0095-1137/05/$08.00
⫹0 doi:10.1128/JCM.43.6.2668–2673.2005
Copyright © 2005, American Society for Microbiology. All Rights Reserved.
Use of the Duplex TaqMan PCR System for Detection of Shiga-Like
Toxin-Producing Escherichia coli O157
Ching-Fang Hsu, Tsung-Yu Tsai, and Tzu-Ming Pan*
Institute of Microbiology and Biochemistry, National Taiwan University, Taipei, Taiwan 106, Republic of China
Received 2 August 2004/Returned for modification 13 September 2004/Accepted 19 January 2005
Real-time PCR assays have been applied for the detection and quantification of pathogens in recent years.
In this study two combinations of primers and fluorescent probes were designed according to the sequences of
the rfb
Escherichia coli O157and stx
2genes. Analysis of 217 bacterial strains demonstrated that the duplex real-time
PCR assay successfully distinguished the Escherichia coli O157 serotype from non-E. coli O157 serotypes and
that it provided an accurate means of profiling the genes encoding O antigen and Shiga-like toxin 2. On the
other hand, bacterial strains that lacked these genes were not detected by this assay. The quantitative ranges
of the real-time PCR assay for these two genes were linear for DNA concentrations ranging from 10
3to 10
9CFU/ml of E. coli O157:H7 in pure culture and milk samples. The real-time PCR allowed the construction of
standard curves that facilitated the quantification of E. coli O157:H7 in feces and apple juice samples. The
detection sensitivity of the real-time PCR assay ranged from 10
4to 10
9CFU/g (or 10
4to 10
9CFU/ml) for feces
and apple juice and 10
5to 10
9CFU/g for the beef sample without enrichment. After enrichment of the food
samples in a modified tryptic soy broth, the detection range was from 10
0to 10
3CFU/ml. The real-time PCR
assays for rfb
E. coli O157and stx
2proved to be rapid tests for the detection of E. coli O157 in food matrices and
could also be used for the quantification of E. coli O157 in foods or fecal samples.
Escherichia coli is a gram-negative bacterium that generally
inhabits the intestinal tract of humans and animals. However,
some of isolates of this organism are pathogenic, and these
enterovirulent E. coli isolates are important food-borne
patho-gens associated with severe gastrointestinal and circulatory
system diseases, such as hemorrhagic colitis (HC),
hemorrhag-ic-uremic syndrome (HUS), and thrombotic thrombocytopenic
purpura, in humans (17, 19). E. coli O157:H7 is a major strain
which causes these kinds of food-borne outbreaks all over the
world. In 1975, E. coli O157:H7 was first isolated from clinical
samples, but it was not reported in association with outbreaks
until 1982 (18). In 1996 there were some large outbreaks in
Japan which originated in Sakai City, Osaka (22). These
out-breaks affected more than 17,000 people. A total of 106
chil-dren developed HUS, and 13 of these chilchil-dren died (18).
Similar outbreaks have been reported in Australia, Canada,
the United States, various European countries, and Africa (6,
8, 10, 22, 26, 28).
The pathogenicity of E. coli O157:H7 is associated with a
number of virulence factors, including Shiga-like toxins 1 and
2 (encoded by the stx
1and stx
2genes, respectively) and intimin
(encoded by the eaeA gene). Shiga-like toxins are believed to
play a major role in the pathogenesis of HC and HUS through
a cytopathic effect on the vascular endothelial cells of the
kidneys and intestines (29). Strains isolated from patients with
HC usually produce both Shiga-like toxins 1 and 2, and strains
that produce only stx
1are uncommon (11).
In Taiwan, infection with E. coli O157:H7 is a reportable
infectious disease. No cases were reported in Taiwan until
2001. In the summer of 2001, a patient presented with
symp-toms that included bloody diarrhea, HC, and HUS. This
pa-tient’s diarrhea stools, other suspected stools, and
environmen-tal samples were collected. We analyzed and confirmed that
the infectious strain was E. coli O157:H7. This was the first
infectious case caused by E. coli O157:H7 in Taiwan (35).
Cattle are generally considered the major reservoir for this
organism, although it has also been isolated from sheep (20),
goats (3), dogs, deer, horses, and seagulls (18). An important
aspect of this organism is the fact that the ingestion of 10 to 100
of these organisms may be sufficient to cause an infection (33).
Among the most important sources of human infection are
direct contact with cattle and other ruminants, contaminated
bathing water, beef products, unpasteurized milk, vegetables,
fruits, and drinking water (7). The detection and correct
iden-tification of this strain are important parts of food hygiene.
Traditional methods for the identification of E. coli O157:H7,
such as biochemical and serotype tests, used to take 5 to 7 days.
In recent years, some molecular methods were developed to
detect and identify this food-borne pathogen, such as PCR and
enzyme-linked immunosorbent assay. PCR is a rapid and
easy-to-use method and can provide a preliminary characterization
(5, 9). The use of the PCR method to detect pathogens,
how-ever, has some shortcomings, such as some false-positive or
false-negative results for more complex samples and a low
sensitivity with more primer sets. At the same time, the
ethidium bromide used to stain the electrophoresis gel after
PCR is a harmful chemical and its application is
time-consum-ing. The TaqMan detection system (Applied Biosystems,
Fos-ter City, Calif.) is a new qualitative and quantitative system
that uses a fluorogenic hybridization probe to detect the target
genes; and it has previously been demonstrated to be a rapid,
high-throughput, semiautomatic PCR scheme for the
identifi-cation of E. coli (23, 29), Salmonella (28), and Listeria spp. (1).
