Abstract—In order to provide feasible primer set for
performing a Polymerase chain reaction (PCR) experiment, many primer design methods have been proposed. However, the majority of these methods require a long time to obtain an optimal solution since quantities of template DNA need to be analyzed, and the designed primer sets usually do not provide a specific PCR product size. In recent years, evolutionary computation has been applied to PCR primer design and appeared to yield good results. In this paper, a memetic algorithm is proposed to solve primer design problems associated with providing a specific product size for PCR experiments. A simple accuracy for the primer design of the gene CYP1A1, associated with a heightened lung cancer risk, calculated and the sequence NM_000499. Five hundred runs of MA and GA primer design methods were performed with different PCR product lengths, and with different methods of calculating Tm ways are performed. A comparison of the accuracy results for GA and MA primer design showed that the MA for primer design yields better than the GA for primer design. The results further indicated that the proposed method finds optimal or near-optimal primer sets and effective PCR products from a large amount of template DNA in a relatively short time.
I. INTRODUCTION
OLYMERASE chain reaction (PCR) is a method for fast mass duplication of DNA sequences [1]. It is a common technology applied in the biomedical and biotechnology field. By repeating some cycles of the denaturation reactions, annealing and extension, it allows a small amount of DNA to be amplified exponentially. Typically 25–45 of these cycles are performed [2]. Amplification of specific regions of the genome is determined by specific primer sets with forward and reverse orientation. The length of the PCR product is also depended on the primer set. In the past, feasible primer sets were usually found manually through trial and error, but this method is time consuming, because many constraints have to be satisfied at the same time. Typically considered primer design constraints are the length of the primer, the GC content, the melting temperature and GC clamp. The length of primers
Manuscript received February 15, 2008.
Cheng-Hong Yang is with the Department of Electronic Engineering, National Kaohsiung University of Applied Sciences, Taiwan (e-mail: chyang@cc.kuas.edu.tw).
Yu-Huei Cheng is with the Department of Electronic Engineering, National Kaohsiung University of Applied Sciences, Taiwan (e-mail: yuhuei.cheng@gmail.com).
Hsueh-Wei Chang is with the Faculty of Biomedical Science and Environmental Biology, Kaohsiung Medical University, Taiwan (e-mail: changhw@kmu.edu.tw).
Li-Yeh Chuang is with the Department of Chemical Engineering, I-Shou University, Kaohsiung, Taiwan (email: chuang@isu.edu.tw).
should be within 16 bps~28 bps, while the difference of the primer pair length should not exceed 3 bps. The GC content of primers should be within 40%~60%. The melting temperature of primers should be within the range of 50oC~62oC, while the difference of the melting temperature of a primer pair should not exceed more than 5oC. Finally, the 3’ end of primers should be G or C.
In past decades, secondary structures were applied to primer design, such as dimer, hairpin and specificity of a primer pair. Today, various approaches for primer design have been proposed. Kämpke et al. (2001) use dynamic
programming to design primers. This allows for designing multiple primers for multiple target DNA sequences [2, 3]. Wu et al. (2003) proposed a genetic algorithm (GA) imitating
nature’s process of evolution and genetic operations on chromosomes in order to achieve optimal solutions [3]. Chen
et al. (2003) used a GA to develop a web-based tool (PDA)
for primer design [4]. Hsieh et al. (2003) developed an
efficient algorithm using automatic variable fixing and automatic redundant constraint elimination to tackle the binary integer programming problem associated with the minimal primer set (MPS) selection problem [5]. Wang et al.
employed a greedy algorithm to generate the MPS specifically annealed to all open reading frames (ORFs) in a given microbial genome to improve the hybridization signals of microarray experiments [6].
In this research, an evolutionary algorithm for primer design is proposed and primer design with specific PCR product size is performed. The proposed algorithm is a Memetic algorithm (MA). PCR and primer constraints, such as primer length, difference of primer pair length, GC proportion, PCR product length, melting temperature (Tm), difference of melting temperature (Tm-diff), GC clamp, dimer of primer pair (including cross-dimer and self-dimer), hairpin and specificity are used to design optimal primer sets. A simple accuracy calculation was performed to compare the MA with GA primer design methods with different PCR product lengths and different methods of calculating the melting Tm temperature.
MAs were inspired by Dawkins’ notion of a meme [7]. They are similar but superior to GAs. MAs progress through a local search before being involved in the evolution process [8]. MAs assure that all chromosomes and offspring gain some experiences. The MA algorithm proposed for the primer design correctly and quickly identifies an optimal primer pair required for a specific PCR experiment. Our experimental results further indicate that MAs applied to the design of primer sets performed better than GAs when applied the gene CYP1A1, which has been associated with a
Primer design with specific PCR product size using Memetic
algorithm
Cheng-Hong Yang,
Member, IEEE, Yu-Huei Cheng, Hsueh-Wei Chang, and Li-Yeh Chuang
P
2008 IEEE Conference on Soft Computing in Industrial Applications (SMCia/08), June 25-27, 2008, Muroran, JAPAN
-heightened lung cancer risk, and the sequence NM_000499, which is defined in NCBI as “Homo sapiens cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1), mRNA”.
