國立交通大學
生物科技研究所
碩 士 論 文
探討大毒性質體
pLVPK 在克雷白氏肺炎桿菌致
病過程中扮演的角色
Role of pLVPK in Klebsiella pneumoniae
pathogenesis
研究生:陳 育 聖
學號:9328502
指導教授:彭 慧 玲 博士
中文摘要
已知克雷白氏肺炎桿菌臨床分離株 CG43 中帶有一個 219 kb 與毒性相關 的大型質體。為了了解此大型質體在克雷氏肺炎桿菌中致病過程中所扮演的角 色,我們分析了 127 株克雷白氏肺炎桿菌臨床分離株的普及率。研究發現許多 克雷白氏肺炎桿菌帶有大型質體,普及率為56.69% (72/127)。利用 rmpA, iutA, iroB, silS, terA 此五個基因作為探針,以及螯鐵分子 siderphore 分泌,我們發現 在所有的肝膿瘍臨床分離株之中均帶有一個與 pLVPK 相關的大型質體。然而兩 者之間是否具有關係仍需要進一步研究。 在 pLVPK 之中,發現兩個相似的轉錄因子 rmpA 與 rmpA2。過去研究已 知在克雷白氏肺炎桿菌 CG43 中,轉錄因子 rmpA2 的缺失會造成莢膜多醣體 生合成減少,此現象與另一個轉錄因子 rcsB 無關。本實驗首先利用 RT-PCR 以 及南方墨點法證明與 rmpA2 核酸相似度高達 78% 的 rmpA 基因是個活化基 因。為了進一步研究 rmpA 基因的功能,我們建構 rmpA 缺失突變株。比較 rmpA 以及 rmpA2 缺失突變株,發現此兩種突變株具有類似的表現型, 例如在 黏性、莢膜多醣體合成能力均會下降,而生物膜形成的能力會上升。 此外, 利 用 LacZ 報導蛋白,分析莢膜多醣體基因的啟動子在這些突變株的表現差異。 結果顯示:莢膜多醣體基因組開放骨架 1-2 或 16-17 的啟動子在 rmpA 突變株 下,活性較野生株下降約 30%。然而,也發現 rmpA 的啟動子在 rcsB 突變株 下,活性只有野生株的一半,因此我們推論 RcsB 可以活化 rmpA 的啟動子, 此點與 rmpA2 啟動子不同。
Abstract
The large plasmid pLVPK, of 219-kb in size, has been shown to be required for the virulence of Klebsiella pneumoniae CG43, a highly virulent clinical isolate. To assess the role of pLVPK in K. pneumoniae pathgogenesis, the prevalence of large plasmids in 127 K. pneumoniae isolates of various origins was analyzed. Consistent with the previous findings indicating that many clinical isolates harbor large plasmids of 200 kb in size, the analysis revealed 56.69% (72/127) prevalence of large plasmids. Interestingly, by probing with the pLVPK specific genes rmpA, iutA, iroB, silS, and terA, together with the assays of siderphore synthesis, we have found all the liver abscess isolates harbored a pLVPK related plasmid. Nevertheless, whether the presence of the pLVPK like plasmid could be correlated with K. pneumoniae liver abscess requires more studies.
On pLVPK, two transcription factor encoding genes, rmpA and rmpA2 were identified. In K. pneumoniae CG43, deletion of rmpA2 has been reported to reduce the production of capsular polysaccharides (CPS) at transcriptional level in RcsB-independent manner. In the mutant CG43S3 rmpA2-, expression of rmpA, an rmpA2 homolog sharing 78% nucleotide sequence identity, could be demonstrated by RT-PCR and Southern blotting analysis. An rmpA deletion mutant was subsequently generated and the mutant was found to exert similar phenotypes to that of CG43S3 rmpA2-, which including decrease of colony mucoidy, and reduction of glucouronic acid content, but increase of biofilm formation capability. In addition, promoter activity measurements using the lacZ as the reporter indicated that rmpA deletion, as well as the rmpA2 deletion, reduce the activity of Porf1-2::lacZ and Porf16-17::lacZ
approximately 30% in comparing with that of wild type. Nevertheless, activity of PrmpA in rcsB- strain reduced to a half of that in wild type suggesting a positive
致謝
兩年來碩士生活的點點滴滴,不論是研究上或者日常生活中都學習了很多, 感謝大家一路走來的支持、照顧,讓我順利完成這段人生最重要的旅途。 這篇論文的完成,最感謝我的指導老師 彭慧玲 老師,在研究方向提供許多 想法、意見,並給予充分的討論,指引我找出正確的研究方向。特別是在完成這 篇論文期間,讓老師費心費力,在此致上最深的感激。更感謝兩位口試委員 楊 昀良 與 賴怡琪 老師,以不同的觀點切入問題,並細心指證及建議,讓此篇論 文更加趨向完善。 感謝實驗室所有同伴,給予許多的幫助以及美好回憶。特別是靖婷、盈蓉兩 位學姊,這兩年來不論在實驗或者處事方面給予我很多建議,短短的幾句話是無 法完全道出心中的感謝,希望你們能早日找到屬於自己的幸福;感謝健誠、小新 學長,兩位對於實驗的想法以及對於實驗執著的態度,讓我充分了解當研究生必 備的條件,希望你們能繼續將此精神發揚下去;智凱,細心的你,希望能早日找 到適合你的工作還有屬於你的幸福;心瑋,祝你博士班能順利;至於三位學弟, 格維、朝陽、登魁,謝謝這段時間的幫忙,讓這段求學期間增添許多歡樂,謝謝 你們。 最後感謝爸爸、媽媽、姐姐以及我的室友盛雄,在我困惑的時候適時給予我 幫助以及照顧,讓我能順利畢業,僅此論文獻給我的家人以及各位。Table of contents
Abstract (Chinese)………..….…...ii
Abstract……….……iii
致謝……….………...iv
Table of contents………..…...v
Abbreviation………..……vi
List of the tables and figures………....vii
Introduction………..………..…1
Materials and methods
1. Bacterial strains, and growth conditions……….………52. Genomic DNA extraction and plasmid typing………5
3. Dot blotting hybridization and PCR detection of the rmpA, terA, iutA, iroB, and silS genes………..5
4. Assessment of the siderphore biosynthesis activity………6
5. Definition of mucoviscosity and colony size of 19 liver abscess clinical isolates……….6
6. RT-PCR analysis………...………..7
7. Bioinformatics analysis………...………7
8. Recombinant DNA technique……….………7
9. Construction of the gene-deletion mutant and complement strains…………7
10. Construction the complement strains……….……….8
11. Southern blotting analysis………...………8
12. Quantification of biofilm formation………8
13. Extraction and quantification of CPS………..9
15. β-galactosidase activity assay………..………….10
Results
Part 1 :
Prevalence of pLVPK in Klebisella pneumoniae clinical isolates 1-1. Plasmid typing of 127 K. pneumoniae clinical isolates………..111-2. Distribution of rmpA, terA, iutA, iroB, and silS genes in clinical isolates………11
1-3. The liver abscess isolates harbor pLVPK-like plasmid………..12
1-4. The siderophore biosynthesis activity of the K. pneumoniae isolates....12
1-5. The capsular polysaccahride biosynthesis activity and biofilm formation of the K. pneumoniae isolates………13
Part 2 :
Identification of the regulatory role of RmpA 2-1. RT-PCR demonstrated the expression of rmpA in K. pneumoniae CG43………..142-2. Biological role of RmpA………14
2-3. RmpA regulates positively the expression of cpsorf1-2 and cpsorf16-17….15 2-4. Expression of rmpA and rmpA2 in K. pneumoniae CG43……….15
Discussions ………...………...17
References ………...………21
Tables ………...………...25
Figures ………...……….30
Abbreviation
BLAST basic local alignment search tool bp base pair
CAS chrome azurol S CFU colony forming unit CPS capsular polysaccharide CV crystal violet
LB Luria-Bertani
ONPG ο-nitrophenyl-β-D-galactopyranoside PCR polymerase chain reaction
PFGE pulsed field gel electrophoresis rpm revolutions per minute
SDS sodium dodecyl sufate
List of the tables and figures
Table 1. Bacterial strains used and constructed in this study………25
Table 2. Plasmids used and constructed in this study………26
Table 3. Primers used in this study………...27
Table 4. Prevalence analysis of pLVPK in 127 K. pneumoniae clinical isolates…….28
Table 5. String test and colony size of 19 liver abscess isolates………..29
Figure. 1. Plasmid profile analysis of the 127 K. pneumoniae isolates by PFGE……30
Figure 2. Dot-blotting analysis of 127 K. pneumoniae clinical isloates respectively probing with rmpA, iutA, iroB, silS, and terA gene………..31
Figure 3. PCR detection of (A) rmpA, (B) iutA ,(C) iroB ,(D) silS, and (E) terA gene………..32
Figure 4. The presence of pLVPK in the liver abscess isolates………33
Figure 5. Siderophore synthesis activity of K. pneumoniae isolates in CAS solution……….34
Figure 6. Sedimentation test of the K. pneumonia isolates………..35
Figure 7. Biofilm formation of wild-type and rmpA mutant strain………..36
Figure 8. RT-PCR analysis of rmpA expression………...……37
Figure 9. Construction of rmpA deletion mutant………..38
Figure 10. Phenotype analysis of the rmpA- and rmpA2- strains………..39
Figure 11. The biofilm formation in the wild type and mutant strain at time course (A) 12 H ,(B) 24 H, and (C) 36 H.………..40
Figure 12. Promoter activity analysis of cps genes in wild type, rmpA-, and rmpA2- stains………...………..41
Figure 13. Activity of PrmpA::lacZ and PrmpA2::lacZ inK. pneumoniae LacZ16 in LB medium…………...……….……….42
Figure 14. Nucleotide sequence alignment of the rmpA and rmpA2 genes promoters………..43
Figure 15. Activity of PrmpA and PrmpA2 in K. pneumoniae LacZ16, rcsA-Z16, and rcsB-Z16………...……….44
Figure 16. A proposed model of the regulation of RmpA and RmpA2………45
Introduction
Klebsiella pneumoniae is a common pathogen of community-acquired and nosocomial infections. It causes a wide spectrum of infections, including septicemia, pneumonia, urinary tract infection, meningitis, and purulent abscess at various sites, especially liver abscess. In particular, a distinctive clinical syndrome, which is characterized by community-acquired K. pneumoniae bacteremia with primary liver abscess, metastatic meningitis, and endophthalmitis, has been recognized in Taiwan (19, 20, 39, 47). There are several virulence factors identified to participate in K. pneumoniae infections, which include capsular polysaccharides (CPS), lipopolysaccharides (LPS), adhesins, and iron acquisition systems (12, 19, 27, 32). In addition to these virulence factors, very little is known about the roles of other genes that may be associated with its pathogenesis. Most recently, the prevalence of multiple drug-resistant strains has significantly restricted the availability of antibiotics for effective treatment of the bacterial infection (34). Therefore, investigation the molecular mechanism involved in K. pneumoniae pathogenesis to provide novel drug targets brooks no delay.
