Purification and characterization of two cellulase free xylanases from
an alkaliphilic Bacillus firmus
Min-Jen Tseng
a,*, Mee-Nagan Yap
b, Khanok Ratanakhanokchai
c, Khin Lay Kyu
c,
Shui-Tein Chen
baGraduate Institute of Cell and Molecular Biology, Taipei Medical University, Taipei, Taiwan bInstitute of Biological Chemistry, Academia Sinica, Taipei, Taiwan
cSchool of Bioresources and Technology, King Mongkut’s University of Technology, Thonburi, Bangkok, Thailand Received 27 November 2000; received in revised form 3 September 2001; accepted 1 October 2001
Abstract
Two xylanases from Bacillus firmus were purified to homogeneity by gel filtration and ion-exchange chromatography. These enzymes have molecular weights of 45 kDa and 23 kDa, respectively, and both show enzymatic activity over the pH range of 5.0 –11.0 at 37°C. These enzymes hydrolyzed xylan from birchwood to release mainly the products of xylose, xylotriose and xylohexose, thus indicating that the xylanases act preferentially toward the internal glycosidic bonds of xylo-oligosaccharides. However, the two xylanases show different modes of action, and a combination of both is likely to lead to concerted degradation of xylan down to the mono- and disaccharides. © 2002 Elsevier Science Inc. All rights reserved.
Keywords: Xylanases; Bacillus firmus; Enzyme purification; Xylanase action
1. Introduction
Xylan is the most abundant of the hemicelluloses which are heteropolysaccharides having a chain of -1,4-linked xylopyranose residues. The complete hydrolysis of xylan requires the combined action of various enzymes such as endoxylanase (EC 3.2.1.8), exoxylanase (-D-xylan xylo-hydrolase), and-D-xylosidase (EC 3.2.1.37) etc. [1]. Xy-lanases randomly hydrolyze the -1,4-glycosidic bonds of xylan to produce several xylo-oligomers. In recent years, xylanases have received attractable research interest due to their potential industrial applications [2]. However, such applications require xylanase(s) with particular properties, the bio-bleaching of paper pulp requires a xylanase that remains active even above pH 9.0 and lacks all cellulase activity.
The bacterium used in this study, Bacillus firmus, was previously isolated from a wastewater treatment plant of pulp and paper industry at Bang-Pre-In at Prankornsriayut-taya province, Thailand [3,4]. Bacillus firmus is capable of
growth at pH values, ranging from 10 –12. Normally growth is at temperature of 37°C; the cultures were thermolabile at temperatures above 55°C. Moreover, this strain produces two major extracellular xylanases, with molecular weights of 45 kDa and 23 kDa, respectively. No other hemicellu-lose-degrading enzyme activities were detected in the cul-ture medium with xylan, CMC or avicel as the sole carbon source, suggesting that this strain produces xylanase mainly. The mode of action of xylanase and cellulase has been reported. It was considered that the cellulose/xylan binding domain (CBD/XBD) was an important factor in the degra-dation process of insoluble cellulosic materials [5,6]. For xylanases the concept of substrate recognition and induc-tion, i.e. the control of transcription by the cooperative actions of an activator and a repressor, have generally been accepted. Xylanase synthesis is induced by natural xylan and other -1,4-xylo-oligosaccharides isomers [7,8].
In this study, we describe the purification and character-ization of two major xylanases from Bacillus firmus bacte-ria. These enzymes are active over a wide range of pH. In addition other properties presented here suggest that these xylanases could be of commercial interest.
* Corresponding author. Tel: 27361661; fax: ⫹886–2-23778620.
E-mail address: cmbtseng@tmu.edu.tw (M.-J. Tseng).