* Corresponding author. Mailing address: Institute of Microbiology
and Biochemistry, National Taiwan University, Taipei, Taiwan 106,
Republic of China. Phone: 886-2-2363-0231, ext. 3813. Fax:
886-2-2362-7044. E-mail: tmpan@ntu.edu.tw.
2668
at NATIONAL TAIWAN UNIV MED LIB on May 27, 2009
jcm.asm.org
The objective of this study was to assess the utility of the
TaqMan PCR system for the detection and identification of
the stx
2gene (which is responsible for the biosynthesis of
Shi-ga-like toxin 2) and the rfb
E. coli O157gene (which is responsible
for the biosynthesis of the O antigen) (31). We developed
some modifications to improve the pretreatment and
purifica-tion of food samples for PCR. These improvements were
de-signed to increase the specificity and the accuracy of the PCR.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions. E. coli O157:H7 strain Tw04 was used throughout this study. Strain Tw04 was isolated from a clinical sample in Taiwan in 2001 by our laboratory (35). In addition, a further 217 bacterial strains were used to evaluate the specificity of the real-time PCR assay for E. coli O157 detection in this study. These strains were E. coli O157:H7, clinical E. coli strains, and other non-E. coli strains. They were purchased from the Bioresources Collection and Research Center (Hsinchu, Taiwan) and the American Type Culture Collection, and some came from among the clinical isolates kept in our laboratory and the Centers for Disease Control, Taiwan. Before testing of the bacterial strains, they were retrieved from frozen storage and cultured in nutrient broth (Difco Laboratories, Detroit, Mich.) at 37°C overnight. The cultures were then transferred to tryptic soy broth (TSB; Difco) and incubated at 37°C until the optical density at 600 nm reached 1.2 to 1.4. The cells were subsequently harvested and used for DNA preparation and other tests. In addition, before they were tested they were checked for their genotype (stx2, rfbE. coli O157) by PCR (24).
Genomic DNA preparation.Two different methods were used for the prepa-ration of genomic DNA. In one method the genomic DNA was prepared by direct purification of boiled cells lysed by the double-distilled water method. In the other method the genomic DNA was prepared with a Wizard Genomic DNA purification kit (Promega Corporation, Williamsburg, Iowa). The direct boiled cell method was performed as follows: 1 ml of log-phase cultured bacterial broth was centrifuged at 15,000⫻ g for 10 min. After centrifugation, the cell pellets were resuspended in 250l sterile distilled water and boiled for 10 min. The lysed cell debris was then removed by centrifugation (15,000⫻ g for 5 min), and the DNA in the supernatant was transferred to a fresh and sterile Eppendorf tube.
The preparation with the kit was performed according to the manufacturer’s instructions. Bacterial cultures were centrifuged at 12,000⫻ g and mixed with nucleic lysis buffer and proteinase K at 37°C for 15 to 60 min to lyse the cell wall. The protein precipitation buffer was then used to bind the protein and precipi-tate it. After centrifugation, the supernatant was mixed with isopropanol to precipitate the DNA. The extracted DNA was then washed with ethanol and resuspended in double-distilled water. All DNA samples were used immediately or stored at⫺30°C.
Sequencing of the Shiga-like toxin 2 gene of E. coli O157:H7 isolated from Taiwan.Five primers were used to sequence the Shiga-like toxin 2 gene of E. coli O157:H7 strains which were isolated in Taiwan. The sequences of the primers
are shown in Table 1. The PCRs were carried out in a total volume of 25l which included 1⫻ reaction buffer, 200 nM of deoxynucleoside triphosphates, 200 nM of each of the primers, and 1.5 U of Taq polymerase (Takara, Tokyo, Japan). The PCR products were analyzed with a genetic analyzer (Applied Biosystems).
Cloning.To determine the detection limit of the TaqMan PCR assay, the 862-bp PCR product from the stx2gene of E. coli strain Tw04, obtained with primers NS5 and NS7, was cloned into E. coli strain DH5␣ by using the pGEM-T and pGEM-T Easy Vector systems kit (Promega Corporation) (25).
Plasmid preparation.Plasmid DNA was purified by using the Plasmid Mini-prep Purification Kit II (Genemark Technology Corporation, Tainan, Taiwan). The extracted plasmid DNA was then assayed on 1.5% (wt/vol) agarose gels to make sure that the product was pure. Following the gel assay, the product was repurified with a Gene-Spin 1-4-3 DNA extraction kit (Protec Technology En-terprise Corporation, Taipei, Taiwan). The concentration of plasmid was deter-mined by a fluorescent dye, bisBenzimide Hoechst 33258 (Sigma, St. Louis, Mo.), and with a fluorescence meter (Bio-Tek, Winooski, Vt.).
Design of primers and fluorogenic probes.The nucleotide sequences of the primers and fluorogenic probes are listed in Table 1. The primers and fluoro-genic probes were designed according to the sequences listed in the instructions of the BLAST kit by the Primer Express software (v1.5; Applied Biosystems).
