國立交通大學生物科技學系
博士論文
克雷白氏肺炎桿菌中第三型纖毛的特性分析
Characterization of type 3 fimbriae in Klebsiella pneumoniae
研究生: 黃盈蓉
Student: Ying-Jung Huang
指導教授: 彭慧玲博士
Advisor: Hwei-Ling Peng, Ph.D.
中華民國九十六年一月
中文摘要
附著寄主細胞,是細菌達成感染的第一步驟。已知,第三型纖毛的表現與克 雷白氏肺炎桿菌感染寄主細胞的部位密切相關。本論文主題在探討第三型纖毛的 表現與克雷白氏肺炎菌致病過程的關聯:首先,我們以PCR-RFLP的技術分析第 三型纖毛黏附蛋白基因型,結果在十七株腦膜炎菌株中發現四種新的黏附蛋白基 因型,分別命名為mrkDV1,mrkDV2, mrkDV3和mrkDV4。接著,我們以大腸桿菌 JM109 為表現載體構築了表現第三型纖毛的重組質體,分別帶有mrkDV1, mrkDV2,mrkDV3和mrkDV4基因型。而藉穿透式電子顯微鏡來觀察這些重組大腸桿 菌表現的纖毛,我們發現mrkD基因型的變異會影響第三型纖毛的長度和型態。 同時,測試這些重組細菌黏附第四型和第五型膠原蛋白的能力、生物膜的形成與黏附細胞的活性,結果顯示,E. coli JM109[pMrkABCDV3F]在這些測試中,活性
皆高於帶有其他三種黏附蛋白基因型的大腸桿菌。我們也發現MrkDv3 黏附蛋白
上的RGD序列可以決定E. coli JM109[pMrkABCDV3F]黏附HCT-8 細胞的專一
性。其次,我們也探討第三型纖毛在克雷白氏肺炎桿菌CG43 中的表現調控:核 酸序列分析顯示第一型纖毛和第三型纖毛的基因群相連。在LB或GCAA培養液 中,都可以偵測到第三型纖毛的表現;相反的,只有在mrkA缺損或是fimB大量表 現的情況下,才能偵測到第一型纖毛的表現。而在fimB大量表現時,第三型纖毛 的表現會明顯下降,這樣的結果暗示著這兩種纖毛的表現有互相調控的關係。而 在 第 一 型 纖 毛 和 第 三 型 纖 毛 基 因 群 之 間 , 我 們 發 現 有 兩 個 基 因 和 Erwinia chrysathemi調控毒性因子的pecS和pecM相似,分別命名為phgS和phgM。在測量 啟動子活性時,我們發現PS-mrkA的活性在phgS或phgM缺損株中,都明顯降低,這
組蛋白PhgS可以結合PS-mrkA也顯示PhgS具有轉錄調控的功能。另外,我們還找到
一段不受PhgS/PhgM影響的啟動子PL-mrkA,顯示第三型纖毛的表現調控可能並不
Abstract
As generally known, attachment of pathogens to their host is a prerequisite step
of infection. The study reports the involvement of type 3 fimbriae, which has been
shown as the primary adhesion factor in Klebsiella pneumoniae, in the bacterial
pathogenesis. Firstly, four novel mrkD alleles namely mrkDV1, mrkDV2, mrkDV3 and
mrkDV4, were identified in seventeen K. pneumoniae meningitis strains. A type 3
fimbriae display system in Escherichia coli was subsequently constructed to
determine the effect of MrkD allelic variation on the fimbrial activity. The TEM
analysis indicated that the proper growth of the filament and fimbrial morphology
were MrkD adhesin dependent. The assessments via measurements of collagen IV and
V binding activity, biofilm formation, and cell adherence revealed that the E. coli
JM109[pmrkABCDV3F] had the highest level of fimbriae activity and the adhesion to
HCT-8 cells is probably through the interaction of the RGD sequence on MrkDv3 with
integrin. Secondly, regulation of expression of type 3 fimbriae in K. pneumoniae
CG43 is investigated. Sequence analysis revealed that type 1- and type 3-fimbriae
gene clusters are physically linked in the genome of K. pneumoniae CG43. The
expression of type 3 fimbriae in LB or GCAA medium could be readily demonstrated
in the bacteria. In contrast, the expression of type 1 fimbriae was evident only in the
overexpression of fimB diminished the expression of type 3 fimbirae suggesting a
cross regulation is present for the expression of the two fimbriae. In-between the two
gene clusters, homologues of Erwinia chrysanthemi virulence regulatory genes pecS
and pecM were identified and named phgS and phgM respectively. The promoter
activity measurement revealed that the deletion of phgS or phgM reduced the activity
of the putative promoter PS-mrk, which suggesting a PhgS/PhgM-dependent expression
of type 3 fimbriae. The binding of the recombinant PhgS to PS-mrk demonstrated by
EMSA also supported a transcriptional regulation of PhgS on the expression of type 3
fimbriae. Nevertheless, a PhgS/PhgM-independent promoter PL-mrk was also identified