II.PROBLEM DEFINITION
In the section, we define the problem for primer design. Let
D
T be the template DNA sequence, which is make up by base-nucleic acid sequence of DNA. TD is defined as
follows: } , G' ' or C' ' or T' ' or A' ' | { ∀ ∈ ∈Ζ+ = B B i TD i i (1)
The primer design problem is to find a pair of subsequences of corresponding constraint fromTD. The pair
of subsequences one is called forward primer and another is called reverse primer. The forward primer and reverse primer are defined as follows:
} , of index the , G' ' or C' ' or T' ' or A' ' | { e F i F T i B B P s D i i f ≤ ≤ ∈ ∈ ∀ = (2) } , of index the , G' ' or C' ' or T' ' or A' ' | { e r R i R T i B B P s D i i ≤ ≤ ∈ ∈ ∀ = (3)
where Pf is the forward primer, Fs and Fe are denoted as the start index and the end index of Pf in TD. Pr is the forward primer, Rs and Re are denoted as the start index and the end index of Pr in TD. The Pf and Pr are called primer pair.
In the Fig. 1, we define the length of template DNA is ι, the minimum PCR product length is ρ, the maximum PCR product length is σ, the start position of forward primer is Fs,
the length of forward primer is Fl, PCR product length is Pl
between forward primer and reverse primer, the length of reverse primer is Rl, the random range of F is η, the length s from Fs to the template DNA end is δ. Now, a vector with
s
F , Fl, Pl and Rl can determine a primer pair. We defined
the vector as follows:
) , , , ( v Fs Fl Pl Rl P = (4) By the P , we can calculate the reverse primer start index, v
that is
s
R = Fs+Pl-Rl (5)
Consequently, the forward primer and reverse primer can be got from Pv. The Pv is the prototype of individual in MA and later section will use Pv to do evolutionary computation. Table I lists all parameters in Fig. 1.
s F l F l P l R l P
Fig. 1. The parameters between template DNA and primer set TABLE I.THE PARAMETERS IN FIGURE1.
Parameter Description
s
F the start position of forward primer l
F the length of forward primer l
P PCR product length between forward primer and reverse primer
l
R the length of reverse primer η the random range of Fs
ρ the minimum PCR product length σ the maximum PCR product length δ the length from Fs to the template DNA end ι the length of template DNA
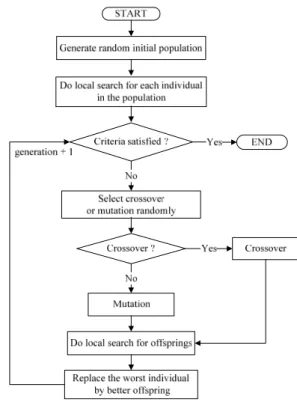
III. MEMETIC ALGORITHM FOR PRIMER DESIGN The flowchart of proposed algorithm for the PCR primer design is shown as Fig. 2. In order to explain how to design optimal primer pair, we described some processes in detail, contain the generation of random initial population, fitness evaluation, local search, crossover and mutation, and termination conditions. Finally, the calculation of accuracy is mentioned. They are described as follows:
Fig. 2. The flowchart of the proposed algorithm for PCR primer design. 1. Generate random initial population
-However, they are still not generally used in primer design problem.
TABLE II.THE ACCURACY AND RUNNING TIME FOR GA AND MA PRIMER DESIGN USING WALLACE FORMULA AND BOLTON AND MCCARTHY FORMULA WITH PCR PRODUCT LENGTH BETWEEN 150~300 BPS,500~800 BPS AND 800~1000 BPS.THE NUMBER OF ITERATIONS IS 100 TIMES FOR BOTH, THE MA AND GA. A,
ACCURACY (%); T, RUNNING TIME (MS).BOLDFACE INDICATES HIGHEST VALUES.
Wallace's formula Bolton and McCarthy formula
GA MA GA MA Tm formula and primer design methods PCR product length a (%) t (ms) a (%) t (ms) a (%) t (ms) a (%) t (ms) 150~300 bps 72.60 325843 99.60 1167562 24.40 475421 88.20 1152250 500~800 bps 70.19 347094 97.80 1180594 26.00 456125 89.40 1201156 800~1000bps 75.20 346109 97.60 1168875 25.60 435562 89.20 1242563 average 72.66 339682 98.33 1172344 25.33 455703 88.93 1198656
TABLE III.THE ACCURACY AND RUNNING TIME FOR GA AND MA PRIMER DESIGN USING WALLACE FORMULA AND BOLTON AND MCCARTHY FORMULA WITH PCR PRODUCT LENGTH BETWEEN 150~300 BPS,500~800 BPS AND 800~1000 BPS.THE NUMBER OF ITERATIONS IS 500 AND 100 TIMES FOR GA AND MA,
RESPECTIVELY. A, ACCURACY (%); T, RUNNING TIME (MS).BOLDFACE INDICATES HIGHEST VALUES.
