Enhancement of gene transactivation activity of androgen receptor by
hepatitis B virus X protein
Yanyan Zheng
a, Wen-ling Chen
a, W.-L. Maverick Ma
b,
Chawnshang Chang
b, J.-H. James Ou
a,⁎
aDepartment of Molecular Microbiology and Immunology, University of Southern California, Keck School of Medicine, 2011 Zonal Avenue,
HMR-401, Los Angeles, CA 90033, USA
bGeorge Whipple Lab for Cancer Research, Department of Pathology, Urology, Radiation Oncology and the Cancer Center,
University of Rochester Medical Center, Rochester, NY, USA
Received 8 January 2007; returned to author for revision 22 January 2007; accepted 30 January 2007 Available online 28 February 2007
Abstract
Hepatitis B virus (HBV) X protein (HBx) is a regulatory protein that is required for efficient replication of HBV in its natural host. In this report, we demonstrate by co-immunoprecipitation experiments that HBx can physically bind to the androgen receptor (AR), which is a nuclear hormone receptor that is expressed in many different tissues including the liver. This observation is further supported by confocal microscopy, which reveals that HBx can alter the subcellular localization of the AR both in the presence and in the absence of dihydrotestosterone (DHT). Further studies indicate that HBx can enhance the gene transactivation activity of AR by enhancing its DNA binding activity in a DHT-dependent manner. However, HBx does not remain associated with AR on the DNA. As AR can regulate the expression of a number of cellular genes, our results raise the possibility that HBV pathogenesis may be mediated in part via the interaction between HBx and AR.
© 2007 Elsevier Inc. All rights reserved.
Keywords: Hepatitis B virus; HBV X protein; Androgen receptor; Dihydrotestosterone; Gene transactivation; Hepatocellular carcinoma; Chromatin immunoprecipitation
Introduction
Hepatitis B virus (HBV) is an important human pathogen that can cause severe liver diseases including hepatocellular carcinoma (HCC). This virus has a small DNA genome that contains four genes: the S gene encodes the viral envelope proteins; the C gene encodes the viral core protein and a serum e antigen; the P gene encodes the viral DNA polymerase; and the X gene encodes a 16.5-kDa regulatory protein with multiple functions.
The HBV X protein (HBx) does not bind to DNA directly
(Yen, 1996). However, it can bind to transcription factors
including CREB, ATF-2, AP-2 and HNF1 to modify their activities (Li et al., 2002; Maguire et al., 1991; Seto et al., 1990;
Williams and Andrisani, 1995). It can also bind to RBP5, a
subunit of all three mammalian RNA polymerases (Cheong et al., 1995), to the proteasome (Fischer et al., 1995; Hu et al., 2006), p53 (Wang et al., 1994), a component of the DNA repair complex UV-DDB (Becker et al., 1998), and a member of the human voltage-dependent ion channel family HVDAC3 (Rahmani et al., 2000). HBx can also stimulate cellular calcium signaling pathways, resulting in the activation of focal adhesion kinase (FAK), prolin-rich tyrosine kinase 2 (Pyk2) and Src, and the downstream mitogen-activated protein kinase (MAPK) signaling pathway and the cyclin A–cdk2 complex (Bouchard
et al., 2001, 2006; Klein and Schneider, 1997). HBx can also
activate NFκB (Su et al., 2001). Studies on the related woodchuck hepatitis B virus (WHV) indicate that the X gene is required for efficient replication of WHV in woodchucks (Chen et al., 1993; Zhang et al., 2001; Zoulim et al., 1994). Further studies using transgenic mice carrying the entire HBV genome demonstrate that HBx can enhance HBV gene expression and the subsequent viral DNA replication (Xu et al., 2002).
⁎ Corresponding author. Fax: +1 323 442 1721.
E-mail address:jamesou@hsc.usc.edu(J.-H. James Ou).
0042-6822/$ - see front matter © 2007 Elsevier Inc. All rights reserved. doi:10.1016/j.virol.2007.01.040
HBV can induce HCC, which is male preponderant, suggesting a possible role of androgen and/or its receptor in HBV pathogenesis. Androgen receptor (AR) is a member of the nuclear receptor superfamily. This protein is approximately 110 kDa in size. It contains several functional domains including the ligand binding domain, the DNA binding domain, the transcription modulation domain and a nuclear localization signal (Lee and Chang, 2003). After binding to its ligand testosterone or
5α-dihydrotestosterone (DHT), AR is activated and transported into the nucleus where it binds to the promoters of its target genes to activate their expression (Lee and Chang, 2003). AR is expressed in many different tissues including the liver and has a wide range of activities. In the liver, it can upregulate the expression of transforming growth factorβ1 (TGF-β1), sex-limited protein (Slp) and various drug- and steroid-metabolizing enzymes including specific cytochrome P450 isozymes (Chatterjee et al., 1996; Yoon
et al., 2006). AR is believed to play an important role in
hepatocellular carcinogenesis, as there is an increased risk for hepatocellular carcinoma (HCC) in patients treated with androgens (Touraine et al., 1993), and a positive correlation between the HCC risk and the serum testosterone level (Tanaka et al., 2000; Yu et al.,
2000, 2001). Furthermore, a lower number of the CAG
trinucleotide repeats in exon 1 of the AR gene, which increases the gene transactivation activity of AR (Beilin et al., 2000; Chamberlain et al., 1994; Irvine et al., 2000), is also associated with an increased risk for HCC (Yu et al., 2000, 2001).