0141-0229/02/$ – see front matter © 2002 Elsevier Science Inc. All rights reserved. PII: S 0 1 4 1 - 0 2 2 9 ( 0 1 ) 0 0 0 1 8 - 2
2. Materials and methods
2.1. Bacterial strain and culture conditions
The bacterial strain, used in this study, was isolated from a wastewater treatment plant of a pulp and paper manufac-turer [3,4] and identified by the Micro-IS System as Bacillus firmus. It was grown at 37°C in Berg’s mineral salts me-dium [9] supplemented with 0.2% NaNO3, 0.1% yeast
ex-tract, 0.05% K2HPO4, 0.02% MgSO4.7H2O, 0.002%
MnSO4.H2O, 0.002% FeSO4.7H2O, 0.002% CaCl2.2H2O
and 0.5% birchwood xylan (⬎90% xylose residues, Sigma Chemical Co., St. Louis, Mo.). The pH was initially ad-justed to 10.5 with 1% Na2CO3.After 4 days of incubation,
the culture supernatant was recovered by centrifugation at 8,000 rpm for 15 min.
2.2. Purification of xylanases
All steps were performed at 4°C unless otherwise noted. Solid ammonium sulfate was added to the culture superna-tant to 65% saturation. The precipitate was recovered by centrifugation and dissolved in an appropriate volume of 10 mM Tris-HCl (pH 8.0). The final solution was dialyzed three times against 5 liters of the same buffer for overnight standing. The dialyzed solution was applied to a DEAE-Toyopearl 650F column (1.6 by 25 cm) previously equili-brated with 10 mM Tris-HCl buffer. The column was first washed with 140 ml of 10 mM Tris-HCl buffer and then eluted with a linear gradient of 0 to 0.5 M NaCl in 500 ml of the same buffer at a flow rate of 1 ml/min. The eluted fractions with xylanase activity were pooled, dialyzed to remove any salt contaminants and lyophilized into powder. This partially purified protein was dissolved in 2 ml of 10 mM Tris-HCl buffer and applied onto a TSK-Fractogel 55F column (2.6 by 140 cm). Elution was carried out with the same buffer containing 0.3 M of NaCl
2.3. Xylanase assay
The assay mixture consisted of 40l of enzyme solution and 160l of a 0.5% birchwood xylan suspension in 100 mM Na2CO3-NaHCO3 buffer (pH 9.0). The reaction was
incubated at 37°C for 10 min and 0.4 ml of DNS reagent (1% dinitrosalicylic acid, 0.2% phenol, 0.05% sodium sul-fite, 1% sodium hydroxide) [10] was added to stop the reaction, then boiled for 5 min. The absorbance at 500 nm was measured after adding 2.4 ml of water. One unit was defined as the amount of enzyme required to produce re-ducing sugars equivalent to 1mol of xylose per min under the conditions described above; 1 U corresponds to 16.7 nkat in Systeme International d’ Unites units [11].
2.4. Effect of pH on activity of xylanases
The pH values of various reaction solution were adjusted with 100 mM of following buffer systems: acetate buffer (pH 4.0 to 5.5), phosphate buffer (pH 6.0 to 6.5), Tris-HCl buffer (pH 7.0 to 9.0), glycine-NaOH buffer (pH 10.0 to 11.4), and Na2HPO4-NaOH buffer (pH 11.0 to 12.0). The
substrate, 0.5% birchwood xylan in various pH buffer, was incubated with 0.5 U of 45 kDa xylanase or 0.15 U of 23 kDa xylanase for 30 min at 37°C and 0.4 ml of DNS reagent was added to stop the reaction, and then boiled for 5 min. The absorbance at 500 nm was measured after adding 2.4 ml of water.
2.5. Enzymatic hydrolysis of xylan
Enzymatic hydrolysis of birchwood xylan was per-formed according to the method of Matte and Forsberg [12]. Separation of the products of hydrolysis was performed by isocratic gradient of acetonitrile:water (78:22) in a HPLC system equipped with Microsorb Amine column and a RI detector 8110. Further characterization of the collected products were carried out by mass spectrometry analysis and monitored by xylo-oligomer standards.