Wallace's formula Bolton and McCarthy formula
GA MA GA MA Tm formula and primer design methods PCR product length a (%) t (ms) a (%) t (ms) a (%) t (ms) a (%) t (ms) 150~300 bps 76.60 1179390 99.60 1167562 30.80 1654391 88.20 1152250 500~800 bps 72.39 1167531 97.80 1180594 33.20 1746156 89.40 1201156 800~1000bps 75.80 1171125 97.60 1168875 33.20 1653312 89.20 1242563 average 74.93 1172682 98.33 1172344 32.40 1684619 88.93 1198656 V. CONCLUSION
Primer design has become an important issue over the last decade. The quality of primers always influences whether a PCR experiment is successful or not. To date, many primer design methods and tools have been developed, but most of these are inefficient or do not design optimal primers pair for use in the PCR experiments. In this study, we designed optimal primer pairs using an MA, and performed PCR with primer constraints to appraise the fitness values, such as primer length, difference of primer pair length, GC proportion, PCR product length, melting temperature (Tm), difference of melting temperature (Tm-diff), GC clamp, dimer of primer pair (including cross-dimer and self-dimer), hairpin and specificity. Based on their significance each constraint was given a corresponding weight. Through the design of a fitness function, feasible primer sets could always be obtained using the MA algorithm. The primer design results show that different Tm calculation methods affect the size of the primer length and the melting temperature. Using Wallace formula for calculating Tm acquires a shorter primer length and lower temperature value, but using Bolton and McCarthy formula for calculating Tm yields longer primer lengths and a higher temperature value. The performance of a GA has been shown to be superior to SFS (sequential forward search), PTA (plus and take away) and SFFS (sequential forward floating search) [9]. At present, GAs are widely applied to solve various optimization problems. However, GAs tend to get trapped in a local optimum easily. The study applied MAs to design primers with specific PCR product lengths. The results indicate that the proposed method can develop optimal or near-optimal primer sets for use in PCR experiments.
ACKNOWLEDGMENTS
This work is partly supported by the National Science Council in Taiwan under grants NSC96-2622-E-151-019-CC3, NSC96-2221-E-214-050-MY3, and NSC96-2622-E214-004-CC3.
REFERENCES
[1] Mullis,K. and Faloona,F. Specific synthesis of DNA in vitro via a polymerase catalyzed chain reaction. Methods Enzymol., vol. 155, pp. 335–350, 1987.
[2] Kämpke,T., Kieninger,M. and Mecklenburg,M. Efficient primer design algorithms. Bioinformatics, vol. 17, pp. 214–225, 2001. [3] Jain-Shing Wu, Chungnan Lee, Chien-Chang Wu and Yow-Ling
Shiue. Primer design usig genetic algorithm, Bioinformatics, pp. 1710-1717, 2004.
[4] Lin, C. Y., Chen, S. H, Lo, C.Z., Cho, C.S., Hsiung, C. A., Primer Design Assistant (PDA): a web-based primer design tool, Nucleic Acids Research, vol.31, no.13, pp. 3751-3754, 2003.
[5] Hsieh, M.-H., W. Hsu, S. Chiu, and C. Tzeng. An Efficient Algorithm for Minimal Primer Set Selection. Bioinformatics, vol.19, pp. 285-286, 2003.
[6] Wang J, Li KB, Ken Wing Kin SUNG. G-PRIMER: greedy algorithm for selecting minimal primer set. Bioinformatics (Applications note), vol. 20(15), pp. 2473-2475, 2004.
[7] Dawkins R. The selfish gene. Oxford: Oxford University Press; 1976. [8] Merz P, Freisleben B. A genetic local search approach to the quadratic assignment problem. In: Ba¨ck CT, editor. Proceedings of the 7th international conference on genetic algorithms. San Diego, CA: Morgan Kaufmann. pp.465–72, 1997.
[9] Oh, I.-S. Lee, J.-S., and Moon, B.-R., “Hybrid Genetic Algorithms for Feature Selection”, IEEE Trans. Pattern Analysis and Machine Intelligence, vol.26, no.11, Nov. 2004.
[10] E. Elbeltagi, T. Hegazy, and D. Grierson, Comparison among five evolutionary-based optimization algorithms, Advanced Engineering Informatics, vol. 19, pp. 43-53, 2005.
[11] Moscato P, Norman MG. A memetic approach for the traveling salesman problem—implementation of a computational ecology for combinatorial optimization on message-passing systems. In: Valero M, Onate E, Jane M, Larriba JL, Suarez B, editors. International conference on parallel computing and transputer application. Amsterdam, Holland: IOS Press; 1992. pp. 177–86.