In this report, we have investigated the possible interaction between the HBV regulatory protein HBx and AR. Our results indicated that HBx could bind to and enhance the gene transactivation activities of AR in the presence of DHT. This enhancement of the AR activity by HBV in a DHT-dependent manner may explain the gender difference of HCC incidence among HBV carriers.
Results
Co-immunoprecipitation of HBx and AR
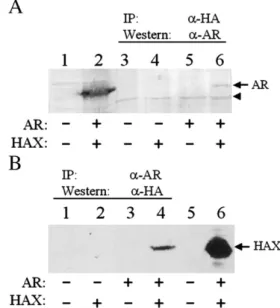
To investigate whether HBx and AR can bind to each other, we performed the co-immunoprecipitation experiments. Huh7 cells, which are human hepatoma cells that do not express androgen receptor (Xie et al., 2001), were co-transfected with the AR expression plasmid and the expression plasmid for HA-tagged HBx. Cells were lysed 48 h after transfection and immunoprecipitated with the anti-HA antibody, followed by the Western blot analysis using the anti-AR antibody. As shown in Fig. 1A, AR could be immunoprecipitated by the anti-HA antibody in the presence of HA-tagged HBx (lane 6) but not in the absence of it (lane 5). This result indicated that AR and HBx could physically bind to each other. This result was confirmed by a reciprocal experiment, in which the cell lysates were first immunoprecipitated with the AR anti-body followed by the Western blot analysis using the anti-HA antibody. As shown inFig. 1B, the HA-tagged protein could be immunoprecipitated by the anti-AR antibody only in the presence of AR (lane 4) and not in the absence of it (lane 2), confirming that HBx and AR could bind to each other.
Co-localization of HBx and AR in cells
To further investigate how HBx and AR may interact with each other in the cell, we also performed the confocal microscopy. Huh7 cells were first transfected with either the HA-tagged HBx expression plasmid or the AR expression plasmid. As shown in
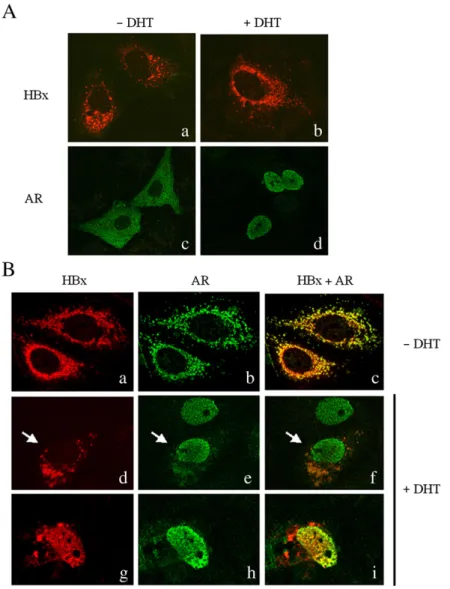
Fig. 2A, HBx displayed a predominantly cytoplasmic
localiza-tion whether or not the cells were treated with dihydrotestoster-one (DHT) (panels a and b). The staining pattern of HBx in the cytoplasm was granular, which is consistent with the previous report (Rahmani et al., 2000). In contrast, AR displayed a more diffuse cytoplasmic localization in the absence of DHT (panel c). In agreement with the previous report (Tomura et al., 2001), AR was localized predominantly to the nucleus after cells were treated with DHT (panel d).
If HBx and AR can indeed bind to each other in the cell, they will likely have similar subcellular localizations when they are expressed in the same cell. For this reason, we also co-transfected Huh7 cells with the HA-tagged HBx expression plasmid and the AR expression plasmid. As shown inFig. 2B, in the absence of DHT, HBx and AR displayed the same granular cytoplasmic staining pattern (panels a–c), indicating the co-localization of HBx and AR in the cytoplasm. In the presence of DHT, most AR was localized to the nucleus. However, a fraction of AR was also found co-localized with
Fig. 1. Co-immunoprecipitation of HBx and AR. Huh7 cells were co-transfected with HA-tagged HBx expression plasmid (pCMV-HAX), AR expression plasmid (pSG5-AR) and their control vectors in different combinations as indicated under the gels. (A) Cell lysates were immunoprecipitated using the anti-HA antibody and Western blotted using the anti-AR antibody. Lanes 1 and 2 were total cell lysates prior to immunoprecipitation. Cells in lane 1 were transfected with pRc/CMV and pSG5 control vectors whereas cells in lane 2 were transfected with pCMV-HAX and pSG5-AR. The arrow denotes the AR protein band and the arrowhead denotes a non-specific protein band, which served as an internal loading control. (B) Cell lysates were immunoprecipitated using the anti-AR antibody and Western blotted using the anti-HA antibody. Lanes 5 and 6 were total cell lysates. Cells in lane 5 were transfected only with the control vectors whereas cells in lane 6 were transfected with both HAX and AR expression plasmids. The arrow denotes the HA-tagged HBx (HAX) protein band.