2.6. Zymogram analysis for xylanase activity
This analysis was performed by the method of Morag et al. with slightly modification [6]. Samples were subjected to electrophoresis on a SDS-PAGE containing 0.1% xylan. After electrophoresis, the gel was washed three times for 30 min at 4°C in 100 mM Na2CO3-NaHCO3 buffer (pH 9.0)
containing 25% isopropanol for the first two washes to remove SDS, then incubated in the same buffer for 10 min at 37°C. The zymogram was prepared by soaking the gel in 0.1% Congo red solution for 15 min at room temperature, then washed with 1 M NaCl and introduced 0.5% acetic acid to expose two xylanase active bands that contrasted the dark background
2.7. General protein techniques
Protein concentration was estimated by the method of Brad-ford [13] using bovine serum albumin as a standard.
SDS-Table 1
Summary of purification of xylanases from Bacillus firmus
Source Specific activity
(U/mg protein)
Purification
Crude filtrate 1.75 1
45 kDa xylanase 34.08 19.5
23 kDa xylanase 3.67 2.1
Unit definition: 1 U was defined as the amount of enzyme which produced reducing sugars equivalent to 1mol of xylose per minute under the conditions described.
PAGE was performed by the method of Laemmli [14]. Iso-electric focusing was done on PhastGEL IEF (pH 3 to 9) by automatically controlled of PhastSystem (Pharmacia). Purified xylanases were subjected to N-terminal microsequencing using automated Edman degradation in an Applied Biosystem model 467A sequencer under standard conditions.
3. Results and discussion
3.1. Purification of xylanases
After 4 days of cultivation of Bacillus firmus in an alkaline medium with the presence of birchwood xylan as the sole carbon source, the extracellular xylanase was de-tected at 1.75 U/mg protein in the culture supernatant (Table 1). The crude extract from the culture medium was taken through the two-step purification of ion-exchange chroma-tography and gel filtration. Two purified xylanase proteins appeared as single bands on SDS-PAGE and had the mo-lecular masses of 45 kDa and 23 kDa, respectively (Fig. 1). Both proteins also showed relatively clear bands on the zymogram gel, detected by Congo red staining, indicating they were active xylanases (Fig. 1). However, the xylanase activity band for purified 23 kDa protein was very faint. A summary of the purification is presented in Table 1. Final purification of the xylanases increased their specific
activi-Fig. 1. (A) SDS-PAGE analysis of xylanases from Bacillus firmus. Lane 1, molecular mass markers: phosphorylase b, 94 kDa; albumin, 67 kDa; ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa; trypsin inhibitor, 20.1 kDa;␣-lactalbumin, 14.4 kDa. Lane 2, 45 kDa xylanase (10 g); lane 3, 23 kDa xylanase (10 g). (B) Zymogram of the corresponding clear band showing xylanase activity. Lane 1, crude filtrate (50g); lane 2, 45 kDa xylanase (10g); lane 3, 23 kDa xylanase (10 g).
Fig. 2. IEF-PAGE of purified xylnase from Bacillus firmus. Lane 1, 45 kDa xylanase; lane 2, 23 kDa xylanase; lane M, pI markers.
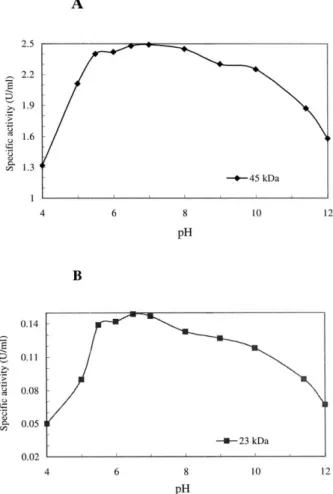
Fig. 3. The enzymatic activity of the purified 45 kDa (A) and 23 kDa (B) xylanases measured under different pH conditions. The reaction pHs were adjusted to 4.0 to 12.0 with various buffers at 100 mM. The substrate was incubated with purified enzymes for 30 min at 37°C. For the assay of 45 kDa xylanase each reaction contained 0.5 U of enzyme which correspond-ing to 14g of protein. The amount of 23 kDa xylanase used in each reaction was 0.15 U which corresponding to 40g of protein.