Large plasmids associated with virulence, heavy metal resistance, and multiple antibiotic resistance have been reported in Bacillus anthracis, Salmonella, E. coli, K. pneumoniae, Shigella, and Yersinia pestis (9, 17, 18, 23, 25, 30, 38). For example, B. anthracis harbors the 181.7-kb plasmid pXO1, which is essential for the manifestation of the disease anthrax (17). In Shigella, the virulence plasmids pWR100, pMYSH6000, and pSS120 are determinants for invasiveness and the ability to cause disease. These plasmids carry the genes for invasions, molecular chaperons, motility, regulation, and a type III secretion pathway with a common 31-kb region (23). In K.
pneumoniae, the correlation of large plasmid and virulence has also been reported (24, 25). The large plasmid encodes the production of aerobactin which has been reported a virulence factor (25). Recently, a large plasmid named pLVPK (Large Virulence Plasmid of K. penumoniae) was found in K. pneumoniae CG43, a clinical isolate K2 serotype with LD50 as low as 10 C.F.U (18). Removal the 200-kb plasmid from K. pneumoniae CG43 resulted in a loss of colony mucoidy, a loss ability of secreting aerobactin, and 1000 fold decrease in virulence (18).
Sequence analysis of pLVPK showed that this plasmid consists of 219-kb containing 251 ORFs, and approximately 37% of the ORFs are functionally identified (5). Several gene clusters that may attribute to the bacterial virulence were noted, which include iut, iro, pbr, pco, sil, and ter gene clusters. The gene cluster iucABCDiutA, which is carried on a mobile region similar to the SHI-2 PAI of Shigella flexneri (23), encodes the proteins for aerobactin synthesis upon iron deprivation. The iroBCDEN gene cluster, firstly described in S. enterica, is known to participate in the uptake of catecholate-type siderophores, enterobactin (13). Secretion of aerobactin and enterobactin are essential for bacteria to survive in host cells (42). The regulation of siderophore synthesis is via Fur protein. Fur is a transcriptional repressor, which displays iron-dependentbinding to conserved DNA sequences (Fur boxes) located in the promoters of iron-regulated genes (42). In most bacteria, including K. pneumoniae, the iron-complexed form of Fur binds topromoters of iron-uptake genes, thus repressing iron uptakein iron-replete conditions. The other four gene clusters encode the proteins respectively for lead-resistance genes (3), copper-resistance (4), silver-resistance (10), and tellurite resistance (38). Both the lead and silver resistance gene clusters, pbrABC and silCBAP, encode the proteins to form the efflux system, and PbrRS and SilRSE regulate respectivly the expression of pbrRSABC and silCBAPsilRS (3, 37).
In K. pneumoniae, CPS is an important virulence factor as a barrier against the antibacterial reactions in host cell (20). K. pneumoniae CPS has been classified into at least 77 serotypes (18). On basis of the mouse lethality assay, strains belonging to serotypes K1 and K2 are the most virulent (18). Klebsialla K2 CPS has be determined as [→]4-Glc-(1→3)-α-Glc-(1→4)-β-Man-(3←1)-α-GlcA]-(1→)n (21), which is
produced by a similar biosynthetic pathway of Group 1 CPS in E. coli and made from three transcription units (2), including the orf1-2, involved in the synthesis CPS precursor, the orf3-15, encoding the core elements for synthesis CPS repeat units, and the orf16-17, encoding the enzymes involved in the biosynthesis of CPS precursors (2, 46, 47).
The regulatory strategy of E. coli group 1 CPS biosynthesis may serve as a model (43, 44), in which the cps gene cluster is regulated by RcsC/RcsD/RcsB and by Lon protease for degradation of RcsA (21). RcsC is a sensior protein, RcsB is a response regulator, and RcsD is a histidine-containing phosphotransfer protein that transfers the phosphoryl group from RcsC to RcsB. The activated RcsB together with the auxiliary protein, RcsA, subsequently coordinate the activation of the cps operon transcription (8, 41). In aligning many of the genes under the control by RcsAB, an RcsAB binding box TaAGaatatTcctA was identified (17, 44). Many reports suggest that Rcs system could sense alterations of cell envelop (such as osmotic shock) and metal signals (Zn2+ concentration) to positively control the genes involved in CPS synthesis and negative control the genes involved in flagellar synthesis, motility, and chemotaxis suggesting the Rcs system regulate the biofilm formation (14, 21, 40). In E. coli, curli encoding by csgBADEFG were necessary for biofilm formation. Rcs system negatively controlled the expression of csgBADEFG to affect the biofilm formation indirectly (40).
associated with the mucoidy phenotype of K. pneumoniae were also identified on pLVPK (5). The RmpA encoding gene firstly reported by Nassif et al. (24), is a positive regulator for the production of K2 CPS in K. pneumoniae and colonic acid, Group 1 CPS in E. coli (24). In E. coli, rmpA gene has been shown to be able to complement the deficiency of rcsA-lon mutation, but not an rcsA mutation, indicating that rmpA expressed an rcsA-like activity (24). The rmpA2, which was named by the sequence identity with rmpA, has lately been reported (1, 42). The major difference between these two gene products is that the RmpA2 has an extended N-terminal region (42). In the previous studies, RmpA2 as well as RcsB were able to confer E. coli JM109 or HB101 the ability to produce Klebsiella K2 capsular in the presence K2 cps gene cluster (1, 24). Furthermore, RmpA2 could directly bind to the cps gene promoter via its C-terminal DNA binding motif. We have also shown that the expression of rmpA2 was negatively autoregulated (18).
In this study, the relationship of pLVPK and the types of diseases was investigated. In addition, we attempt to elucidate the functional role of RmpA by using comparative analysis of the properties of wild type, the rmpA deletion mutant, and rmpA2 mutant. Finally, the putative promoters of rmpA, rmpA2 and cps genes were cloned respectively upstream to the promoterless lacZ to demonstrate a differential expression of rmpA and rmpA2 in K. pneumoniae CG43.
Materials and methods
1. Bacterial strains, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1 and Table 2. K. pneumoniae CG43 is a clinical isolate recovered from a patient in the Chang Gung Memorial Hospital, Linkou and K. pneumoniae NTUH-K2044 of K1 serotype, was isolated at National Taiwan University Hospital. Other clinical isolates of K. pneumoniae used were recovered from different tissue specimens of patients with a variety of infections at the Veteran General Hospital, Taipei, from 1991 to 1998. The bacteria were propagated at 37℃ in Luria-Bertani (LB) broth or the medium supplemented with appropriate antibiotics which include kanamycin (25 µg/ml), ampicillin (100 µg/ml) and tetracycline (20 µg/ml).
2. Genomic DNA extraction and plasmid typing
Genomic DNAs of 127 Klebsiella pneumoniae isolates were extracted by the method of Kado and Liu (15). Cells from a single colony scraped from an agar plate were suspended in 50 μl lysis buffer (in 3% SDS, 50 mM Tris-HCl, pH 12.6). The mixture was incubated at 65℃ for 30 min and extracted with unbuffered phenol-chloroform (1:1). The genomic DNAs were separated by pulsed field gel electrophoresis (PFGE) (Rotaphor○R TypV, Biometra).
3. Dot blotting hybridization and PCR detection of the rmpA, terA, iutA, iroB, and silS genes
The genomic DNAs were transferred to Hybond-N+ membrane (Amersham-Pharmacia, Piscataway, NJ) respectively by dot blotting. The membrane
was then subject to denaturation, and hybridization with the appropriate probe for 16 h at 65℃. Finally, the signals on the membrane detected with CDP-star reagent (Amersham-Pharmacia, Piscataway, NJ). The specific primers used for PCR detection of iutA, iroB, silS, and terA, are listed in table 3.