455 Y. Zheng et al. / Virology 363 (2007) 454–461
HBx in the cytoplasm (panels d–f, denoted by an arrow), indicating a partial retention of AR in the cytoplasm by HBx. On rare occasions, HBx was found to co-localize with AR in the nucleus (panels g–i). These results further indicate the physical interaction of these two proteins in Huh7 cells.
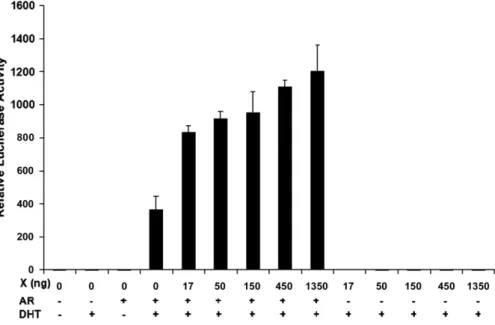
Enhancement of gene transactivation activity of AR by HBx To further investigate whether the interaction between HBx and AR affects the biological activities of AR, we conducted a gene transactivation assay using the firefly luciferase reporter, which has been linked to a promoter that contains the androgen responding element. Huh7 cells were transfected with this re-porter in the absence or presence of the AR expression plasmid and the HA-tagged HBx expression plasmid, and with or without the DHT treatment. As shown inFig. 3, this reporter displayed
only basal luciferase activities after its transfection into Huh7 cells, whether the cells were treated with DHT or not. These results were not surprising, as Huh7 cells do not express androgen receptor (Xie et al., 2001). The co-transfection of this reporter with the androgen receptor into Huh7 cells did not significantly increase the firefly luciferase reporter expression. However, if the co-transfected cells were treated with DHT for 24 h, the expression of the reporter was increased approximately 300-fold. This result is consistent with the previous observations that DHT can activate AR (Kang et al., 2002). The inclusion of the HBx expression plasmid in the co-transfection studies led to a further 2- to 4-fold increase of the firefly luciferase activities in a dose-dependent manner. This effect of HBx on the reporter was observed only in the presence of DHT and not in the absence of it (data not shown). HBx by itself also had no effect on the expression of the reporter whether or not Huh7 cells were treated
Fig. 2. Confocal microscopy of the subcellular localization of HBx and AR in Huh7 cells. (A) Subcellular localization of HBx and AR. Panels a and b, cells expressing HA-tagged HBx; panels c and d, cells expressing AR. Panels a and c, without DHT treatment; panels b and d, with DHT treatment. (B) Co-localization analysis of HBx and AR. Huh7 cells were co-transfected with the HA-tagged HBx expression plasmid and the AR expression plasmid. Red color, HBx staining; green color, AR staining; yellow or orange color, co-localization of HBx and AR. Panels a–c, without DHT treatment; and panels d–i, with DHT treatment. The arrow in panels d–f denotes a cell that co-expressed AR and HBx. The partial colocalization of AR and HBx in the cytoplasm of this cell was clearly visible. Most of HBx and AR double-positive cells displayed this staining pattern (unpublished observation). The unmarked cell in panels d–f expressed only AR, which was detectable only in the nucleus. On rare occasions, HBx and AR co-localized in the nucleus (g–i).
with DHT (Fig. 3, also data not shown). These results indicated that the effect of HBx on the reporter required DHT and was mediated by AR.
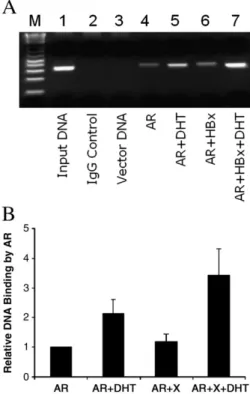
Enhancement of the DNA binding activity of AR by HBx To further investigate how HBx enhanced the gene trans-activation activity of AR, we also conducted the chromatin immunoprecipitation (ChIP) assay. In this study, Huh7 cells were transfected with the pMMTV-luciferase reporter plasmid with or without the AR expression plasmid and the HBx expression plasmid, and with or without the DHT treatment. The pMMTV-luciferase contains only one copy of the androgen responding element and was used in this particular study. After cell lysis, the AR–DNA complex was immunoprecipitated with the anti-AR antibody and the DNA was quantified by the polymerase chain (PCR) reaction. The PCR results are shown inFig. 4A and the quantification results are shown inFig. 4B. As shown inFig. 4A, no reporter DNA was detected in this ChIP assay if this reporter was co-transfected with the AR expression plasmid and immunoprecipitated with a control antibody (lane 2) or if this reporter was co-transfected with the control pRc/CMV expres-sion vector and immunoprecipitated with the anti-AR antibody (lane 3). These two experiments served as the negative controls. In contrast, a basal level of the reporter DNA was detected if this reporter was co-transfected with the AR expression plasmid into Huh7 cells and immunoprecipitated with the anti-AR antibody (lane 4). The amount of the reporter DNA precipitated by the anti-AR antibody was doubled if cells were treated with DHT for 16–18 h (Fig. 4A, lane 5; alsoFig. 4B). This increase was further enhanced by HBx (Fig. 4A, lane 8; alsoFig. 4B). HBx had little effect on the DNA binding activity of AR in the absence of DHT
(Fig. 4A, lane 6; alsoFig. 4B). Thus, these results indicated that HBx could enhance the DNA binding activity of AR only in the presence of DHT.