The sequence accession numbers in the GenBank database are X07865 for stx2 and AF049343 for rfbE. coli O157. 6-Carboxyfluorescein and 6-carboxy-4 ⬘,5⬘-di-chloro-2⬘,7⬘-dimethoxyfluorescein were used as fluorescent reporter dyes and were conjugated to the 5⬘ ends of the probes to detect amplification products specific for stx2and rfbE. coli O157, respectively. The quencher dye 6-carboxytet-ramethylrhodamine was attached at the 3⬘ ends of these probes. The primers were synthesized by the Purigo Biotech Corporation (Taipei, Taiwan), and the probes were synthesized by the MWG Biotech Corporation (Ebersberg, Ger-many).
TaqMan PCR assay for detection of E. coli O157.PCR was performed in a reaction mixture with a total volume of 25l containing 1 l of extracted DNA, 0.5 mM of stx2and rfbE. coli O157primers, 0.2 mM of each fluorogenic probe, and TaqMan Universal Master Mix (Applied Biosystems). The Master Mix contained AmpErase uracil-N-glycosylase (UNG), deoxynucleoside triphosphate with dUTPs, 6-carboxyrhodamine as an internal passive fluorogenic reference, and an optimized buffer component. Amplification and detection were carried out in optical-grade 96-well plates in an ABI Prism 7700 sequence detection system (Applied Biosystems) with an initial step of 50°C for 2 min, which is the required optimal AmpErase UNG enzyme activity, and then at 95°C for 10 min, to activate the AmpliTaq Gold DNA polymerase and to deactivate the AmpErase UNG enzyme, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The reaction conditions for amplification and the parameters for fluorescence data collection were programmed into a Power Macintosh 4400/20 computer (Apple Computer, Santa Clara, Calif.) linked directly to the ABI Prism 7700 sequence detection system by using the SDS 1.6 application software, according to the manufacturer’s instructions. After real-time data acquisition, the threshold, which was defined as being 10-fold higher than the baseline, was determined; and the cycle threshold (CT) value was manually set so that it intersected the ampli-fication curves in the linear region of the semilog plot. The quantity of target gene copies in the PCR sample is predicted by the CTvalue (12).
TABLE 1. Sequences of primers and probes used in this study
Primer name Sequence (5⬘33⬘) Source orreference
Stx2F
aACC ACA TCG GTG TCT GTT ATT AAC C
This study
Stx2R
aCGG TAG AAA GTA TTT GTT GCC GTA TT
This study
Stx2P
aTTT GCT GTG GAT ATA CGA GGG CTT GAT GTC TAT
This study
rfbF
bATG CTG CCC ACA AAA ATA ATG TAAA
This study
rfbR
bTTC CAT AAT CGG TTG GTG TGC TAA
This study
rfbP
bAAC TGC TTT TCC TCG GTT CGT CGT GTA T
This study
NS5
cCTT CGG TAT CCT ATT CCC GG
34
NS7
cCGC TGC AGC TGT ATT ACT TTC
34
VT
2S
4F
dATC CAG TAC AAC GCG CCA
4
VT
2S
4R
d
CAC AGA CTG CGT CAG TGA GG
4
aPrimers Stx2F, Stx2R, and Stx2P were developed to amplify the Shiga-like toxin 2 gene, and 6-carboxyfluorescein was added as a label to the 5⬘ end of Stx2P. bPrimers rfbF, rfbR, and rfbP were developed to amplify the rfb gene, which was part of the O157 antigen, and 6-carboxy-4⬘,5⬘-dichloro-2⬘,7⬘-dimethoxyfluorescein was added as a label to the 5⬘ end of rfbP.
cPrimers NS5 and NS7 were designed to amplify Shiga-like toxin 2. dPrimers VT
2S4R and VT2S4F were designed to amplify Shiga-like toxin 2.
at NATIONAL TAIWAN UNIV MED LIB on May 27, 2009
jcm.asm.org
Food sample preparation.The pasteurized milk, beef, and apple juice were obtained from a local supermarket. All of the samples were initially tested by culture on sorbitol-MacConkey agar plates and by the PCR method. The micro-organisms were prepared from 10-fold serial dilutions of a log-phase culture of E. coli O157:H7 strain Tw04, which contained 109to 100CFU/ml viable cells, for mixing with the food samples. The direct boiling cell lysis method and modified QIAamp DNA stool mini kit (QIAGEN Companies) method were used to extract DNA from samples which contained different concentrations of patho-gens. The direct boiling cell lysis method was used for preparation of pasteurized milk samples and was performed as follows. One milliliter of milk sample was prepared, and cells were allowed to lyse for 10 min at 100°C. After boiling of the suspension, it was cooled to room temperature, spun at 16,000⫻ g for 30 s, and transferred to a new tube and spun again. The supernatant was then used as the PCR template in this study. The modified QIAamp DNA stool mini kit method used for the apple juice sample was performed as follows: 1 ml of intermixed apple juice sample was spun at 16,000⫻ g for 10 min, and then 850 l of the supernatant was removed. After this step, all the follow-up steps were the same as those described in the original protocol for the kit until the wash step, when 2⫻ washing buffer was added and the mixture was eluted with 50 l of double-distilled water. The DNA was then extracted from imitated beef samples by the QIAamp DNA stool mini kit method, which was modified in this study. The constructed plasmid, the concentration of which was determined by use of the fluorescent dye bisBenzimide (Hoechst 33258), was placed in the extracted DNA as an internal standard in order to contrast the results of quantification.