4. Assessment of the siderphore biosynthesis activity
The K. pneumoniae liver abscess isolates were cultured in M9 minimal medium overnight, and 100 μl of bacteria diluted 1/100 in iron deficient medium containing 0.2 mM 2,2-dipyridyl in M9, was incubated at 37℃ for 30 min. after centrifugation 6000 rpm for 5 min, the supernatant was then mixed with chrome azurol S (CAS) solution containing in 100 ml with 1.5 ml iron solution (1 mM FeCl3‧6H2O), 7.5 ml
2 mM aqueous CAS solution, and 4.307 g piperazine dissolved in 6.25 ml 12 M hydrochloric acid (35).
5. Definition of mucoviscosity and colony size of 19 liver abscess clinical isolates
Strains with the mucoviscosity phenotype were defined as being virulent. To determine mucoviscosity phenotype, a standard bacteriologic loop was used to stretch a mucoviscous string from the colony. Strains were defined as mucoid positive and large colony when viscous strings were > 0.5 cm (a positive string test result) and colony diameter were > 3 mm (a large colony result). We have previously noted that the degree or strength of viscosity seems to be slightly diminished for colonies at their first subculture from frozen isolates, but their phenotype is well recovered at the second subculture and is in accordance with that from primary culture plates. Therefore, all isolates required 2 subcultures to get well-grown colonies before rmucoviscosity testing.
6. RT-PCR analysis
Total RNA was isolated from the K. pneumoniae cells in mid-log phased (OD600
= 0.6~0.8) by extraction with the 1 ml TRI reagent (Molecular Research Center, Cincinnati, OH). Contaminating DNA was eliminated from the RNA samples with RQ1 RNase-Free DNase (Promega, Madison, WI). The cDNA products were amplified with the specific primers rmpA07/rmpA08.
7. Bioinformatics analysis
Homology search analysis and gene annotation were performed with BLAST analysis in NCBI (http://www.ncbi.nlm.nih.gov) and VectorNTI (Invitrogen Vector NTI™ Advance). Promoter prediction was carried out by website (http://www.softberry.com/all.htm).
8. Recombinant DNA technique
The recombinant DNA experiment was carried out by standard procedures as described (33). Plasmid DNA was prepared by VIOGENE Miniprep Kit (Gene-Spin
TM-V2). Restriction endonucleases and DNA modifying enzymes were purchased form
MBI (Fermentas, Hanover, MD), and were used according the recommendation of the suppliers.
9. Construction of the gene-deletion mutant and complement strains
The individual deletions in rmpA were introduced into chromosome of K. pneumoniae CG43 by allelic exchange strategy. The primer sets used for PCR amplification are listed in Table 3. Approximately 1000 bp sequence flanking both sides of the deleted region were cloned into plasmid pKAS46 (18), a suicide vector containing rpsL, which allows positive selection with streptiomycin for the loss of the
vector, to generate an in frame deletion plasmid, pRmpA1-4. The resulting plasmids were then mobilized to K. pneumoniae CG43S3 through conjugation from E. coli S17-1 λpir. The transconjugants, carrying with pRmpA1-4 integrated in the chromosome via homologous recombination, were selected by ampocollin and kanamycin on minimal medium. One of colonies was grown in LB at 37℃ for overnight and then spread onto a LB plate containing 500 μg/ml streptomycin. The streptomycin-resistant and kanamycin sensitive colonies were selected and deletion of rmpA was verified by PCR and Southern blotting with a gene specific probe. The resulting bacteria with mutation in rmpA were named rmpA-.
10. Construction the complement strains
A 1.1 kb fragment including the promoter and coding sequence was amplified with the specific primers rmpA05/ rmpA06 and then cloned to yT&A (PCR cloning vector), named pRmpA02. A HindIII/EcoRI fragment of pRmpA02 was subcloned to pRK415 and resulted in pRmpA03.
11. Southern blotting analysis
Chromosome DNA was prepared from overnight cultures of K. pneumoniae CG43S3 or the derived bacteria grown in LB medium. After the DNA digested with restriction endonuclease EcoRV, the DNA fragments were resolved on 1% agarose gels by electrophoresis and transferred to Hybond-N+ membrane (Amersham-Pharmacia, Piscataway, NJ). Finially, the membranes were hybridized with rmpA probes to confirm the mutation of rmpA-.
12. Quantification of biofilm formation
of 96-well microtire dishes made of PVC (TPP 96 flat) with some modification of the reported protocol. Essentially, the indicator medium (200 μl/well) contained an aliquot of 1:10 diluted overnight bacteria culture and the plat was incubated at 37℃ for 36 h for biofilm formation. The unadherent bacteria was washed triply with 200 μl ddH2O and then 200 μl of 1% crystal violet (CV) was added to each well. After the
plate was placed at room temperature for 30 min, ddH2O was utilized to wash the
plate for three times again. Finally, the CV-strained biofilm was solubilized in 200 μl of 0.1% SDS and absorbance determined at OD595nm using spectrophotometer
(ELx800, BIO-TEK).
13. Extraction and quantification of CPS
CPS was extracted by using the method described (7). Briefly, bacteria were collected from 500 μl of culture medi and mixed with 100 μl of 1% Zwittergent 3-14 detergent (Sigma-Aldrich, Milwaukee, WI) in 100 mM citric acid (pH2.0). The mixture was incubated at 50℃ for 20 min. After centrifugation, 250 μl of the supernatant was transferred to a new tube and CPS was precipitated with 1 ml of absolute ethanol. The pellet was then dissolved in 200 μl distilled water and a 1,200 μl of 12.5 mM borax (Sigma-Aldrich, Milwaukee, WI) in H2SO4 was added. The
mixture was vigorously vortexed, boiled for 5 min, cooled, and then 20 μl 0.15% 3-hydroxydiphenol (Sigma-Aldrich, Milwaukee, WI) was added and the absorbance at 520 nm was measured. The uronic acid content was determined from a standard curve of glucuronic acid (Sigma-Aldrich, Milwaukee, WI) and expressed as μg per 109 CFU.
14. Construction of the reporter gene fusion
rmpAp01/rmpAp02 (Table 3) was cloned into yT&A (PCR cloning vector), and resulted in the plasmids pRmpA04. The BamHI/BglII fragment of pRmpA04 was then subcloned in front of the plasmidless lacZ gene, and resulted in the plasmid pRmpAZ15.
15. β-galactosidase activity assay
β-galactosidase was assayed according to the method of Miller (22). The bacteria in the early or late logarithmic growth phase (optical density at 600 nm 0.4 or 0.7) were taken 100 μl, and mixed with 900 μl Z buffer (60 mM Na2HPO4, 40 mM
NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol), 17 μl of 0.1%
SDS and 35 μl chloroform and incubated for 10 min at 30℃. Subsequently, 200 μl of 4 mg/ml ο-nitrophenyl-β-D-galactopyranoside (ONPG) was added and the mixture vortexed for 10 s, then incubated at 30℃ until yellow color was apparent. Finally, the reaction was stopped by adding 500 μl of stop solution (1 M Na2CO3) and the
absorbance of the supernatant was measured OD420. One unit of β-galactosidase is
Results
Part 1 : Prevalence of pLVPK in Klebisella pneumoniae clinical
isolates
1-1. Plasmid typing of 127 K. pneumoniae clinical isolates.
To identify the association of large plasmid and diseases, a total of 127 K. pneumoniae isolates respectively from liver abscess (TVH1-19), bile (TVH20-28), urine (TVH29-44), ascites (TVH45-58), sputum (TVH-59-78), wound (TVH79-102), and blood (TVH103-128) were collected. By using PFGE analysis, 106 clinical isolates were found to harbor large plasmids of at least 200 kb in size (Fig. 1). Among the 106 clinical isolates, 19 (100%) liver abscess isolates, 8 (88.89%) bile isolates, 12 (75%) urine isolates, 11 (78.57%) ascites isolates, 16 (70.37%) sputum isolates, 19 (79.17%) wound isolates, and 12 (80.77%) blood isolates were identified. This indicated that many of the K. pneumoniae clinical isolates harbor large plasmid and all of liver abscess isolates carry large plasmid. In addition, some of the isolates carry more than one large plasmid, such as K. pneumoniae TVH6, TVH9, and TVH16.
1-2. Distribution of rmpA, terA, iutA, iroB, and silS genes in clinical isolates.