As HBx could bind to AR, we also investigated whether HBx remained associated with AR on the reporter DNA. As HBx was fused to the HA-tag, the anti-HA antibody was used to immunoprecipitate HBx for the ChIP assay. As shown inFig. 5, except for the positive control, almost no reporter DNA could be detected in this ChIP assay, indicating that HBx did not remain associated with AR on the reporter DNA.
Discussion
HBx is a regulatory protein with multiple functions. By conducting the co-immunoprecipitation experiments, we now demonstrate that HBx can also bind to the steroid hormone receptor AR (Fig. 1). Our observation was further confirmed by confocal microscopy. AR had a diffused cytoplasmic localiza-tion in the absence of DHT (Fig. 2A). However, when it was co-expressed with HBx, it became co-localized with HBx, which had a more granular staining pattern in the cytoplasm (Fig. 2B). AR is translocated into the nucleus after its activation by DHT (Fig. 2A) (Tomura et al., 2001). In the presence of HBx, it was partially retained by HBx in the cytoplasm (Fig. 2B). In-terestingly, HBx was also found on rare occasions to co-localize with AR in the nucleus (Fig. 2B). It is possible that a small amount of HBx, which is not detectable by confocal micro-scopy, is also co-localized with AR in the nucleus in the presence of DHT.
Upon its activation by DHT, AR is translocated into the nucleus where it will bind to the androgen responding element of its target genes to activate their expression (Kang et al., 2002;
Fig. 3. Enhancement of gene transactivation activity of AR by HBx. Huh7 cells were transfected with the reporter plasmid Probasin-luc. This plasmid contains the firefly luciferase reporter, which has been linked to androgen responding elements derived from the rat probasin promoter (Jia et al., 2003). This reporter plasmid was co-transfected with control vectors or the AR and/or the HBx expression plasmid into Huh7 cells, with or without further DHT treatment as indicated under the chart. The renila luciferase expression plasmid pRL-SV40 was included in all the transfection studies to monitor the transfection efficiency. Cells were lysed 48 h after transfection for the dual luciferase assay. The experiments were repeated at least three times and the results shown represent the mean. The relative firefly/renila luciferase activity of cells transfected with Probasin-luc alone without DHT treatment was arbitrarily defined as one.
457 Y. Zheng et al. / Virology 363 (2007) 454–461
Lee and Chang, 2003). Indeed, as shown inFig. 3, we were able to detect approximately 300-fold increase of the expression level of the luciferase reporter after the activation of AR by DHT. Due to the ability of HBx to bind to AR, we have investigated whether HBx could affect the gene transactivation activity of AR. As shown inFig. 3, HBx could further enhance the gene transactivation activity of AR 2- to 4-fold in a dose-dependent manner. This effect of HBx on AR was DHT dependent, as HBx could not enhance the activity of AR in the absence of DHT (data not shown).
HBx is not a DNA-binding protein. However, it has been shown to enhance the DNA binding activity of a number of transcription factors (Yen, 1996). Our ChIP results shown inFig. 4demonstrated that HBx could also enhance the DNA binding activity of AR. However, as shown inFig. 5, HBx did not appear to remain associated with AR on the DNA. This effect of HBx on AR is reminiscent of its activities on other transcription factors such as CREB, ATF-2 and HNF1 (Li et al., 2002; Maguire et al.,
1991; Williams and Andrisani, 1995). HBx also binds to these
transcription factors to enhance their DNA binding activities, but it does not remain associated with those transcription factors after their binding to their target DNA sequences.
It is unclear whether it is the cytoplasmic form or the nuclear form of HBx that is responsible for the activation of the AR activity. If it is the cytoplasmic form, then the binding of HBx to AR may sensitize AR to the activation induced by DHT. This can explain why HBx enhanced the DNA binding activity of AR without becoming associated with AR on its target DNA sequence. It is equally likely that the nuclear form of HBx activates the AR activity. In this case, however, HBx will enhance the DNA binding activity of AR after its activation by DHT, as nuclear transport of AR is DHT dependent.