Human stool sample preparation.A stool sample was collected from a 24-year-old healthy man. We screened the stool sample for E. coli O157:H7 by culture on sorbitol-MacConkey agar plates and PCR before the sample was seeded. The microorganisms were prepared from 10-fold serial dilutions of a log-phase culture of E. coli O157:H7 strain Tw04 containing 109to 100CFU/ml viable cells for mixing with the stool sample. The QIAamp DNA stool mini kit method was used to extract DNA from samples which contained different con-centrations of the pathogen. The constructed plasmid, the concentration of which was determined by use of the fluorescent dye bisBenzimide (Hoechst 33258), was placed in the extracted DNA as the internal standard in order to contrast the results of quantitation.
Enrichment of food samples in mTSB medium.Different concentrations of freshly grown and enumerated cultures of E. coli O157:H7 strain Tw04 were used to inoculate 1 ml of apple juice or raw milk samples. The inoculated samples were then added to the enrichment medium, modified TSB (mTSB; Merck, Germany), to 10% (vol/vol) and incubated at 37°C.
RESULTS
Nucleotide sequences of Shiga-like toxin 2 in E. coli strains
Tw02, Tw03, and Tw04.
In this study we used five primers,
including primers NS5 and NS7 (34), primers VT
2S
4F and
VT
2S
4R (4), and primer stx2P, to identify DNA sequences of
about 1,274 bp of Shiga-like toxin 2 in E. coli strains Tw02,
Tw03, and Tw04. These E. coli O157:H7 strains were isolated
from environmental samples (strains Tw02 and Tw03) and a
clinical sample (strain Tw04) in Taiwan. The sequence
data-bases were aligned by use of the BLAST program (available at
http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). We
com-pared the results obtained in our laboratory for E. coli
O157:H7 strain Tw01 (GenBank accession number AF291819)
in 2000, as shown in Table 2. The range of similarities of the
stx
2gene sequences between each pair of strains was 95% to
99%.
Specificity of TaqMan PCR assay.
Genomic DNA from E.
coli O28ac (stx
2⫹and O157
⫺), O157:H7 (stx
2⫺and O157
⫹),
O78:H11 (stx
2⫺and O157
⫺), and O157:H7 (stx
2⫹and O157
⫹)
was initially tested to evaluate the primers and probes used in
the PCR assay for their abilities to amplify and detect PCR
products specific for the stx
2and rfb
E. coli O157:H7genes. A
fluorescent signal 10 times higher than the standard deviation
of the mean baseline emission was indicative of a positive
detection result. Stx
2- and rfb
E. coli O157-specific probes
pro-duced exponential increases in fluorescence only when the
DNA from strains containing these genes was used as a
tem-plate in the PCR assay. The amplified products generated in
the samples were also analyzed on a 4% agarose gel by
stan-dard horizontal gel electrophoresis. Those samples that
re-sulted in an exponential increase in fluorescence with a
partic-ular probe also contained an amplicon that was of the
predicted size and that corresponded to the gene detected by
the probe (data not shown).
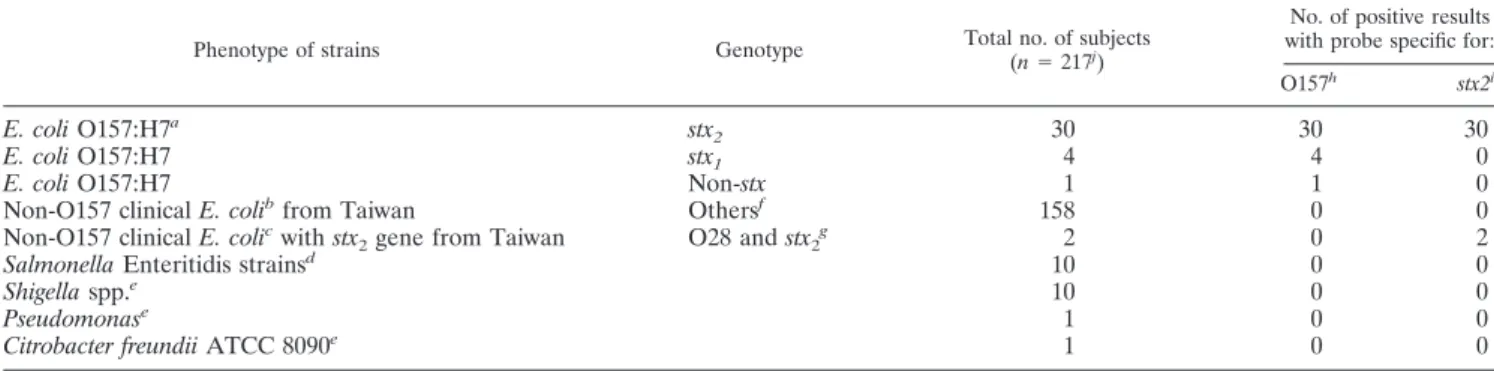
Based on detection of the characteristics of toxin and
O-antigen genes of E. coli O157, a duplex TaqMan PCR assay
was developed. The specificity of the assay was tested against a
panel of bacterial templates from 217 E. coli or non-E. coli
strains, including 35 E. coli O157:H7 strains, 2 stx
2and 158
non-stx
2E. coli strains isolated from clinical cases in Taiwan,
and 22 strains of other bacterial species. All Shiga-like toxin
2-producing E. coli O157:H7 strains were detected by the
du-plex TaqMan PCR assay, whereas the other strains of bacteria
were negative, as shown in Table 3. The positive samples gave
C
Tvalues lower than 16 and a high endpoint fluorescence
usually of
⬎1.0 (data not shown).