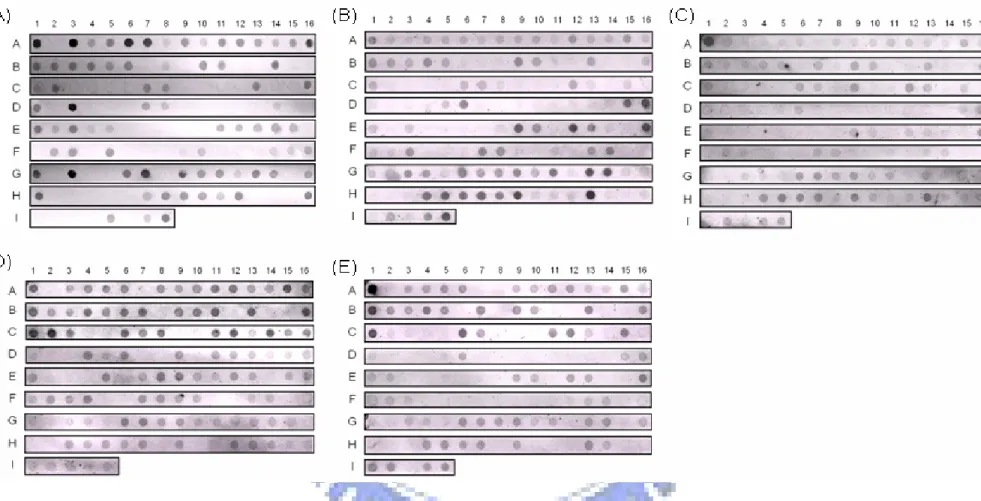
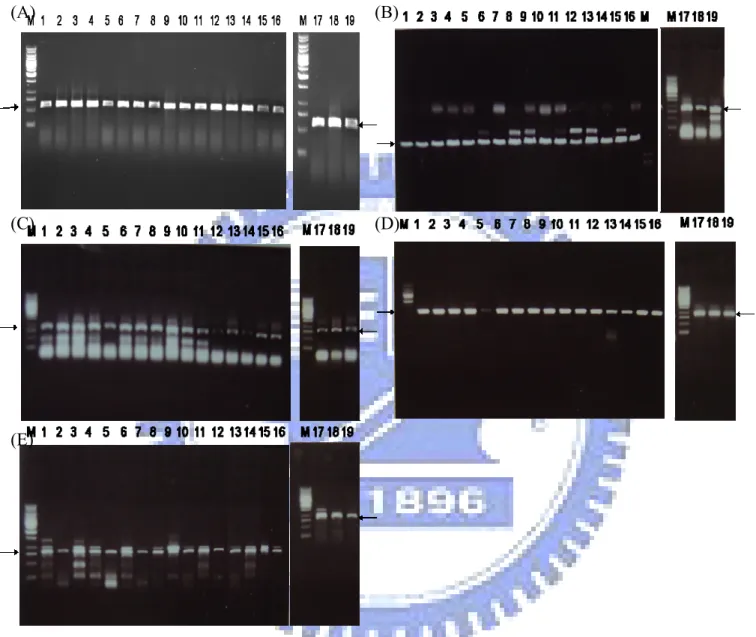
Genomic DNAs of 127 Klebsiella pneumoniae isolates were extracted by standard techniques, and the DNAs used for dot blotting detection are rmpA, terA, iutA, iroB, and silS genes. As shown in Fig. 2 and Table 4, many of the isolates harbored rmpA, terA, iutA, iroB, and silS genes. Interestingly, all liver abscess isolates appear to carry the genes, rmpA, iutA, and iroB. In order to further confirm the results of the dot blotting, PCR analysis of 19 K. pneumoniae liver abscess isolates were performed. As shown in Fig. 3, the PCR products of expected size were obtained in all
the liver abscess isolates which further confirmed the finding of the dot blotting analysis. But the other isolates carried the 5 genes on the genome were respectively 33.33 % (bile), 31.25 % (urine), 21.43 % (ascites), 42.11 % (sputum), 45.83 % (wound), and 42.31 % (blood). These results suggested a close association of pLVPK with liver abscess isolates.
1-3. The liver abscess isolates harbor pLVPK-like plasmid.
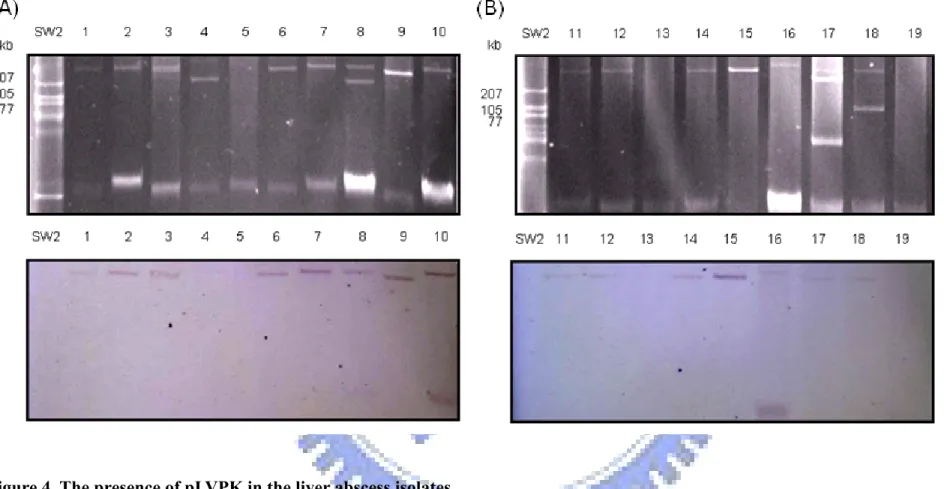
To determine whether pLVPK plasmid is indeed carried on all the liver abscess isolates, the plasmids of these liver abscess isolates were isolated and resolved by PFGE, and the gel was subjected to Southern blotting analysis with an rmpA probe. As shown in Fig. 4, rmpA gene could be detected on both chromosome and the large plasmid of TVH10 and TVH16. These plasmids appeared to contain rmpA except those of TVH 5, TVH 13, and TVH 19. However, the discrepancy is not consistent with the finding of the PCR detection (Fig. 2). But these results indicated the pLVPK-like plasmids identified in liver abscess isolates may play a role in the Klebsiella infection process.
1-4. The siderophore biosynthesis activity of the K. pneumoniae isolates.
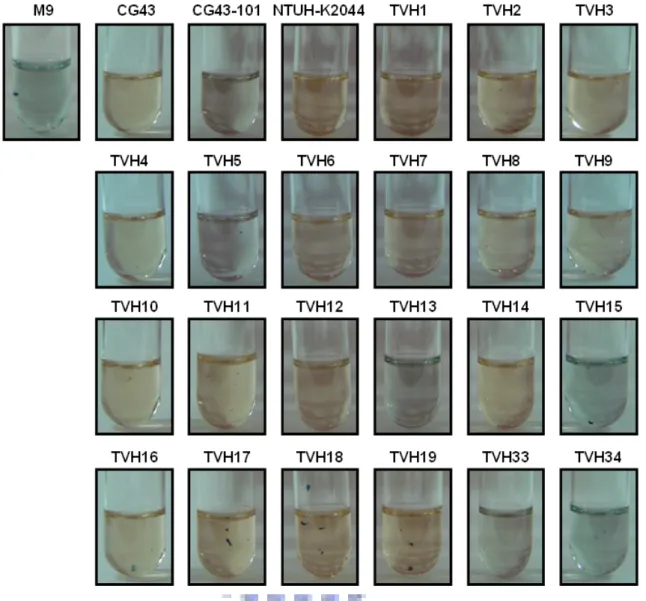
Iron is generally an essential element for all the microorganisms. To survive under an iron-deficient condition, bacteria produce siderophores which are low molecular mass of iron chelating compounds capable of binding iron with high affinity (41). Since both enterobactin and aerobactin iron acquisition gene clusters are contained in pLVPK, siderophore synthesis activity of the isolates were determined using CAS assay solution and compared. As shown in Fig. 5, comparing to K. pneumoniae CG43-101 curing of pLVPK, most of the liver abscess isolates produced siderophore in the iron deficient broth. However, no siderophore synthesis activity
could be detected in TVH13 and TVH15, as well as the plasmidless isolates TVH33, and TVH34.
1-5. The capsular polysaccahride biosynthesis activity and biofilm formation of the K. pneumoniae isolates.
In previous study, mucoid phenotype of K. pneumoniae was a large plasmid-encoded virulence factor (24, 25). As a reporter for pLVPK activity, biofilm formation activity of these isolates were also determined and compared. As shown in Fig. 6 and Table 5, all the liver abscess isolates displayed different levels of mucoidy, implying that pLVPK or the pLVPK-like plasmids affected differently the level of capsular polysaccharide biosynthesis. As shown in Fig. 7, biofilm formation capability of isolates appeared to be variable, which revealed that, in addition to the virulent plasmid, other factors encoding by the genes reside in bacterial chromosome also play roles in regulation of the biofilm formation and biosynthesis of the capsular polysaccharide.
Part 2 : Identification of the regulatory role of RmpA
2-1. RT-PCR demonstrated the expression of rmpA in K. pneumoniae CG43. In previous study, the rmpA2 were transcribed in K. pneumoniae CG43, but we
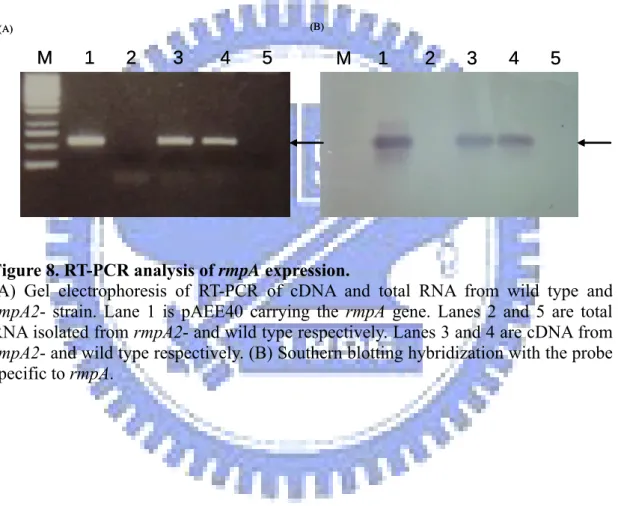
didn’t identity the present of rmpA gene. After pLVPK sequencing, we confirmed the existence of rmpA and rmpA2 genes. To demonstrate expression of the rmpA encoding gene in K. pneumoniae CG43 and the derived rmpA2-, total RNA were isolated and subject to RT-PCR and Southern blotting hybridization. As shown in Fig. 8 the rmpA transcript could be detected in both the K. pneumoniae CG43 and rmpA2-, including an active expression of the rmpA gene.