AR is expressed in many different tissues including the liver and has a wide range of activities. In the liver, it can upregulate the expression of many genes including TGF-β1 (Chatterjee
et al., 1996; Yoon et al., 2006) and has been implicated in
hepatocarcinogenesis (Tanaka et al., 2000; Touraine et al., 1993;
Yu et al., 2000, 2001). Our observation that HBx can enhance
the activity of AR in a DHT-dependent manner indicates that HBx can affect the gene expression profiles in hepatocytes of male HBV carriers via AR. This effect of HBx is expected to be lower in the hepatocytes of female HBV carriers due to a lower level of androgen in women. HCC is male preponderant among HBV carriers. It is conceivable that this gender difference of HCC incidence is due in part to the differential effect of HBx on AR in the hepatocytes of male and female HBV carriers. In this regard, it is interesting to note that the difference of HCC risks is much less pronounced between male and female HCV patients (Chen et al., 2006; Kirk et al., 2004; Lu et al., 2006; Stroffolini et al., 2006). It is likely that, unlike HBV, HCV does not possess a factor that can enhance the activity of AR. Further research in
Fig. 4. Enhancement of DNA binding activity of AR by HBx. (A) The ChIP assay. Huh7 cells were co-transfected with DNA plasmids and lysed 48 h after transfection for the ChIP assay (see Materials and methods for details). Lane M, DNA molecular weight markers; lane 1, the input DNA control; lane 2, cells co-transfected with the reporter and pCMV-AR and then immunoprecipitated with a control antibody; lane 3, cells co-transfected with the reporter and the control vector pRc/CMV; lane 4, cells co-transfected with the reporter and pCMV-AR; lane 5, cells co-transfected with the reporter and pCMV-AR and then treated with DHT on the second day after transfection; lane 6, cells co-transfected with the reporter, pCMV-AR and pCMV-HAX; and lane 7, cells co-transfected with the reporter, pCMV-AR and pCMV-HAX and then treated with DHT. In lanes 3–7, the cell lysates were immunoprecipitated with the anti-AR antibody. In lanes 2–5, the control vector pRc/CMV was used for co-transfection to replace pCMV-HAX to ensure that the same amount of DNA was used for transfection. (B) Quantification of the ChIP assay results. The relative DNA binding activity of AR was determined by Scion Imaging software. The DNA binding activity of AR in the absence of HAX and DHT was arbitrarily defined as 1. The results represent the mean of three independent experiments.
Fig. 5. Association of HBx with AR on the reporter DNA. Huh7 cells were co-transfected with DNA plasmids of different combinations as shown under the gel and lysed 48 h after transfection for the ChIP assay (see Materials and methods for details). Lane M, DNA molecular weight markers; lane 1, cells co-transfected with the reporter, pCMV-HBx and pCMV-AR and then immuno-precipitated with a control antibody; lane 2, the input DNA control; lane 3, cells co-transfected with the reporter and the control vector pRc/CMV; lane 4, cells co-transfected with the reporter and pCMV-HAX; lane 5, cells co-transfected with the reporter and pCMV-AR; lane 6, cells co-transfected with the reporter, pCMV-AR and pCMV-HAX; and lane 7, cells co-transfected with the reporter, pCMV-AR and pCMV-HAX and then treated with DHT. In lanes 3–7, the cell lysates were immunoprecipitated with the anti-HA antibody. Except for the input DNA control (lane 2), no significant amount of the reporter DNA could be detected in other lanes.
this area will likely generate interesting results for under-standing HBV pathogenesis in different genders.
Materials and methods Cell lines and DNA plasmids
Huh7 cells were maintained in Dulbecco's modified essential medium (DMEM) containing 10% fetal bovine serum (FBS). Depending on the experiments, the FBS might be treated with dextran-coated charcoal (Sigma-Aldrich) for the removal of small molecules. pCMV-HAX (Li et al., 2002), which expresses the HA-tagged HBx under the control of the immediate early promoter of cytomegalovirus, and pSG5-AR
(Litvinov et al., 2004), which expresses the human androgen
receptor under the SV40 early promoter control, have been previously described. The expression vector pSG5 was from Stratagen. Probasin-Luc is a plasmid that contains the firefly luciferase reporter linked to the herpesvirus thymidine kinase (tk) promoter and three copies of the rat probasin promoter fragment (nucleotides−244 to −96) that contains the androgen responding elements (Jia et al., 2003). The plasmid pMMTV-Luciferase, which contains 1 copy of the androgen responding element, has been described before (Nordeen, 1988).
Co-immunoprecipitation experiments
Huh7 cells in a 10-cm Petri dish were transfected with pCMV-HAX, pSG5-AR and their control expression vectors in different combinations using the calcium phosphate precipita-tion method. Forty-eight hours after transfecprecipita-tion, cells were rinsed twice with TBS (10 mM Tris–HCl, pH 7.0, 150 mM NaCl) and lysed in 1 ml TBS containing 0.5% Nonidet P-40 (NP40). The cell lysates were briefly centrifuged to remove cell debris. The supernatant was mixed 1:1 with RIPA (10 mM Tris– HCl, pH 7.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate) and split into two aliquots. The first aliquot was incubated with 1μl mouse anti-HA monoclonal antibody (USC Norris Cancer Center Cell Culture Core) and the second aliquot was incubated with 1μl rabbit anti-AR antibody (Affinity Bioreagents). After incuba-tion at 4 °C overnight, the immune complex was precipitated with 20μl Gammabind Plus (Amersham). The immunopreci-pitates were washed four times with a 3:1 mixture of RIPA and the TBS lysis buffer, followed by electrophoresis in an 8–15% gradient gel. The protein gel was Western blotted to nitro-cellulose membrane, which was hybridized first with either the rabbit anti-AR antibody (first aliquot of cell lysates) or the mouse anti-HA antibody (the second aliquot of cell lysates) and then with the respective alkaline phosphatase-conjugated goat anti-mouse or anti-rabbit antibody. The protein bands were visualized using the BioRad AP color kit (BioRad).