Sensitivity of TaqMan PCR assay.
Genomic DNA prepared
from 10-fold serial dilutions of a log-phase culture of E. coli
O157:H7 strain Tw04 was used as the template to determine
the detection sensitivity of this multiplex real-time PCR and to
construct standard curves by plotting the numbers of CFU
versus the C
Tvalues produced for both of the target genes.
Standard curves showed a linear relationship between the log
10input CFU and the C
T(the PCR cycle at which the
fluores-cence intensity rises above the threshold). The lowest detection
limit of the PCR assay for the rfb
E. coli O157and stx
2genes was
approximately 10
3CFU/ml (10
3copies/ml), as shown in Table
4.
Sensitivity of TaqMan PCR with food samples.
In this study,
we also tried to detect E. coli O157:H7 DNA in three kinds of
food samples, including beef, apple juice, and raw milk, which
were associated with E. coli O157:H7 infections in the past.
The DNA isolated from food samples was mixed with 10-fold
serial dilutions of E. coli O157:H7 strain Tw04 and was used in
this assay to construct a standard curve for the stx
2gene. This
quantification was linear over a range of initial target
concen-trations from 10
3to 10
9CFU/ml in a raw milk sample. In apple
juice samples it was linear over a range of initial target
con-centrations from 10
4to 10
9CFU/ml, as shown in Table 4. In
beef samples it was linear over a range of initial target
con-centrations from 10
5to 10
9CFU/g.
Sensitivity of TaqMan PCR with stool samples.
Human
fe-ces containing different concentrations of E. coli O157:H7
strain Tw04 (stx
2⫹and O157
⫹) were tested to evaluate the
TABLE 2. Similarities of stx
2gene sequences between each pair of
strains isolated in Taiwan
Strain % Similarity Tw01 Tw02 Tw03
Tw01
Tw02
97
Tw03
99
97
Tw04
97
95
97
at NATIONAL TAIWAN UNIV MED LIB on May 27, 2009
jcm.asm.org
utility of this real-time PCR assay for the detection of E. coli
O157:H7 in humans. The DNA isolated from stool samples
was mixed with 10-fold serial dilutions of E. coli O157:H7
strain Tw04 and was used in this assay to construct standard
curves for the stx
2and rfb
E. coli O157genes. The numbers of
CFU of E. coli O157:H7 present per gram of feces were
inter-polated from these standard curves and then compared to the
bacterial counts determined by plating the same fecal samples
on sorbitol-MacConkey agar-streptomycin plates. The
num-bers of CFU per gram of feces estimated by this real-time PCR
assay by using stx
2- and rfb
E. coli O157-specific probes were
sim-ilar to those determined by plating the same samples on
sor-bitol-MacConkey agar. This quantification was linear over a
wide range of initial target concentrations (10
4to 10
9CFU/g),
as shown in Table 4. On the other hand, we detected E. coli
O157:H7 strain Tw04 in seeded samples for evaluation of the
assay methods that we had set up. In all experiments, the
estimated values and the actual values were similar. The results
are shown in Table 5.
Enrichment of food samples in mTSB.
In this study, the
effects of the length of the enrichment time on the sensitivity of
the multiplex real-time PCR for detection of E. coli O157 in
milk and apple juice samples were investigated. The results are
shown in Table 6. After being enriched for 4 h, the sensitivity
for the detection of E. coli O157 in milk samples was 10
0TABLE 3. Strains used in this study and associated multiplex real-time PCR results
Phenotype of strains Genotype Total no. of subjects
(n⫽ 217j )
No. of positive results with probe specific for:
O157h stx2i
E. coli O157:H7
astx
2
30
30
30
E. coli O157:H7
stx
14
4
0
E. coli O157:H7
Non-stx
1
1
0
Non-O157 clinical E. coli
bfrom Taiwan
Others
f158
0
0
Non-O157 clinical E. coli
cwith stx
2
gene from Taiwan
O28 and stx
2g2
0
2
Salmonella Enteritidis strains
d10
0
0
Shigella spp.
e10
0
0
Pseudomonas
e1
0
0
Citrobacter freundii ATCC 8090
e1
0
0
aAll strains were isolated from the United States (10 strains), Canada (12 strains), Japan (9 strains), and Taiwan (4 strains). bAll strains were isolated from clinical samples of diarrhea cases in Taiwan.
cAll strains were isolated from clinical samples in Taiwan by the Centers for Disease Control, Taiwan. dAll strains were isolated from food-borne cases by the Centers for Disease Control, Taiwan. eAll strains were from Bioresource Collection and Research Center, Hsinchu, Taiwan. fAll genotypes are non-stx
2; and their phenotypes include the following: O1, O3, O6:H?, O6:H12, O8:H?, O8; O14; O15:H21 O15:NM, O18, O20:H20, O23, O25:H51, O25:K98:NM, O26:H?, O26:H11, O27, O28:H?, O28ac:H?, O28ac, O29:H?, O29:NM, O30, O38, O44, O48, O55:H?, O55:H12, O75, O78:H11, O78:H2, O78:K80:H12, O86a, O90, O111ab:H21, O112ac:H?, O114, O115, O123, O124:H?, O125, O127a:H1, O127a:H16, O136:H19, O138, O142:H6, O144, O146:NM, O148, O153:NM, O158, O159:H21, O166, O168, O169:H28, and unknowns.
gAll genotypes are stx
2, and the serotype was O28ac.
hNumbers of positive results by the multiplex real-time PCR assay for the rfbE. coli O157:H7gene. iNumbers of positive results by the multiplex real-time PCR assay for the stx
2gene. jTotal numbers of strains in this study.