2-2. Biological role of RmpA.
To investigate the function of RmpA, the rmpA deletion mutant was constructed
by the allelic exchange and the deletion confirmed by using Southern blotting hybridization. The deleted DNA fragments included about 527 bp in rmpA. As shown in Fig. 9, 800 bp and 1327 bp signals were obtained respectively in the rmpA- and K. penumoniae CG43S3. The 4083 bp signals in the rmpA- and K. penumoniae CG43S3 were the fragments including the rmpA2 gene. Both rmpA- and rmpA2- strain displayed similar colony in size with the K. pneumoniae CG43S3. In addition, the growth rate of rmpA- and rmpA-rmpA2- appear to grow faster than both wild type and rmpA2- in LB medium, but not in M9 medium (Fig. 10A). Mucoidy of rmpA- and rmpA2- were significantly reduced as assessed using low-speed centrifugation. Introducing the plasmid carrying either rmpA or rmpA2 into the mutants appeared to compensate for the deficiency (Fig. 10B).
The amount of CPS produced in wild type and mutant strains were further quantified by measuring the uronic acid content, which serves as an indicator of
Klebsiella K2 CPS. As shown in Fig. 10C, the rmpA-, as well as the rmpA2-, produced much less CPS than wild type strain and the deficiency could be restored by the complements. In previous studies, expression of CPS has been correlated with the regulation of biofilm formation. As shown in Fig. 11, capability of biofilm formation in rmpA- and rmpA2- like that in rcsB- were increased. Transformation of the plasmid carrying rmpA or rmpA2 into the mutants appeared to be able restore the capability of biofilm formation. These results suggested that RmpA as well as RmpA2 could positively regulate the CPS biosynthesis.
2-3. RmpA regulates positively the expression of cpsorf1-2 and cpsorf16-17.
The reducing production of CPS by deletion of rmpA or rmpA2 suggested a regulatory role on cps genes expression, while the three lacZ reporter fusion constructs (Fig. 12A), Porf1-2, Porf3-15, and Porf16-17, were analyzed in the rmpA-. Activity
of Porf1-2 and Porf16-17 appeared to decrease 30 % in comparing with wild type bacteria,
but in rmpA2- was only reduced 20 %. Compared the activity of Porf3-15 in wild type
with either rmpA- or rmpA2- strain, no obvious change of Porf3-15 activity in rmpA- or
rmpA2- strains was noted (Fig. 12B).
2-4. Expression of rmpA and rmpA2 in K. pneumoniae CG43.
To analysis the expression of rmpA and rmpA2, the putative promoters of rmpA and rmpA2 were isolated and respectively fusion with the promoterless lacZ in pLacZ15, and be transformed into K. pneumoniae LacZ16. As shown in Fig. 13, activity of PrmpA2 appeared to be much higher than PrmpA. As shown in Fig. 14, 49.2%
identity in nucleotide sequence lower than that of coding region. Analysis the putative promoters of rmpA and rmpA2 showed Fur binding box and RcsAB binding box on PrmpA. Alignment of the putative RcsA-RcsB binding site with 40-bp region of the
Erwinia amylovora ams (amylovoran biosynthesis), E. coli cps (16), and the rmpA promoter using VectorNTI (Invitrogen Vector NTI™ Advance) identified a 28 bp region of RcsAB box. To examine whether RcsAB affect rmpA expression, PrmpA
activity was analyzed in either rcsA- or rcsB-. As shown in Fig. 15, rcsB deletion appeared to reduced PrmpA activity to half of the wild type strain. Interestingly, the
deletion of rcsB conferred no effect on the activity of PrmpA2. But no obvious change
of PrmpA and PrmpA2 in rcsA- was noted. This indicated a differential regulation of the
Discussion
Most of the work on pathogenesis of K. pneumoniae has been limited to studying the polysaccharidic capsule, which protect against serum bactericidal activity and phagocytosis. Recently, many reports indicated the large plasmid is associated with virulence. In our study, many K. pneumoniae clinical isolates were also shown to be carried large plasmids. Furthermore, all liver abscess isolates (19 strains) and some other isolates (39 strains) could carry the pLVPK-like plasmid.
In Southern blotting analysis, the large plasmid in many liver abscess isolates carried rmpA gene could be detected except those of K. pneumoniae TVH 5, TVH 13, and TVH 19. However, the discrepancy, which is not consistent with the finding of the PCR detection (Fig. 3), is likely due to the concentration of the plasmids was too low to be detected. But the rmpA gene could be detected on both chromosome and the large plasmid of K. pneumoniae TVH10 and TVH16. To analyze sequence around the rmpA gene on pLVPK showed many insertion sequences (IS) around the rmpA gene suggesting rmpA gene could be located on a pathgenocity island by horizontal transfer to chromosome, such as SHI-2 island (23). The possibility is being investigated.
All liver abscess isolates carried the pLVPK-like plasmid suggesting the plasmids on those isolates have similar ability with pLVPK. In CAS assay, many liver abscess isolates could synthesis siderophore, except K. pneumoniae TVH13 and TVH15, indicating that siderophore synthesis pathway in some of the pLVPK carrying isolates may be deficient. In sedimentation test, all the liver abscess isolates containing pLVPK-like plasmid displayed more mucoid than the plasmidless isolates K. pneumoniae TVH33, and TVH34, suggesting the large plasmid is associated with the mucoid phenotype. Biofilm formation capability of isolates appeared to be
variable, which revealed that, in addition to the virulent plasmid, other factors encoding by the genes reside in bacterial chromosome also play roles in regulation of the biofilm formation, such as rcsA and rcsB reported to repress the biofilm formation (40).
Fang et al. has reported that the strains carrying rmpA were significantly associated with the hypermucoviousity phenotype and purulent tissue infections, such as liver abscess (20, 48). In the study, we have shown that the rmpA gene located on pLVPK or pLVPK-like plasmid could be correlated with liver abscess suggesting the RmpA located on pLVPK could regulate other virulence factors involved in the K. pneumoniae pathogenesis.
To investigate the functional role of rmpA, the rmpA deletion mutant was generated. The rmpA deletion mutant reduced the CPS production and increased the growth rate. Although rmpA2- also affected negatively the CPS production, no effect on the bacterial growth indicating a differential role of the two transcription factors suggesting RmpA play more important role than RmpA2 on CPS synthesis. Nevertheless, analysis of expression of the cps gene revealed that RmpA as well as RmpA2 appeared to reduce the expression of cps gene orf1-2 and orf16-17. Analysis of the ORF showed that orf1 is a homolog of S. typhimurium LT2 galF and E. coli galU, which encodes the enzyme UDP-glucose pyrophosphorylase that regulates the supply of UDP-galactose and UDP-glucose, two major precursors for the biosynthesis of CPS (45, 46). While the orf16 and orf17 encoding respectively by the manC and manB genes, which encode mannoas-1-phosphate guanylytransferase (GDP-mannose pyrophosphorylase) and phosphomannomutase respectively that have been reported to be involved in the biosynthesis of mannose (2, 45, 46).
Notably, the cps expression was regulated by many factors, such RcsAB and RmpA2 (18). Previous study was determined that the E. coli co-transformed with
Klebsiella K2 cps gene cluster, rmpA, and rcsB could produce Klebsiella K2 CPS. But E.coli co-transformed with only K2 cps gene cluster and rmpA could not produce Klebsiella K2 CPS (24). Analysis of RmpA revealed a LuxR-like C-terminus as RcsA (21, 24), implying that RmpA in analogy with the role of RcsA interacts with RcsB to activate cps gene in K. pneumoniae. However, this hypothesis awaits to be investigated.
The activity of PrmpA and PrmpA2 after time course showed that the expression of
RmpA and RmpA2 were under different regulation. Analysis of the putative promoter of rmpA was found a putative RcsAB box and Fur box, but not rmpA2, suggesting the expression of rmpA could be regulated by RcsAB complex and Fur. This hypothesis was demonstrated by analysis the activity of PrmpA in wild type and rcsB- strain. But
comparison of the activity of PrmpA or PrmpA2 under iron-limit and iron-rich medium
were not obvious difference. We presumed that rmpA belonged to Rcs regulon is under the phosphoryl-RcsB control, but not rmpA2. To further identify the RcsAB binding region on PrmpA, the deletion fragment of PrmpA could be fused with LacZ
reporter and analyzed.
In previous study, the RmpA2 could be autoregulated (18). Whether the RmpA could be autoregulated or cross-regulated with RmpA2, the activities of PrmpA and
PrmpA2 in rmpA- and rmpA2- strain were analyzed. The activities of PrmpA in LacZ16,
rmpA- and rmpA2- strain or PrmpA2 in LacZ16, rmpA- and rmpA2- strain were no
obvious difference (Data not show) suggesting RmpA or RmpA2 could not directly cross-regulate the expression of RmpA or RmpA2 and RmpA could not be autoregulated. However, this hypothesis awaits to be investigated.
In conclusion, as shown in Fig. 16, we propose a regulatory circuit that RmpA belong to Rcs regulon, but not RmpA2. RcsB activates the expression of rmpA. The RmpA could interact with RcsB to activate CPS synthesis. But RmpA2 could directly
bind to cps genes without RcsB. In addition, expression of rmpA could be activated with osmotic shock or metal ion. The signal to activate the expression of rmpA2 is unknown. However, this pathway is yet to be identified.
