynthetic gene cluster can
provide important information about the biosynthesis of
mo-nacolin K (lovastatin) between Monascus and Aspergillus. It is
interesting that polyketide synthases between mok genes and
loV genes are othologues and also related in compactin
biosyn-thesis by P. citrinum. Thus, there are three othologous gene
clusters in different fungi, which are useful to study evolution
of genes for secondary metabolism. Moreover, this suggests that
the structural variety of polyketides produced by fungi
ac-companies the enzymatic variation (7, 11). Studies on the
regulation of fungal secondary metabolism and the development
of novel polyketides have great potential for screening effective
medications.
ACKNOWLEDGMENT
Support of the Ministry of Economic Affairs (Taiwan, ROC)
(Grant 94-EC-17-A-17-R7-0563) to the Food Industry Research
and Development Institute (FIRDI) is appreciated.
LITERATURE CITED
(1) Endo, A. Monacolin K, a new hypocholesterolemic agent produced by a Monascus species. J. Antibiot. (Tokyo) 1979, 32, 852–854. Figure 5. Identification of monacolin K produced by M. pilosus BCRC38072. (A) UV light absorption spectrum of monacolin K between the wavelengths of 210 and 380 nm. (B) Liquid chromatography-mass spectroscopy analysis of monacolin K. m/z, mass-to-charge ratio. (C) HPLC analysis of monacolin K produced from M. pilosus BCRC38072 and BCRC38135 on the eighth day of cultivation.
(2) Endo, A.; Hasumi, K.; Nakamura, T.; Kunishima, M.; Masuda, M. Dihydromonacolin L and monacolin X, new metabolites which inhibit cholesterol biosynthesis. J. Antibiot. (Tokyo) 1985, 38, 321– 327.
(3) Endo, A.; Komagata, D.; Shimada, H. Monacolin M, a new inhibitor of cholesterol biosynthesis. J. Antibiot. (Tokyo) 1986,
39, 1670–1673.
(4) Shimizu, T.; Kinoshita, H.; Ishihara, S.; Sakai, K.; Nagai, S.; Nihira, T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. EnViron. Microbiol. 2005, 71, 3453–3457.
(5) Hendrickson, L.; Davis, C. R.; Roach, C.; Nguyen, D. K.; Aldrich, T.; McAda, P. C.; Reeves, C. D. Lovastatin biosynthesis in
Aspergillus terreus: Characterization of blocked mutants, enzyme
activities and a multifunctional polyketide synthase gene. Chem.
Biol. 1999, 6, 429–439.
(6) Tobert, J. A. Lovastatin and beyond: The history of the HMG-CoA reductase inhibitors. Nat. ReV. Drug DiscoVery 2003, 2, 517– 526.
(7) Hutchinson, C. R.; Kennedy, J.; Park, C.; Kendrew, S.; Auclair, K.; Vederas, J. Aspects of the biosynthesis of non-aromatic fungal polyketides by iterative polyketide synthases. Antonie Van
Leeu-wenhoek 2000, 78, 287–295.
(8) Sorensen, J. L.; Auclair, K.; Kennedy, J.; Hutchinson, C. R.; Vederas, J. C. Transformations of cyclic nonaketides by
Aspergil-lus terreus mutants blocked for lovastatin biosynthesis at the loVA
and loVC genes. Org. Biomol. Chem. 2003, 1, 50–59.
(9) Sorensen, J. L.; Vederas, J. C. Monacolin N, a compound resulting from derailment of type I iterative polyketide synthase function en route to lovastatin. Chem. Commun. 2003, 13, 1492–1493. (10) Kennedy, J.; Auclair, K.; Kendrew, S. G.; Park, C.; Vederas, J. C.;
Hutchinson, C. R. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 1999,
284, 1368–1372.
(11) Nicholson, T. P.; Rudd, B. A.; Dawson, M.; Lazarus, C. M.; Simpson, T. J.; Cox, R. J. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 2001,
8, 157–178.
(12) Peterson, D. G.; Tomkins, J. P.; Frisch, D. A.; Wing, R. A.; Paterson, A. H. Construction of plant bacterial artificial chromo-some (BAC) libraries: An illustrated guide. J. Agric. Genomics 2000, 5.
(13) Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186– 194.
(14) Gordon, D.; Abajian, C.; Green, P. Consed: A graphical tool for sequence finishing. Genome Res. 1998, 8, 195–202.
(15) Bingle, L. E. H.; Simpson, T. J.; Lazarus, C. M. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 1999, 26, 209–223. (16) Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A
Laboratory Manual; Cold Spring Harbor Laboratory Press: New
York, 1989, Vol. 1, p 7.39.
(17) Pfeifer, B. A.; Khosla, C. Biosynthesis of polyketides in heter-ologous hosts. Microbiol. Mol. Biol. ReV. 2001, 65, 106–118. (18) Abe, Y.; Suzuki, T.; Ono, C.; Iwamoto, K.; Hosobuchi, M.;
Yoshikawa, H. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium
citri-num. Mol. Genet. Genomics 2002, 267, 636–646.
(19) Kuraishi, H.; Itoh, M.; Katayama, Y.; Hasegawa, A.; Sugiyama, J. Ubiquinone systems in fungi. V. Distribution and taxonomic implications of ubiquinones in Eurotiales, Onygenales and the related plectomycete genera, except for Aspergillus, Paecilomyces,
Penicillium, and their related teleomorphs. Antonie Van Leeuwen-hoek 2000, 77, 179–186.
(20) Duffy, B.; Keel, C.; De´fago, G. Potential role of pathogen signaling in multitrophic plant-microbe interactions involved in disease protection. Appl. EnViron. Microbiol. 2004, 70, 1836–1842. (21) Kroken, S.; Glass, N. L.; Taylor, J. W.; Yoder, O. C.; Turgeon,
B. G. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci.
U.S.A. 2003, 100, 15670–15675.
(22) Abe, Y.; Suzuki, T.; Mizuno, T.; Ono, C.; Iwamoto, K.; Hosobu-chi, M.; Yoshikawa, H. Effect of increased dosage of the ML-236B (compactin) biosynthetic gene cluster on ML-ML-236B pro-duction in Penicillium citrinum. Mol. Genet. Genomics 2002, 268, 130–137.
(23) Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. EVol. 1987, 4, 406–425.
(24) Kagan, R. M.; Clarke, S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltrans-ferases suggests a common structure for these enzymes. Arch.
Biochem. Biophys. 1994, 310, 417–427.
Received for review February 27, 2008. Revised manuscript received May 9, 2008. Accepted May 14, 2008.