TABLE 4. Sensitivity of the multiplex real-time PCR assay for
detection of E. coli O157:H7 strain Tw04 in milk, apple juice,
beef, and feces
Sample Result of this
studya R2b Previous result (reference)c
Pure culture
10
3CFU/ml
0.992
10
3CFU/ml (15)
Feces
10
4CFU/g
0.992
10
4CFU/g (27), 3.5
⫻ 10
4CFU/g (15)
Apple juice
10
4CFU/ml
0.997
10
8CFU/ml (9)
Milk
10
3CFU/ml
0.998
10
3CFU/ml (21)
Beef
10
5CFU/g
0.990
aThe limit of detection for E. coli O157:H7 in each sample by real-time PCR, for which the combinations of primers and probes were designed in this study.
bThe results of linear regression by each concentration of each strain. cThe limit of detection for E. coli O157:H7 in each sample by real-time PCR in other studies.
TABLE 5. Detection of E. coli O157:H7 strain Tw04 in
seeded samples
Sample
Real-time PCR result
with probe specific for: Plate count O157 stx2
Plasmid
P1
cND
a6.9
b7.1
bP2
cND
5.1
4.3
Milk
M1
d8.0
8.0
8.0
M2
d3.6
3.6
4.0
Apple juice
A1
e7.8
7.8
8.0
A2
e5.1
5.1
5.1
Human feces
F1
f6.7
6.7
7.0
F2
f4.9
4.9
5.1
aND, not detectable; stx2-specific probe has only the stx2gene. bLog
10CFU/ml or log10CFU/g of sample.
cP1 and P2 were different concentrations of the plasmid in the imitated samples.
dM1 and M2 were different cells of E. coli O157:H7 strain Tw04 in the imitated milk samples.
eA1 and A2 were different cells of E. coli O157:H7 strain Tw04 in the imitated apple juice samples.
fF1 and F2 were different cells of E. coli O157:H7 strain Tw04 in the imitated fecal samples.
at NATIONAL TAIWAN UNIV MED LIB on May 27, 2009
jcm.asm.org
CFU/ml. In the apple juice samples, the same sensitivity of 10
0CFU/ml was obtained after a 10-h enrichment period.
DISCUSSION
Traditional methods based on biochemical characteristics
are labor-intensive, and the total time required for
determina-tion of the identities of the pathogens is typically about 72 h.
Rapid detection techniques directed at similar immunological
and genetic targets are therefore of great interest, especially
since it is very difficult to determine the total number of
bac-teria present in foods or fecal samples. Immunological
meth-ods based on the detection of Shiga-like toxins have been
developed. However, these methods cannot differentiate E.
coli O157:H7 from other less virulent enterohemorrhagic and
enteropathogenic E. coli strains. Other methods, based on the
detection of O157 somatic and H7 flagellar antigens, are
equally inadequate because of their lack of specificity.
Real-time PCR offers the ability to determine the absolute
and relative amounts of pathogens in complex matrices, and
assays that were recently developed for the identification of E.
coli O157:H7 are based on the detection of genes encoding
Shiga-like toxins (2, 21), intimin (23), and O antigen (9).
How-ever, those single real-time PCR methods sometimes lack
spec-ificity. For example, the targets for the H7 flagellar antigen
genes may cross-react with E. coli O55:H7 and so will fail to
identify E. coli O157:NM (where NM is nonmotile). Although
some published reports have shown that a real-time PCR
de-signed for use with two or three combinations of primers and
probes could provide good specificity and sensitivity for the
detection of enterovirulent E. coli or E. coli O157:H7 (15, 27),
we decided that less effort for detection would be required by
use of combinations of primers and probes which amplified the
O antigen and Shiga-like toxin. The rfb
E. coli O157gene encodes
an enzyme that is necessary for O-antigen biosynthesis and that
is highly conserved among isolates of the E. coli O157 serovar.
According to previous reports, strains of E. coli O157 isolated
from patients with HC usually produce both Shiga-like toxins 1
and 2. Isolates that produce only stx
1are uncommon (11).
Therefore, in this study we designed two combinations of
prim-ers and probes to detect and identify E. coli O157 isolates
producing Shiga-like toxin 2 by targeting the particular genes
stx
2and rfb
E. coli O157.
The specificity of the real-time PCR was evaluated with 217
strains, including E. coli O157:H7 strains and other isolates.