ties considerably, to 34.08 and 3.67 U/mg protein for the 45 kDa and 23 kDa xylanases, respectively. The former showed an activity that appeared to be 10-fold higher than the latter. This may also reflect the barely detected band on the zymogram gel for the 23 kDa xylanase (Fig. 1)
3.2. General properties of xylanases
Purified xylanases were homogeneous when further ex-amined by isoelectrofocusing (IEF)(Fig. 2). The 45 kDa xylanase had a pI value of 5.8 whereas a pI value of 6.8 for the 23 kDa xylanase.
The activities of both xylanases at various pH values were measured by using birchwood xylan as the substrate. The reaction pHs were adjusted to 4.0 to 12.0 with various buffers at 100 mM. Both xylanases showed enzyme activity over a broad pH range of 4.0 to 12.0 at 37°C (Fig. 3). The pH optimum is at 6 – 8 for 45 kDa xylanase and at 6 –7 for 23 kDa xylanase, respectively. The 45 kDa xylanase was slightly more alkaline resistant than that of 23 kDa enzyme with higher relative activity remained at pH 11. The pH profile of the purified enzymes indicated that the xylanase activities of both proteins remained considerable in the alkaline pH range. The properties of these two enzymes may be advantageous in the application of prebleaching of kraft pulps. 3.3. Analysis n-terminal sequences
The N-terminal amino acid sequences of the purified xylanases were compared with protein sequence data avail-able at National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) and European Bioin-formatics Institute (EBI, http://srs.ebi. ac.uk/) databases
us-Fig. 4. The N-terminal sequences of the purified xylanases compared with those of other xylanases. Ba. C-125, xylanase A from Bacillus sp. C-125 [19]; Ba. NG-27, an alkaline thermostable xylanase from Bacillus sp. NG-27 [20]; Ba. st., xylanase A from Bacillus stearothermophilus [21]; Ch. gr., a xylanase from Chaetomium gracile [22]. The partial sequences of the xylanases were aligned relatively to (A) 45 kDa and (B) 23 kDa xylanase to indicate identical amino acid residues. The numerical number showed the position of amino acid residue in its entire sequence. The X and “⫹” symbols represent undecided and conserved amino acid residues, respectively.
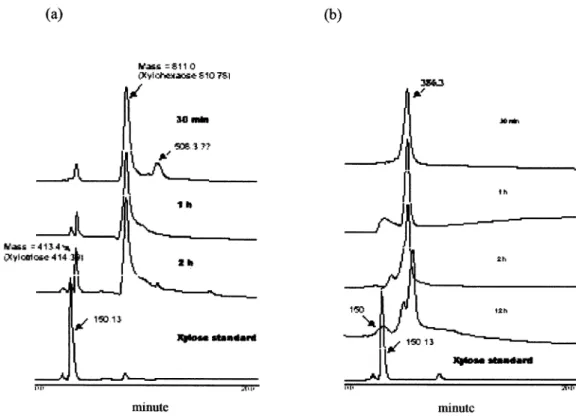
Fig. 5. Hydrolysis of xylan with purified 45 kDa (a) and 23 kDa (b) xylanase. The reaction mixture of 10 ml containing 1 mg of enzyme and 0.2 g birchwood xylan in 100 mM Tris-HCl buffer (pH 8.5), and incubated at 37 C for indicated time. The products were analyzed using a Microsorb Amine column with acetonitrile: water (78:22) as a mobile phase and were measured in the eluent by using a Waters refractive index (RI) detector. The molecular mass and identity of each product were determined by a mass spectrometer.