Reference
1. Arakawa Y, Ohta M, Wacharotayankun R, Mori M, Kido N, Ito H,
Komatsu T, Sugiyama T, Kato N. Biosynthesis of Klebsiella K2 capsular
polysaccharide in Escherichia coli HB101 requires the functions of rmpA and the chromosomal cps gene cluster of the virulent strain Klebsiella pneumoniae Chedid (O1:K2). Infect Immun. 1991 Jun;59(6):2043-50.
2. Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M.
Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995 Apr;177(7):1788-96.
3. Borremans B, Hobman JL, Provoost A, Brown NL, van Der Lelie D.
Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol. 2001 Oct;183(19):5651-8.
4. Brown NL, Barrett SR, Camakaris J, Lee BT, Rouch DA. Molecular genetics
and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol Microbiol. 1995 Sep;17(6):1153-66.
5. Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and
analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004 Aug 4;337:189-98.
6. Der Vartanian M. Differences in excretion and efficiency of the aerobactin and
enterochelin siderophores in a bovine pathogenic strain of Escherichia coli. Infect Immun. 1988 Feb;56(2):413-8.
7. Domenico P, Schwartz S, Cunha BA. Reduction of capsular polysaccharide
production in Klebsiella pneumoniae by sodium salicylate. Infect Immun. 1989 Dec;57(12):3778-82.
8. Ebel W, Trempy JE. Escherichia coli RcsA, a positive activator of colanic acid
capsular polysaccharide synthesis, functions to activate its own expression. J Bacteriol. 1999 Jan;181(2):577-84.
9. Ghosh A, Singh A, Ramteke PW, Singh VP. Characterization of large plasmids
encoding resistance to toxic heavy metals in Salmonella abortus equi. Biochem Biophys Res Commun. 2000 May 27;272(1):6-11.
10. Gupta A, Matsui K, Lo JF, Silver S. Molecular basis for resistance to silver
cations in Salmonella. Nat Med. 1999 Feb;5(2):183-8.
11. Guan S, Clarke AJ, Whitfield C. Functional analysis of the
galactosyltransferases required for biosynthesis of D-galactan I, a component of the lipopolysaccharide O1 antigen of Klebsiella pneumoniae. J Bacteriol. 2001 Jun;183(11):3318-27.
12. Han, S.H.B. Review of hepatic abscess from Klebsiella pneumoniae. An association with diabetes mellitus and septic endophthalmitis. West. J. Med.162:220-224.
13. Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins,
siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003 Apr 1;100(7):3677-82. Epub 2003 Mar 24.
14. Huang YH, Ferrieres L, Clarke DJ. The role of the Rcs phosphorelay in
Enterobacteriaceae. Res Microbiol. 2006 Apr;157(3):206-12. Epub 2006 Jan 4.
15. Kado CI, Liu ST. Rapid procedure for detection and isolation of large and
small plasmids. J Bacteriol. 1981 Mar;145(3):1365-73.
16. Kelm O, Kiecker C, Geider K, Bernhard F. Interaction of the regulator
proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol Gen Genet. 1997 Sep;256(1):72-83.
17. Koehler TM. Bacillus anthracis genetics and virulence gene regulation. Curr
Top Microbiol Immunol. 2002;271:143-64. Review.
18. Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsuleb biosynthesis in
Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol. 2003 Feb;185(3):788-800.
19. Lee, C.C., C. Y. Chen, F. H. Chen, R. A. Zimmerman, and H. S. Hsiao.
Septic metastatic endophthalimitis from Klebsiella pneumoniae liver abscess: CT and MR imaging characteristics-report of three cases. Radiology.
20. Lin JC, Chang FY, Fung CP, Xu JZ, Cheng HP, Wang JJ, Huang LY, Siu
LK. High prevalence of phagocytic-resistant capsular serotypes of Klebsiella
pneumoniae in liver abscess. Microbes Infect. 2004 Nov;6(13):1191-8.
21. Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal
transduction system. Annu Rev Microbiol. 2005;59:379-405.
22. Miller, J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor
Laboratory Press, Cold Spring Harbor, NY.
23. Moss, J.E., Cardozo, T.J., Zychlinsky, A., Groisman, E.A. The
selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol Microbiol. 1999 Jul;33(1):74-83.
24. Nassif X, Honore N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of
colonic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol. 1989 Oct;3(10):1349-59.
25. Nassif X, Fournier JM, Arondel J, Sansonetti PJ. Mucoid phenotype of
1989 Feb;57(2):546-52.
26. Pannucci J, Okinaka RT, Sabin R, Kuske CR. Bacillus anthracis pXO1
plasmid sequence conservation among closely related bacterial species. J Bacteriol. 2002 Jan;184(1):134-41.
27. Peng HL, Wang PY, Wu JL, Chiu CT, Chang HY. Molecular epidemiology of
Klebsiella pneumoniae. Chinese Journal of Microbiology 1991
28. Podschun, R., and U. Ullmann. Klebsiella spp. as nosocomial pathogens:
epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589-603.
29. Pristovsek P, Sengupta K, Lohr F, Schafer B, von Trebra MW, Ruterjans H,
Bernhard F. Structural analysis of the DNA-binding domain of the Erwinia
amylovora RcsB protein and its interaction with the RcsAB box. J Biol Chem. 2003 May 16;278(20):17752-9. Epub 2003 Mar 5.
30. Pullinger GD, Lax AJ. A Salmonella dublin virulence plasmid locus that
affects bacterial growth under nutrient-limited conditions. Mol Microbiol. 1992 Jun;6(12):1631-43.
31. Rahn A, Whitfield C. Transcriptional organization and regulation of the
Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol Microbiol. 2003 Feb;47(4):1045-60.
32. Sahly H, Podschun R, Oelschlaeger TA, Greiwe M, Parolis H, Hasty D,
Kekow J, Ullmann U, Ofek I, Sela S. 2000. Capsule impedes adhesion to and
invasion of epithelial cells by Klebsiella pneumoniae. Infect Immun. Dec; 68(12):6744-9
33. Sambrook, J., Russell, D.W. Molecular coloning: a laboratory manual-3rd
edition. Cold Spring Habor Laboratory Press, Cold Spring Harbor, New YorK.
34. Schaberg DR, Culver DH, Gaynes RP. Major trends in the microbial etiology
of nosocomial infection. Am J Med. 1991 Sep 16;91(3B):72S-75S.
35. Schwartz, D.C., and Cantor, C.R. Separation of yeast chromosome-sized
DNAs by pulsed field gradient gel electrophoresis. Cell 37, pp. 67-75 .
36. Schwyn B, Neilands JB. Universal chemical assay for the detection and
determination of siderophores. Anal Biochem. 1987 Jan;160(1):47-56.
37. Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev
Microbiol. 1996;50:753-89. Review.
38. Taylor DE, Rooker M, Keelan M, Ng LK, Martin I, Perna NT, Burland NT,
Blattner FR. Genomic variability of O islands encoding tellurite resistance in
enterohemorrhagic Escherichia coli O157:H7 isolates. J Bacteriol. 2002 Sep;184(17):4690-8.
community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med. 2002 May 13;162(9):1021-7.
40. Vianney A, Jubelin G, Renault S, Dorel C, Lejeune P, Lazzaroni JC.
Escherichia coli tol and rcs genes participate in the complex network affect affecting curli synthesis. Microbiology. 2005 Jul;151(Pt 7):2487-97.
41. Wacharotayankun R, Arakawa Y, Ohta M, Hasegawa T, Mori M, Horii T,
Kato N. Involvement of rcsB in Klebsiella K2 capsule synthesis in Escherichia
coli K-12. J Bacteriol. 1992 Feb;174(3):1063-7.
42. Wacharotayankun R, Arakawa Y, Ohta M, Tanaka K, Akashi T, Mori M,
Kato N. Enhancement of extracapsular polysaccharide synthesis in Klebsiella
pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect Immun. 1993 Aug;61(8):3164-74.
43. Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to
hemophores. Annu Rev Microbiol. 2004;58:611-47.
44. Wehland M, Bernhard F. The RcsAB box. Characterization of a new operator
essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem. 2000 Mar 10;275(10):7013-20.
45. Whitfield C, Paiment A. Biosynthesis and assembly of Group 1 capsular
polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr Res. 2003 Nov 14;338(23):2491-502. Review.
46. Whitfield C, Roberts IS. Structure, assembly and regulation of expression of
capsules in Escherichia coli. Mol Microbiol. 1999 Mar;31(5):1307-19. Review.
47. Yarng, S.S., C. L. Hsieh, and T. L. Chen. Vitrectomy for endogenous
Klebsiella pneumoniae endophthalmitis with massive subretinal abscess. Ophthalmic. Surg. Lasers 28:147-150.
48. Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang
YC. Association between rmpA and magA genes and clinical syndromes caused
by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006 May 15;42(10):1351-8. Epub 2006 Apr 11.