Confocal microscopy
Huh7 cells grown on coverslips in a six-well dish were transfected with pCMV-HAX, pSG5-AR and their respective
control expression vectors pRc/CMV (Invitrogen) and pSG5 in different combinations by calcium phosphate precipitation. Cells were rinsed twice with DMEM without serum on the second day and further incubated in DMEM with or without 50 nM DHT for 24 h. Cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) and then stained with both the mouse anti-HA and the rabbit anti-AR primary antibodies and the rhodamine-conjugated goat anti-mouse and fluorescein-conjugated goat anti-rabbit secondary antibodies for confocal microscopy.
The luciferase reporter assay
Huh7 cells in six-well dishes were co-transfected with 1.5μg Probasin-luc reporter and either 150 ng pSG5-AR or its control expression vector pSG5. 1.5μg of pCMV-HAX and its control vector pRc/CMV in different combinations were also included in the co-transfection experiment. Cells were incubated in DMEM containing 0.5% FBS after transfection, treated with 50 nM DHT or its control solvent ethanol in DMEM on the second day for 24 h and then lysed for the luciferase assay. In all the transfection experiment, 150 ng pRL-SV40 (Promega), a plasmid that ex-presses renila luciferase, was used for co-transfection to monitor the transfection efficiency. The firefly luciferase and the renila luciferase activities were measured using the dual luciferase assay (Promega). The firefly luciferase activities were then normalized against the renila luciferase activities. All the ex-periments were repeated at least three times.
Chromatin immunoprecipitation (ChIP) assay 2.5–5×105
Huh7 cells in a 6-well dish were transfected with 1.5 μg/well of DNA plasmids with 0.5 μg each of pMMTV-Luciferase, AR and HAX. The plasmids pCMV-AR and pCMV-HAX might be replaced by their control vector pRc/CMV, depending on the experiments. Eight to twelve hours after transfection, cells were treated with 10 nM DHT or its control solvent ethanol for 16–18 h in DMEM that contained 10% charcoal-stripped FBS. Cells were then crosslinked with 1% formaldehyde in PBS at room temperature for 10 min on a platform shaker. The cross-linking reaction was stopped by adding glycine to a final concentration of 125 mM with further shaking at room temperature for 5 min. Cells were then rinsed three times with ice cold PBS, scraped in 1 ml PBS and pelleted by centrifugation at 700×g for 4 min. The cell pellet was then resuspended in the cell lysis buffer (5 mM Pipes, pH 8.0, 85 mM KCl and 0.5% Nonidet-P 40) containing 1 μg/ml leupeptin, 1μg/ml aprotinin and 1 mM PMSF and incubated on ice for 10 min. Nuclei were then pelleted by centrifugation at 5,000 rpm for 5 min in a microcentrifuge and lysed in the nuclear lysis buffer (50 mM Tris–HCl, pH 8.1, 10 mM EDTA, 1% SDS) containing the same protease inhibitors for 10 min on ice. The chromosomal DNA was pulse-sonicated on ice to an average length of 600 bp. The nuclear debris was removed by centrifugation at full-speed in a microfuge for 10 min at 4 °C. The supernatant was then diluted 5-fold in the ChIP dilution buffer (16.7 mM Tris–HCl, pH 8.1, 167 mM NaCl, 1.2 mM
459 Y. Zheng et al. / Virology 363 (2007) 454–461
EDTA, 1.1% Triton X-100 and 0.01% SDS) containing the protease inhibitors. The sample was then pre-treated for 30 min at 4 °C with 80μl 50% slurry of protein A-agarose that had been pretreated with salmon sperm DNA. After the removal of the protein A-agarose by a brief centrifugation, an aliquot of the supernatant was saved to serve as the input DNA control, and the anti-AR antibody NH27 (Wang et al., 1994), the anti-HA antibody (USC Cell Culture Core Facility) or a control IgG (Santa Cruz) was added to the rest of the supernatant. After the incubation at 4 °C overnight with shaking, the sample was treated at 4 °C for 1 h with 60μl protein A-agarose slurry that had been pretreated with salmon sperm DNA. The agarose beads were then precipitated by a brief centrifugation in a microfuge and washed consecutively for 3–5 min with 1 ml each of the following solutions: low salt wash buffer (20 mM Tris–HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS); high salt wash buffer (20 mM Tris–HCl, pH 8.1, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS); LiCl wash buffer (10 mM Tris–HCl, pH 8.0, 0.25 M LiCl, 1 mM EDTA, 1% NP40, 1% Na deoxycholate); and TE (10 mM Tris–HCl, pH 8.0, 1 mM EDTA) twice. The pellet was then vortex in 250μl elution buffer (0.1 M NaHCO3, 1% SDS) for 15 min, pelleted again at 15,000×g for 3 min. The super-natant was transferred to a new tube. The wash of the pellet was repeated one more time and the two supernatants were combined. The formaldehyde cross-linking was reversed by adding 1 μl 10 mg/ml RNase and 5 M NaCl to a final con-centration of 0.3 M followed by incubation at 65 °C water bath for 4–5 h. The DNA was precipitated with 2.5 volumes of 100% ethanol at−20 °C overnight and resuspended in 100 μl water. The input DNA aliquot was treated with the same procedures followed by the addition of 2μl 0.5 M EDTA, 4 μl 1 M Tris– HCl, pH 6.5 and 1 μl 20 mg/ml proteinase K and further incubated at 45 °C for 1–2 h. The DNA was then purified using the QiaQuick spin column (Qiagen) and eluted in 50μl 10 mM Tris–HCl, pH 8.0. Two microliters of the DNA was then used for PCR using the following primers: GGTTCCCAGGGCT-TAAGTAAG (sense) and GGACCCTCTGGAAAGTGAAG (antisense). The PCR was conducted using the following conditions: 94 °C 4 min; 94 °C 45 s, 58 °C 45 s and 72 °C 1 min for 35 cycles; 72 °C 7 min. The PCR products were then analyzed on an agarose gel. The experiments were repeated at least three times to ensure the accuracy of the results.