The results have shown that the combinations of primers and
probes designed in this study correctly detected E. coli
O157:H7 and that there were no false-positive or
false-nega-tive results by any of the tests. Comparison of the detection
limits of these combinations of primers and probes showed
that the results were better or the same as those published in
the literature (9, 15, 21, 27). The detection range of the duplex
real-time PCR assay for stx
2and rfb
E. coli O157was linear when
DNA was prepared from pure culture samples containing from
10
3to 10
9CFU/ml of E. coli O157:H7. This result was the same
in the raw milk sample. However, in the apple juice samples,
the results were linear for pathogen concentrations ranging
from 10
4to 10
9CFU/ml. The same results (10
4to 10
9CFU/g)
were evident for the human stool samples. According to the
results of published papers, some PCR inhibitors are present,
such as polyphenolic compounds in apple juice (30) and bile
salts, heparin, and bilirubins in human stools (14, 32). Because
of the presence of these inhibitors in apple juice and human
stool samples, the DNA extracted from these samples needed
to be purified before the real-time PCR was carried out. On
the other hand, there were no inhibitors in the milk, and
therefore, we could carry out the real-time PCR with lysed
boiled cells. Although the range of linear concentrations in
human stool samples was 10
4to 10
9CFU/g, the stools of
food-poisoning patients usually harbor 10
6CFU/g bacteria and
more (13). We used this combination of probes and primers to
correctly detect E. coli O157 in stool samples.
Traditional culture-based approaches for the detection of E.
coli O157:H7 in beef, such as growth on sorbitol-MacConkey
agar and further screening for the production of Shiga-like
toxin, may take several days (16). In this study, we extracted
the DNA of the pathogen directly from the beef and the
reaction was completed in 3 h. The detection range was linear
from 10
5to 10
9CFU/g.
In summary, we have described a duplex TaqMan real-time
PCR method for the detection of E. coli O157 that showed
good specificity and sensitivity and that also saved substantial
time because of the preparation of samples without preculture.
The key points for correct detection in this study were the
DNA extraction efficiency and the removal of PCR inhibitors
from different materials. In the future, we will try to create
other methods for the isolation of the DNA of pathogens and
for the removal of PCR inhibitors from other, different
sam-ples which are related to E. coli O157:H7. The shortening of
the processing time and the increase in the specificity for
pathogen detection are critical for the safety and sanitation of
our food supply.
REFERENCES
1. Bassler, H. A., S. A. Flood, K. J. Livak, J. Marmaro, R. Knorr, and C. A.
Batt.1995. Use of a fluorogenic probe in a PCR-based assay for the detec-tion of Listeria monocytogenes. Appl. Environ. Microbiol. 61:3724–3728. 2. Belanger, S. D., M. Boissinot, C. Menard, F. J. Picard, and M. G. Bergeron.
2002. Rapid detection of Shiga toxin-producing bacteria in feces by multiplex PCR with molecular beacons on the SmartCycler. J. Clin. Microbiol. 40: 1436–1440.
3. Bielaszewska, M., J. Janda, K. Blahova, H. Minarikova, E. Jikova, M. A.
Karmali, J. Laubova, J. Sikulova, M. A. Preston, R. Khakhria, H. Karch, H. Klazarova, and O. Nyc.1997. Human Escherichia coli O157:H7 infection associated with the consumption of unpasteurized goat’s milk. Epidemiol. Infect. 119:299–305.
4. Chen, L. M. 2000. Rapid identification and molecular typing of
enterohem-TABLE 6. Sensitivity of the multiplex real-time PCR assay for
detection of E. coli O157:H7 strain Tw04 in raw milk and
apple juice after enrichment
No. of CFU/
ml
Sensitivity for detection ina:
Milk Apple juice
0 h 2 h 3 h 4 h 5 h 10 h 0 h 2 h 3 h 4 h 5 h 10 h
10
3⫹
⫹
⫹
⫹
⫹
⫹
⫺
⫺
⫹
⫹
⫹
⫹
10
2⫺
⫺
⫹
⫹
⫹
⫹
⫺
⫺
⫹
⫹
⫹
⫹
10
1⫺
⫺
⫺
⫹
⫹
⫹
⫺
⫺
⫺
⫹
⫹
⫹
10
0⫺
⫺
⫺
⫹
⫹
⫹
⫺
⫺
⫺
⫺
⫺
⫹
0
⫺
⫺
⫺
⫺
⫺
⫺
⫺
⫺
⫺
⫺
⫺
⫺
aThe plus or minus sign indicates whether or not the relative fluorescent intensity after 40 cycles was greater than the threshold value for the negative control.
at NATIONAL TAIWAN UNIV MED LIB on May 27, 2009
jcm.asm.org
orrhagic Escherichia coil O157:H7. Master’s thesis. Institute of Agricultural Chemistry, National Taiwan University, Taipei, Taiwan.
5. Davis, K. C., C. H. Nakatsu, R. Turco, S. D. Weagant, and A. K. Bhunia. 2003. Analysis of environmental Escherichia coli isolated for virulence genes using the TaqMan PCR system. J. Appl. Microbiol. 95:612–620.
6. Effler, E., M. Isaacson, L. Arntzen, R. Heenan, P. Canter, T. Barrett, L. Lee,
C. Mambo, W. Levine, A. Zaidi, and P. M. Griffin.2001. Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerg. Infect. Dis.
7:812–819.
7. Eva, M. N., and T. A. Marianne. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5⬘ nuclease PCR Assay. J. Clin. Microbiol. 41:2884–2893.