ing the on-line BLAST network service. The sequencing data of 45 kDa xylanase showed ⬎50% identical with the N-terminal amino acid sequences of other microbial xyla-nases as well as that for 23 kDa xylanase, as shown in Fig. 4. The sequencing data of 23 kDa xylanase also shown good homology with the N-terminal amino acid sequences fol-lowing signal peptides of other 23 kDa xylanases from Bacillus sp. YA-14 (8/14 identical) [15], Bacillus subtilis (8/14 identical) [16] and Bacillus circulans (8/14 identical) [17](Data not shown). Note that the xylanases which showed high homology with the 23 kDa xylanase of B. firmus all belong to family 11 glycosyl hydrolases as clas-sified by Gilkes et al. [18]. This result indicates that the two purified xylanases possess at least a common signal peptide of 28 –35 amino acid residues long, as shown by the se-quence comparison and the similarity of molecular weights of the xylanases from the compared bacteria. We, therefore, conclude that the purified enzymes were indeed xylanases. 3.4. Hydrolysis of native xylan
In order to better understand the mode of action of the purified xylanases, the hydrolytic products of the insoluble birchwood xylan (Sigma) incubated with xylanases were identified by xylose standards and mass spectrometry. As expected, both xylanases effectively hydrolyzed xylan but showed different modes of action. The hydrolysis by 45 kDa xylanase produced mainly the xylotriose (mass ⫽ 413.4) and xylohexaose (mass ⫽ 811). Under prolonged incuba-tions, a trace amount of xylose (mass⫽ 150) was detected (Fig. 5a). Whereas the 23 kDa xylanase even after long period of incubation produced only an oligosaccharide de-rivative of molecular mass of 386.3 (Fig. 5b) which is presumed to be that of a xylooligosaccharide with side chains. However, there is a trace amount of xylose appeared after 12 h of incubation with 23 kDa xylanase. These data indicating that these purified xylanases were both endoxy-lanases that randomly cleave xylan as a substrate. It was interesting that co-incubation of the two xylanases with the substrate resulted in the releasing of xylose, xylobiose and an unknown product with a molecular mass of 375.3 (Fig. 6). The production of xylose also shown to be increasing with time. This result is quite different from those of indi-vidual action of the xylanases, suggesting a cooperative relationship of the two xylanases in the degradation of the polysaccharide into simple oligosaccharides.
4. Conclusion
Two xylanases with molecular masses of 45 kDa and 23 kDa, respectively, were purified from the culturing medium of an alkalophilic Bacillus firmus which was isolated from a wastewater treatment plant of pulp and paper industry. Both enzymes showed a broad pH activity profile and relatively high activity under alkaline conditions toward birchwood
xylan. Both xylanases had endo character on hydrolyzing substrate but showed different modes of action. They coop-eratively released simple sugars of xylose, xylobiose and an unknown product with a molecular mass of 375.3 that was quiet different from the hydrolyzing pattern of individual xylanase. Thus, the crude xylanases extract from this bac-terium may potentially applicable in enzymatic hydrolysis of xylan especially in kraft pulp prebleaching process.
Acknowledgments
We thank Dr. G. Robert Greenberg for insightful com-ments of this manuscript. This work was supported by National Science Council of Taiwan (to M.-J. T., S.-T. C.) and Taipei Medical University (to M.-J. T.).
References
[1] Coughlan MP. Xylans and Xylanases. In: Visser J, Beldman G, Kusters-van Someren MA, Voragen AGJ, editors. Progress in Bio-technology, vol 7. Amersterdam: Amsterdam. 1992. p. 111–39. [2] Sunna A, Antranikian G. Xylanolytic enzymes from fungi and
bac-teria. Crit Review Biotechnol 1997;17:39 – 67.