Acknowledgments
We wish to thank Dr. Ben Yen for critical reading of this manuscript and Dr. Gerry Coetzee for providing us with the probasin-luc reporter DNA plasmid. This research was sup-ported by grants from the National Institutes of Health to CC and JHJO.
References
Becker, S.A., Lee, T.H., Butel, J.S., Slagle, B.L., 1998. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 72 (1), 266–272. Beilin, J., Ball, E.M., Favaloro, J.M., Zajac, J.D., 2000. Effect of the
androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. J. Mol. Endocrinol. 25 (1), 85–96.
Bouchard, M.J., Wang, L.H., Schneider, R.J., 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294 (5550), 2376–2378. Bouchard, M.J., Wang, L., Schneider, R.J., 2006. Activation of focal adhesion kinase by hepatitis B virus HBx protein: multiple functions in viral rep-lication. J. Virol. 80 (9), 4406–4414.
Chamberlain, N.L., Driver, E.D., Miesfeld, R.L., 1994. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 22 (15), 3181–3186. Chatterjee, B., Song, C.S., Jung, M.H., Chen, S., Walter, C.A., Herbert, D.C.,
Weaker, F.J., Mancini, M.A., Roy, A.K., 1996. Targeted overexpression of androgen receptor with a liver-specific promoter in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 93 (2), 728–733.
Chen, H.S., Kaneko, S., Girones, R., Anderson, R.W., Hornbuckle, W.E., Tennant, B.C., Cote, P.J., Gerin, J.L., Purcell, R.H., Miller, R.H., 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67 (3), 1218–1226.
Chen, C.H., Huang, G.T., Yang, P.M., Chen, P.J., Lai, M.Y., Chen, D.S., Wang, J.D., Sheu, J.C., 2006. Hepatitis B- and C-related hepatocellular carcinomas yield different clinical features and prognosis. Eur. J. Cancer 42 (15), 2524–2529.
Cheong, J.H., Yi, M., Lin, Y., Murakami, S., 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14 (1), 143–150.
Fischer, M., Runkel, L., Schaller, H., 1995. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes 10 (1), 99–102.
Hu, Z., Zhang, Z., Kim, J.W., Huang, Y., Liang, T.J., 2006. Altered proteolysis and global gene expression in hepatitis B virus X transgenic mouse liver. J. Virol. 80 (3), 1405–1413.
Irvine, R.A., Ma, H., Yu, M.C., Ross, R.K., Stallcup, M.R., Coetzee, G.A., 2000. Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum. Mol. Genet. 9 (2), 267–274. Jia, L., Kim, J., Shen, H., Clark, P.E., Tilley, W.D., Coetzee, G.A., 2003.
Androgen receptor activity at the prostate specific antigen locus: steroidal and non-steroidal mechanisms. Mol. Cancer Res. 1 (5), 385–392. Kang, H.Y., Huang, K.E., Chang, S.Y., Ma, W.L., Lin, W.J., Chang, C., 2002.
Differential modulation of androgen receptor-mediated transactivation by Smad3 and tumor suppressor Smad4. J. Biol. Chem. 277 (46), 43749–43756. Kirk, G.D., Lesi, O.A., Mendy, M., Akano, A.O., Sam, O., Goedert, J.J., Hainaut, P., Hall, A.J., Whittle, H., Montesano, R., 2004. The Gambia Liver Cancer Study: infection with hepatitis B and C and the risk of hepatocellular carcinoma in West Africa. Hepatology 39 (1), 211–219.
Klein, N.P., Schneider, R.J., 1997. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol. Cell. Biol. 17 (11), 6427–6436.