Strain Genotype or relevant property Reference or source E. coli:
NovaBlue(DE3) endA1 hsdR17(rk12-mk12+) supE44 thi-1 recA1 gyrA96 relA1 lac[F,
pro AB lacqZ△M15:: Tn10](DE3);Tetr
Novagen JM109 RecA1 supE44 endA1 hsdR17 gyrA96 rolA1 thi △
(lac-proAB)
Laboratory stock S17-1λpir Tpr Smr recA, thi, pro, hsdR-
M+
[RP4-2-Tc::Mu:KmrTn7] (pir)
De Lorenzo et al., 1994
ICC188λpir △(ara-leu) araD △lac×74 galE galK phoA20 Taylor et al.,1989
K. pneumoniae:
CG43 Clinical isolate of K2 serotype Laboratory stock
CG43-101 Curing the large plasmid pLVPK from CG43 Laboratory stock
CG43-S3 △rspl, Smr Laboratory stock
rmpA- CG43-S3△rmpA Smr This study
rmpA2- CG43-S3△rmpA2 Smr Laboratory stock
rcsA- CG43-S3△rcsA Smr This study
rcsB- CG43-S3△rcsB Smr Laboratory stock
LacZ16 CG43S3△lacZ Smr Laboratory stock
rmpA-Z16 LacZ16△rmpA Smr This study
rmpA2-Z16 LacZ16△rmpA2 Smr Laboratory stock
rcsA-Z16 LacZ16△rcsA Smr This study
rcsB-Z16 LacZ16△rcsB Smr Laboratory stock
Plasmids Relevant characteristic Source or reference
yT&A vector PCR cloning vector, Apr Sigma
pKAS46 Suicide vector, Apr Kmr Novagene
pRK415 Shuttle vector, mob+, Tcr Laboratory stock
pLacZ15 A derivative of pYC016, containing lacZ as a reporter, Cmr Laboratory stock
pRmpA1-4 pKAS46 carrying a △rmpA fragment This study
pRmpA02 A 1.1 kb PCR product of the rmpA locus with the putative promoter cloned into yT&A This study
pRmpA03 A HindIII/EcoRI fragment of pRmpA02 cloned into the pRK415 This study
pRmpA04 A 0.5 kb PCR product of the rmpA putative promoter region cloned into yT&A This study
pRmpAZ15 A BamHI/BglII fragment of pRmpA04 cloned into the pLacZ15 This study
pRmpA2Z15 A 0.5 kb fragment of the rmpA2 putative promoter region cloned into the pLacZ15 Laboratory stock
porf1Z15 A 0.8 kb fragment of the cps orf1-2 promoter region cloned into the pLacZ15 Laboratory stock
porf3Z15 A 0.9 kb fragment of the cps orf3-15 promoter region cloned into the pLacZ15 Laboratory stock
porf16Z15 A 0.4 kb fragment of the cps orf16-17 promoter region cloned into the pLacZ15 Laboratory stock
pYC084 A XbaI/HindIII fragment cloned into the pRK415 Laboratory stock
pAEE40 A 7 kb fragment of pLVPK cloned into pUC18
Primer Sequence
rmpA1p-01 5'-GTCGGATCCATCGCCAAATA-3' rmpA1p-02 5'-CAG TCA ACA CGG TGC TTT ACA T-3' rmpA01 5'CTCTAGATAAGGCGGCCTTCG-3’ rmpA02 5’-ATAGTCGACGCTATGCTTTACA-3’ rmpA03 5’-TGGTCGACGAAAGATGGCTC-3’ rmpA04 5’-ATGAGCTCAATGTATGCCAAGG-3’ rmpA05 5’-GGCCGAAAGCAGTTAACTG -3’ rmpA06 5’-TTACCTAAATACTTGGCATGA GC -3’ rmpA07 5’- GGCCGAAAGCAGTTAACTG -3’ rmpA08 5’- TTACCTAAATACTTGGCATGAGC -3’ Yu-05 5’-CCTTCACATCCCCTCCCCTT -3’ Yu-06 5’-GTCGGATCCATCGCCAAATAA-3’ iutA01 5'- GAACAGCACAGAGTAGTTCA -3' iutA02 5'- CAGTACACTGAAAACAAGATTG -3' iroB01 5'- CTTTCTGTACCATCGCGATC -3' iroB02 5'- TCATTCTTCAGCGAAGAGAT -3' silS01 5'- ATCAGCCTGTCCACGATACT -3' silS02 5'- GTCAAACATGACCCTGTCAG terA01 5'- GGGGGCAATGCCCCTTTAATAGCT -3' terA02 5'- AGACGGGCAATCGCACACAG -3'
45.67 (58) 55.52 (70) 77.17 (98) 51.45 (73) 61.42 (78) 55.52 (70) 83.46 (106) Total number 42.31 (11) 53.85 (14) 76.92 (20) 69.23 (18) 80.77 (21) 46.15 (12) 80.77 (21) blood (26) 45.83 (11) 54.17 (13) 70.83 (17) 62.5 (5) 66.67 (16) 58.33 (14) 79.17 (19) wound (24) 42.11 (8) 52.63 (10) 84.21 (16) 42.11 (8) 57.89 (11) 63.15 (12) 84.21 (16) sputum (19) 21.43 (3) 21.43 (3) 71.43 (10) 21.43 (3) 21.43 (3) 28.58 (4) 78.57 (11) ascites (14) 31.25 (5) 56.25 (9) 68.75 (11) 43.75 (7) 31.25 (5) 31.25 (5) 75 (12) urine (16) 33.33 (3) 44.44 (4) 66.67 (6) 44.44 (4) 33.33 (3) 44.44 (4) 88.89 (8) bile (9) 89.47 (17) 89.47 (17) 94.74 (18) 100 (19) 100 (19) 100 (19) 100 (19) liver abscess (19) % of isolates carry mpA, iutA,
iroB, sils, and terA (No. of
isolates) % of isolates
carry terA (No. of isolates) % of isolates
carry silS (No. of isolates) % of isolates
carry iroB (No. of isolates) % of isolates
carry iutA (No. of isolates) % of isolates
carry rmpA (No. of isolates) % of isolates carry large plasmid (No. of isolates) Origin of isolation (No. of isolates) 45.67 (58) 55.52 (70) 77.17 (98) 51.45 (73) 61.42 (78) 55.52 (70) 83.46 (106) Total number 42.31 (11) 53.85 (14) 76.92 (20) 69.23 (18) 80.77 (21) 46.15 (12) 80.77 (21) blood (26) 45.83 (11) 54.17 (13) 70.83 (17) 62.5 (5) 66.67 (16) 58.33 (14) 79.17 (19) wound (24) 42.11 (8) 52.63 (10) 84.21 (16) 42.11 (8) 57.89 (11) 63.15 (12) 84.21 (16) sputum (19) 21.43 (3) 21.43 (3) 71.43 (10) 21.43 (3) 21.43 (3) 28.58 (4) 78.57 (11) ascites (14) 31.25 (5) 56.25 (9) 68.75 (11) 43.75 (7) 31.25 (5) 31.25 (5) 75 (12) urine (16) 33.33 (3) 44.44 (4) 66.67 (6) 44.44 (4) 33.33 (3) 44.44 (4) 88.89 (8) bile (9) 89.47 (17) 89.47 (17) 94.74 (18) 100 (19) 100 (19) 100 (19) 100 (19) liver abscess (19) % of isolates carry mpA, iutA,
iroB, sils, and terA (No. of
isolates) % of isolates
carry terA (No. of isolates) % of isolates
carry silS (No. of isolates) % of isolates
carry iroB (No. of isolates) % of isolates
carry iutA (No. of isolates) % of isolates
carry rmpA (No. of isolates) % of isolates carry large plasmid (No. of isolates) Origin of isolation (No. of isolates)
large + TVH10 large + TVH9 large + TVH8 large + TVH7 large + TVH6 large + TVH5 large + TVH4 large + TVH3 large + TVH2 large + TVH1 Colony size mucoid strains large + TVH10 large + TVH9 large + TVH8 large + TVH7 large + TVH6 large + TVH5 large + TVH4 large + TVH3 large + TVH2 large + TVH1 Colony size mucoid strains large + TVH19 large + TVH18 small -TVH17 large + TVH16 small + TVH15 large + TVH14 large + TVH13 large + TVH12 large + TVH11 large + TVH19 large + TVH18 small -TVH17 large + TVH16 small + TVH15 large + TVH14 large + TVH13 large + TVH12 large + TVH11
Table 5. String test and colony size of 19 liver abscess isolates.
Strains were defined as mucoid positive and large colony when viscous strings were > 0.5 cm (a positive string test result) and colony diameter were > 3 mm (a large colony result).
(A) (E)
(B) (F)
(C) (G)
(D) (H)
Figure. 1. Plasmid profile analysis of the 127 K. pneumoniae isolates by PFGE.
Lanes 1 to 16 of (A)-(H), clinical isolates of K. pneumoniae; the size of Erwinia SW2 plasmids are marked. The boxed regions in (A) and (B) are liver abscess isolates.
Figure 2. Dot-blotting analysis of 127 K. pneumoniae clinical isloates respectively probing with rmpA, iutA, iroB, silS, and terA gene.
The genomic DNAs were extracted and spotted on a Hybond-N+ membrane. The membrane was hybridized respectively with the probe specific to (A) rmpA, (B) iutA, (C)
iroB, (D) silS, and (E) terA. The bacteria strains are: A-1, D-1,G-1 represent CG43S3; A-2, D-2, and G-2 represent CG43-101 (cured of pLVPK); A-3, D-3, and G-3 represent
(A) (B)
(C) (D)
(E)
Figure 3. PCR detection of (A) rmpA, (B) iutA ,(C) iroB ,(D) silS, and (E) terA gene.
M: DNA molecular weight marker; PCR products in lanes 1 to 19 are K. pneumoniae TVH1 to K. pneumoniae TVH19; Arrows mark respectively the specific PCR products
Figure 4. The presence of pLVPK in the liver abscess isolates.