Fig. 6. Hydrolysis of birchwood xylan by purified 45 kDa and 23 kDa xylanases in parallel. The reaction mixture of 10 ml containing 45 kDa:23 kDa:xylan⫽ 1 mg:1 mg:100 mg in 100 mM Tris-HCl buffer (pH 8.5) was incubated at 37°C for the indicated times. Products were analyzed using a Microsorb Amine column with acetonitrile:water (78:22) as a mobile phase and measured as in Fig. 5.
[3] Ratanakhanokchai K, Poonpium P, Kyu KL. Cellulosome structure of thermophilic cellulolytic and alkaliphilic xylanolytic microorganisms. Nakhon Tarchasima, Thailand: The 9th. annual meeting of the Thai Society for Biotechnology at Suranaree University of Technology, 1997. [4] Ratanakhanokchai K, Kyu KL, Tanticharoen M. Purification and properties of a xylan-binding endoxylanase from alkaliphilic Bacillus sp. strain K-1. Appl Environ Microbiol 1999;65:694 –7.
[5] Bajpai P. Application of enzymes in the pulp and paper industry. Biotechnol Prog 1999;15:147–57.
[6] Morag E, Edward AB, Lamed R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocelium to cellulose-degrading enzymes. J Bacteriol 1990;172:6098 –105.
[7] Biely P, Petrakova E. Novel inducers of the xylan-degradation en-zyme system of Cryptococcus albidus. J Bacteriol 1984;160:323– 6. [8] Biely P, Vrsanska M, Kratky Z. Xylan-degrading enzymes of the yeast Crytococcus albidus: Identification and cellular localization. Eur J Biochem 1980;108:313–21.
[9] Berg B, Hofstan BV, Peterson G. Growth and cellulase formation by Cellvibrio fulvus. J Appl Bacterio 1972;35:201–14.
[10] Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 1959;31:426 – 8.
[11] Baily MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 1992;23:257–70.
[12] Matte A, Forsberg CW. Purification, characterization, and mode of action of endoxylanases 1 and 2 from Fibrobacter succinogenes S85. Appl Environ Microbiol 1992;58:157– 68.
[13] Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248 –54.
[14] Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680 –5.
[15] Yu JH, Park YS, Yum DY, Kim JM, Komg IS, Bai DH. Nucleotide sequence and analysis of a xylanase gene (xynS) from alkali-tolerant Bacillus sp. YA-14 and comparison with other xylanases. J Microbiol Biotechnol 1993;3:139 – 43.
[16] Wolf M, Geczi A, Simo O, Borriss R. Genes encoding xylan and beta-glucan hydrolysing enzymes in Bacillus subtilis: characteriza-tion, mapping and construction of strains deficient in lichenase, cel-lulase and xylanase. Microbiology 1995;141:281–90.
[17] Yang RC, MacKenzie CR, Narang SA. Nucleotide sequence of a Bacillus circulans xylanase gene. Nucleic Acids Res 1988;16:7187. [18] Gilkes NR, Benrussat B, Kilbum DG, Miler RC, Warren RAJ. Do-mains in microbial-1,4-glycanases: sequence conversion, function, and enzyme families. Microbiol Rev 1991;55:303–15.
[19] Hamamoto T, Honda H, Kudo T, Horikoshi K. Nucleotide sequence of the xylanase A gene of alkalophilic Bacillus sp. strain C-125. Agric Biol Chem 1987;51:953–5.
[20] Gupta N, Ghosh A. Gene cloning, expression and sequence anal-ysis of gene encoding novel alkali stable thermostable endoxyla-nase from alkalophilic Bacillus sp. NG-27. Accession number AF015445, 1997.
[21] Cho S, Choi Y, Nucleotide sequence analysis of an endo-xylanase gene (xynA) from Bacillus stearothermophilus. J Microbiol Biotech-nol 1995;5:117–24.
[22] Yoshino S, Oishi M, Moriyama R, Kato M. Tsukagoshi N. Two family G xylanase genes from Chaetomium gracile and their expres-sion in Aspergillus nidulans. Curr Genet 1995;29:73– 80.