8. Fegan, N., and P. Desmarchelier. 2002. Comparison between human and animal isolates of Shiga toxin-producing Escherichia coli O157 from Austra-lia. Epidemiol. Infect. 128:357–362.
9. Fortin, N. Y., A. Mulchandani, and W. Chen. 2001. Use of real-time poly-merase chain reaction and molecular beacons for the detection of Esche-richia coli O157:H7. Anal. Biochem. 289:281–288.
10. Galanis, E., K. Longmore, P. Hasselback, D. Swann, A. Ellis, and L. Panaro. 2003. Investigation of an E. coli O157:H7 outbreak in Brooks, Alberta, June-July 2002: the role of occult cases in the spread of infection within a daycare setting. Can. Commun. Dis. Rep. 29:21–28.
11. Guan, J., and R. E. Levin. 2002. Quantitative detection of Escherichia coli O157:H7 in ground beef by the polymerase chain reaction. Food Microbiol.
19:159–165.
12. Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986–994.
13. Hiroshi, F., T. Yoshie, and S. Ryotaro. 2003. Duplex real-time SYBR Green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J. Clin. Microbiol. 41:5134–5146.
14. Holland, J. L., L. Louie, A. E. Simor, and M. Louie. 2000. PCR detection of Escherichia coli O157:H7 directly from stool: evaluation of commercial ex-traction methods for purifying fecal DNA. J. Clin. Microbiol. 38:4108–4113. 15. Ibekwe, A. M., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyons. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853–4862.
16. Jaykus, L. A. 2003. Challenges to developing real-time methods to detect pathogens in foods. ASM News 69:341–347.
17. Jones, D. L. 1999. Potential health risks associated with the persistence of Escherichia coli O157:H7 in agricultural environment. Soil Use Manage.
15:76–83.
18. Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229–234.
19. Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15–38.
20. Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1996. Escherichia coli O157:H7 in microbial flora of sheep. J. Clin. Microbiol. 34:431–433.
21. McKillip, J. L., and M. Drake. 2000. Molecular beacon polymerase chain reaction detection of Escherichia coli O157:H7 in milk. J. Food Prot. 63:855– 859.
22. Michino, H., K. Araki, S. Minami, S. Takaya, N. Sakai, M. Miyazaki, A. Ono,
and H. Yanagawa. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787–796.
23. Oberst, R. D., M. P. Hays, L. K. Bohra, R. K. Phebus, C. T. Yamashiro, C.
Paszko-Kolva, S. J. A. Flood, and J. M. Sargeant.1998. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5⬘ nuclease (TaqMan®
) assay. Appl. Environ. Microbiol. 64:3389–3396.
24. Pan, T. M., L. M. Chen, and Y. C. Su. 2002. Identification of Escherichia coli O157:H7 by multiplex PCR with primer specific to the hlyA, eaeA, stx1, stx2, fliC and rfb genes. J. Formosa Med. Assoc. 101:661–664.
25. Perelle, S., F. Dilasser, J. Grout, and P. Fach. 2003. Development of 5⬘-nuclease PCR assay for detecting Shiga toxin-producing Escherichia coli O154 based on the identification of an⬘O-island 29⬘ homologue. J. Appl. Microbiol. 94:587–594.
26. Rocchi, G., and M. Capozzi. 1999. Enterohemorrhagic Escherichia coli O157:H7 infection. Recenti Prog. Med. 90:613–618.
27. Sharma, V. K., and E. A. Dean-Nystrom. 2003. Detection of enterohemor-rhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and shiga toxins. Vet. Microbiol. 93:247–260. 28. Sharma, V. K., and S. A. Carlson. 2000. Simultaneous detection of
Salmo-nella strains and Escherichia coli O157:H7 with fluorogenic PCR and single enrichment broth culture. Appl. Environ. Microbiol. 66:5472–5476. 29. Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automatic
fluorogenic PCR assays (TaqMan®
) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol. Cell. Probes 13:291–302. 30. Siebert, K. J., N. V. Troukhanova, and P. Y. Lynn. 1996. Nature of
polyphe-nol-protein interactions. J. Agric. Food Chem. 44:80–85.
31. Sima, S. B., C. James, J. R. Vary, F. D. Scott, and I. T. Phillip. 1996. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect. Immun. 64:4795–4810.
32. Teegan, T., J. Fotheringham, E. Topp, H. Schraft, and K. T. Leung. 2003. A comparison of DNA extraction and purification methods to detect Esche-richia coli O157:H7 in cattle manure. J. Microbiol. Methods 54:165–175. 33. Willshaw, G. A., J. Thirwell, A. P. Jones, S. Parry, R. L. Salmon, and M.
Hickey.1994. Vero cytotoxin-producing Escherichia coli O157 in beefburgers linked to an outbreak of diarrhea, haemorrhagic colitis and haemolytic uraemic syndrome in Britain. Lett. Appl. Microbiol. 19:304–307. 34. Wu, F. T., and T. M. Pan. 1999. Rapid detection of pathogenic Escherichia
coli by polymerase chain reaction. J. Chin. Agric. Chem. Soc. 37:179–189. 35. Wu, F. T., T. Y. Tsai, C. F. Hsu, T. M. Pan, H. Y. Chen, and I. J. Su. 2005.
Isolation and identification of Escherichia coli O157:H7 associated with the first case in Taiwan. J. Formos Med. Assoc. 104:206–209.