(A) The plasmids isolated from K. pneumoniae liver abscess isolates were subjected to gel electrophoresis. Lane SW2, Erwinia SW2; lanes 1 to 19 are K. pneumoniae TVH1 to TVH19. (B) Southern blot hybridization with the probe specific to the rmpA gene.
Figure 6. Sedimentation test of the K. pneumonia isolates.
K. pneumoniae TVH1 to 19 are liver abscess isolates. K. pneumoniae TVH33 and TVH34 are plasmidless strains.
0 0.1 0.2 0.3 0.4 0.5 0.6 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 OD 59 5 P 1 2 3 4 5 6 7 8 9 10 11 12 13 14 1516 17 18 19 0 0.1 0.2 0.3 0.4 0.5 0.6 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 OD 59 5 P 1 2 3 4 5 6 7 8 9 10 11 12 13 14 1516 17 18 19
Figure 7. Biofilm formation of wild-type and rmpA mutant strain.
Lane P is Pseudomonas aeruginosa PAO1 as a positive control. Lanes 1 to 19 are K. pneumoniae TVH1 to TVH19.
M 1 2 3 4 5 M 1 2 3 4 5 (A) (B) M 1 2 3 4 5 M 1 2 3 4 5 (A) (B)
Figure 8. RT-PCR analysis of rmpA expression.
(A) Gel electrophoresis of RT-PCR of cDNA and total RNA from wild type and rmpA2- strain. Lane 1 is pAEE40 carrying the rmpA gene. Lanes 2 and 5 are total RNA isolated from rmpA2- and wild type respectively. Lanes 3 and 4 are cDNA from rmpA2- and wild type respectively. (B) Southern blotting hybridization with the probe specific to rmpA.
Figure 9. Construction of rmpA deletion mutant.
Lanes1 and 3 are the genomic DNA of rmpA- strains digested by EcoRV. Lanes 2 and 4 are the genomic DNA digested by EcoRV. Lane 5 is the plasmid DNA of pAEE40.
Growth curve in M9 0 0.2 0.4 0.6 0.8 1 1.2 0 2 4 6 8 H ( ) O D 600 Growth curve in LB 0 0.5 1 1.5 0 2 4 6 8 H ( ) O D 600 CG43S3 rmpA-R2035 rmpA- rmpA2-Wild type rmpA -rmpA2 -rmpA -rmpA2 -Hour(S) Hour(S) Growth curve in M9 0 0.2 0.4 0.6 0.8 1 1.2 0 2 4 6 8 H ( ) O D 600 Growth curve in LB 0 0.5 1 1.5 0 2 4 6 8 H ( ) O D 600 CG43S3 rmpA-R2035 rmpA- rmpA2-Wild type rmpA -rmpA2 -rmpA -rmpA2 -Hour(S) Hour(S) (A)
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rcsB -rm pA -(p YS 01 ) rm pA2 -(pY C0 84)
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rcsB -rm pA -(p YS 01 ) rm pA2 -(pY C0 84)
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rc sB -rm pA -(p YS
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rc sB -rm pA -(p YS CG43S3 rmpA- rmpA -(pRmpA) rmpA2- rmpA2 -(pRmpA2)
CG43S3 rmpA- rmpA2- rmpA2
-(pRmpA2)
rmpA
-(pRmpA) Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rcsB -rm pA -(p YS 01 ) rm pA2 -(pY C0 84)
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rcsB -rm pA -(p YS 01 ) rm pA2 -(pY C0 84)
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rc sB -rm pA -(p YS
Glucuronic acid contenet
0 10 20 30 40 50 60 70 strain μ g /C .F .U . CG 43 S3 rm pA -rm pA 2 -rm pA -rm pA 2 -rc sB -rm pA -(p YS CG43S3 rmpA- rmpA -(pRmpA) rmpA2- rmpA2 -(pRmpA2)
CG43S3 rmpA- rmpA2- rmpA2
-(pRmpA2)
rmpA
-(pRmpA)
(B) (C)
Figure 10. Phenotype analysis of the rmpA- and rmpA2- strains.
(A) Growth analysis in LB and M9 medium, (B) sedimentation test, and (C) quantification of CPS.
(A) (B) 0 0.1 0.2 0.3 0.4 0.5 0.6 1 P rm rm rm rm rc rc C 1 2 3 4 5 6 7 8 Strain (s) OD 595 0 0.1 0.2 0.3 0.4 0.5 0.6 1 P rm rm rm rm rc rc C 1 2 3 4 5 6 7 8 0 0.1 0.2 0.3 0.4 0.5 0.6 1 P rm rm rm rm rc rc C 1 2 3 4 5 6 7 8 Strain (s) OD 595 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 1 PAO rmp rmp rmp rmp rcsA rcsB CG 1 2 3 4 5 6 7 8 Strain (s) OD 59 5 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 1 PAO rmp rmp rmp rmp rcsA rcsB CG 1 2 3 4 5 6 7 8 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 1 PAO rmp rmp rmp rmp rcsA rcsB CG 1 2 3 4 5 6 7 8 Strain (s) OD 59 5 (C) 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 1 PAO rmp rmp rmp rmp rcsA rcsB CG4 1 2 3 4 5 6 7 8 Strain (s) OD 59 5 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 1 PAO rmp rmp rmp rmp rcsA rcsB CG4 1 2 3 4 5 6 7 8 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 1 PAO rmp rmp rmp rmp rcsA rcsB CG4 1 2 3 4 5 6 7 8 Strain (s) OD 59 5
Figure 11. The biofilm formation in the wild type and mutant strain at time course (A) 12 H ,(B) 24 H, and (C) 36 H.
Lane 1: P. aeruginosa PAO1, lane 2: rmpA-, lane 3: rmpA- + pRmpA, lane 4: rmpA2-, lane 5: rmpA2- + pRmpA2, lane 6: rcsA-, lane 7: rcsB-, and lane 8: K. pneumoniae CG43.
orf1 orf2 orf3 orf4 orf5 orf6
1 kb orf7 orf8 orf9 orf10 orf11 orf12 orf13orf14 orf15 orf16 orf17
Porf1-2 Porf3-15 Porf16-17
(A)
Porf1-2 Porf3-15 Porf16-17
(B) promoter activity 0 500 1000 1500 2000 2500 3000 3500 4000 1 2 3 β -g ala ct os id as e a ctiv it y ( M ille r un its ) Z16 AZ05 A2Z01 wild type rmpA -rmpA2 -orf1 orf2 orf3 orf4 orf5 orf6
1 kb orf7 orf8 orf9 orf10 orf11 orf12 orf13orf14 orf15 orf16 orf17
Porf1-2 Porf3-15 Porf16-17
(A)
orf1 orf2 orf3 orf4 orf5 orf6 1 kb
1 kb orf7 orf8 orf9 orf10 orf11 orf12 orf13orf14 orf15 orf16 orf17
Porf1-2 Porf3-15 Porf16-17
(A)
Porf1-2 Porf3-15 Porf16-17
(B) promoter activity 0 500 1000 1500 2000 2500 3000 3500 4000 1 2 3 β -g ala ct os id as e a ctiv it y ( M ille r un its ) Z16 AZ05 A2Z01 wild type rmpA -rmpA2
-Porf1-2 Porf3-15 Porf16-17
(B) promoter activity 0 500 1000 1500 2000 2500 3000 3500 4000 1 2 3 β -g ala ct os id as e a ctiv it y ( M ille r un its ) Z16 AZ05 A2Z01 wild type rmpA -rmpA2
-Figure 12. (A)The putative promoter regions of K. pneumoniae K2 cps gene cluster
(B) Promoter activity analysis of cps genes in wild type, rmpA-, and rmpA2- stains were grown in LB at 37 . T℃ he promoter activities were measured when these strain were cultured to late exponential phase.
PrmpA PrmpA2 RcsAB box Fur box PrmpA PrmpA2 RcsAB box PrmpA PrmpA2 RcsAB box Fur box
0 200 400 600 800 1000 1200 1400 1600 1800 PrmpA Mi ll er U ni t( s) lacZ rcsA lacZ rcsB lacZ 0 2000 4000 6000 8000 10000 12000 14000 PrmpA2 M ille r U nit(s ) lacZ rcsAlac rcsBlac β -g al ac tos idas e act iv it y ( M iller u ni ts ) PrmpA 0 PrmpA2 200 400 600 800 1000 1200 1400 1600 1800 PrmpA M il le r Unit( s) lacZ rcsA lacZ rcsB lacZ rcsA-Z16 rcsB-Z16 lacZ16 0 200 400 600 800 1000 1200 1400 1600 1800 PrmpA Mi ll er U ni t( s) lacZ rcsA lacZ rcsB lacZ 0 2000 4000 6000 8000 10000 12000 14000 PrmpA2 M ille r U nit(s ) lacZ rcsAlac rcsBlac β -g al ac tos idas e act iv it y ( M iller u ni ts ) PrmpA 0 PrmpA2 200 400 600 800 1000 1200 1400 1600 1800 PrmpA M il le r Unit( s) lacZ rcsA lacZ rcsB lacZ rcsA-Z16 rcsB-Z16 lacZ16
Figure 15. Activity of PrmpA and PrmpA2 in K. pneumoniae LacZ16, rcsA-Z16, and
Figure 16. A proposed model of the regulation of RmpA and RmpA2. RcsB
activates the expression of rmpA, but not of rmpA2. RmpA could interact with RcsB to activate CPS synthesis, and RmpA2 could directly bind the promoter of cps genes to activate the expression of cps genes.