Lee, H.J., Chang, C., 2003. Recent advances in androgen receptor action. Cell Mol. Life Sci. 60 (8), 1613–1622.
Li, J., Xu, Z., Zheng, Y., Johnson, D.L., Ou, J.H., 2002. Regulation of hepatocyte nuclear factor 1 activity by wild-type and mutant hepatitis B virus X proteins. J. Virol. 76 (12), 5875–5881.
Litvinov, I.V., Chang, C., Isaacs, J.T., 2004. Molecular characterization of the commonly used human androgen receptor expression vector, pSG5-AR. Prostate 58 (4), 319–324.
Lu, S.N., Su, W.W., Yang, S.S., Chang, T.T., Cheng, K.S., Wu, J.C., Lin, H.H., Wu, S.S., Lee, C.M., Changchien, C.S., Chen, C.J., Sheu, J.C., Chen, D.S., Chen, C.H., 2006. Secular trends and geographic variations of hepatitis B virus and hepatitis C virus-associated hepatocellular carcinoma in Taiwan. Int. J. Cancer 119 (8), 1946–1952.
Maguire, H.F., Hoeffler, J.P., Siddiqui, A., 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein–protein interactions. Science 252 (5007), 842–844.
Nordeen, S.K., 1988. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques 6 (5), 454–458.
protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 74 (6), 2840–2846.
Seto, E., Mitchell, P.J., Yen, T.S., 1990. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature 344 (6261), 72–74.
Stroffolini, T., Sagnelli, E., Mariano, A., Craxi, A., Almasio, P., 2006. Characteristics of HCV positive subjects referring to hospitals in Italy: a multicentre prevalence study on 6,999 cases. J. Viral Hepat. 13 (5), 351–354. Su, F., Theodosis, C.N., Schneider, R.J., 2001. Role of NF-kappaB and myc proteins in apoptosis induced by hepatitis B virus HBx protein. J. Virol. 75 (1), 215–225.
Tanaka, K., Sakai, H., Hashizume, M., Hirohata, T., 2000. Serum testosterone: estradiol ratio and the development of hepatocellular carcinoma among male cirrhotic patients. Cancer Res. 60 (18), 5106–5110.
Tomura, A., Goto, K., Morinaga, H., Nomura, M., Okabe, T., Yanase, T., Takayanagi, R., Nawata, H., 2001. The subnuclear three-dimensional image analysis of androgen receptor fused to green fluorescence protein. J. Biol. Chem. 276 (30), 28395–28401.
Touraine, R.L., Bertrand, Y., Foray, P., Gilly, J., Philippe, N., 1993. Hepatic tumours during androgen therapy in Fanconi anaemia. Eur. J. Pediatr. 152 (8), 691–693.
Wang, X.W., Forrester, K., Yeh, H., Feitelson, M.A., Gu, J.R., Harris, C.C., 1994. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. U.S.A. 91 (6), 2230–2244.
Williams, J.S., Andrisani, O.M., 1995. The hepatitis B virus X protein targets the
basic region-leucine zipper domain of CREB. Proc. Natl. Acad. Sci. U.S.A. 92 (9), 3819–3823.
Xie, X., Zhao, X., Liu, Y., Young, C.Y., Tindall, D.J., Slawin, K.M., Spencer, D. M., 2001. Robust prostate-specific expression for targeted gene therapy based on the human kallikrein 2 promoter. Hum. Gene Ther. 12 (5), 549–561. Xu, Z., Yen, T.S., Wu, L., Madden, C.R., Tan, W., Slagle, B.L., Ou, J.H., 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76 (5), 2579–2584.
Yen, T.S., 1996. Hepadnaviral X protein: review of recent progress. J. Biomed. Sci. 3 (1), 20–30.
Yoon, G., Kim, J.Y., Choi, Y.K., Won, Y.S., Lim, I.K., 2006. Direct activation of TGF-beta1 transcription by androgen and androgen receptor complex in Huh7 human hepatoma cells and its tumor in nude mice. J. Cell Biochem. 97 (2), 393–411.
Yu, M.W., Cheng, S.W., Lin, M.W., Yang, S.Y., Liaw, Y.F., Chang, H.C., Hsiao, T.J., Lin, S.M., Lee, S.D., Chen, P.J., Liu, C.J., Chen, C.J., 2000. Androgen-receptor gene CAG repeats, plasma testosterone levels, and risk of hepatitis B-related hepatocellular carcinoma. J. Natl. Cancer Inst. 92 (24), 2023–2028. Yu, M.W., Yang, Y.C., Yang, S.Y., Cheng, S.W., Liaw, Y.F., Lin, S.M., Chen, C.J., 2001. Hormonal markers and hepatitis B virus-related hepatocellular carcinoma risk: a nested case–control study among men. J. Natl. Cancer Inst. 93 (21), 1644–1651.
Zhang, Z., Torii, N., Hu, Z., Jacob, J., Liang, T.J., 2001. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Invest. 108 (10), 1523–1531.
Zoulim, F., Saputelli, J., Seeger, C., 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68 (3), 2026–2030.
461 Y. Zheng et al. / Virology 363 (2007) 454–461