致 謝 在博士班的生涯中,我要感謝的人非常多。首先要感謝的是我的指導老師彭 慧玲博士,在這段時間,不管是在生活上或是學業上,我常常帶給彭老師很大的 麻煩,幸好有彭老師的包容與教導,才能完成此博士論文。除了彭老師,也要感 謝清華大學張晃猶老師、中興大學黃秀珍老師、交通大學林志生和楊昀良老師在 論文口試時提供的寶貴意見,使論文能更加完整。 接著要感謝的是實驗室的夥伴:盈璁學長、榕華學姊、靖婷、怡欣、騰逸、 巧韻、致翔、定宇、美甄、婉君、佩瑄、平輝、健誠、小新、祐俊、智凱、育聖、 心瑋、登魁、格維、朝陽、秉熹、嘉怡…你們就像我的家人一樣,陪伴著我一路 走過來,讓我的博班生活很多采多姿。在實驗上的進行方面,我也得到很多人的 幫助:怡琪學姊在分生技巧上的教導、雲龍學長和慧中學姊在TEM 方面的協助、 博瑞和佳翰在雷射聶夾方面的幫助,讓我很多的實驗都可以順利進行。另外,我 也要感謝我的好友們:像我姐姐一樣照顧我的靖婷、常常讓我耍賴的何狗、默默 支持我的鋒哥、很厲害的政男、溫柔的千婷、善良的建良、一起泡老人茶的程翔, 我不會忘記我們一次次互相鼓勵、一次次互相扶持的感動。最後我要感謝我的家 人,有你們的支持,我才能無後顧之憂的完成我的學業,謝謝你們。 其實還要感謝的人很多,我沒有一一的列舉出來,可是還是要跟你們說一 聲,謝謝你們的幫忙。這各階段的完成並不是代表結束,而是代表著另外一各階 段的開始,我會更加努力的去完成下個階段的任務,不會辜負你們支持與鼓勵唷。
Contents
中文摘要………. ii Abstract……… iv Chapter I 1 Introduction……….. 2 Chapter II 9Materials and methods………. 10
Chapter III
Allelic variation of type 3 fimbrial adhesin MrkD affects the fimbriation and
adhesion activity 21
3.1 Abstract... 22 3.2 Results and discussion………... 23
Chapter IV
Regulation of expression of type 3 fimbriae in Klebsiella pneumoniae CG43 32 4.1 Abstract……….. 33 4.2 Results and discussion………... 34
Chapter V 45
Summary 46
References……… 49 Tables……….... 64 Figures……….. 68
List of Table
Table 1. Table 1. Bacterial strains and plasmids used in this study………. 64
Table 2. Primers used in this study………..……… 65
Table 3. Mr/K hemagglutination assay……… 66
Table 4. Effect of GRGDSP or anti-α5β1 integrin antibody on binding of
JM109[pmrkABCDV3F] ………...
List of Figure
Fig. 1. Amino acid sequence comparison of MrkD1P, MrkDV2, MrkDV3, and
MrkDV4……… 68
Fig. 2. (A) Cellular adherence activity of the meningitis isolates ………. 69 (B) Western blotting analysis of the type 3 fimbrial expression in the 17
K. pneumoniae meningitis isolates……….. 70
Fig. 3. Western blotting analysis of MrkA expression in the recombinant E.
coli…..………. 71
Fig. 4. Transmission electron micrographs of the type 3 fimbriae displaying on
the recombinant E. coli……… 72
Fig. 5. Sequence comparison of MrkA and MrkF……… 76 Fig. 6. RT-PCR analysis to assess an operon structure of mrkABCDF………… 77 Fig. 7. The activity assessments of the recombinant type 3 fimbriae…………..
(A) Cell adhesion assay……… 78 (B) Collagen binding activity………... 78 (C) Biofilm formation of the recombinant E. coli……… 79
(D) Autoaggregation phenotype of E. coli JM109[pmrkABCDV3F]…… 79
Fig. 8. A schematic presentation describing the adhesive force measurement… 81 Fig. 9. Organization of the mrk-fim gene clusters……… 82 Fig. 10. (A) Western blotting analysis of the expression of type 3 fimbriae..…. 83
(B) TEM observation of type 3 fimbriae labeled with 1:50 dilution of thepolyclonal anti-MrkA antiserum and gold particle (10 nm)-
conjugated anti-mouse immunoglobulin……… 83 (C) Diagrammatic representation of the promoter analysisand the
DNA electrophoresis shows the patterns corresponding to phase ON
andOFF for the expression of type 1 fimbriae……….. 84 (D) The expression of type 1 fimbriae was detected with anti-FimA
antibody.………. 84
(E) Yeast agglutination assay. 85
Fig. 11. (A) Analysis of PfimA switch………. 86
(B) Yeast agglutination activity of K. pneumoniae CG43S3
transformed with plasmid pETQ33 or the fimB-expression plasmid
(C) Total proteins isolated from each of the bacteria were resolved on SDS-12.5% polyacrylamide gel by electrophoresis (C)-1, and the gel transferred into PVDF and labeled with anti-FimA antibody (C)-2 and
anti-MrkA antibody (C)-3……….. 87
Fig. 12. (A) Analysis of the deletion effect of phgS or phgM on the activity of PfimB, PfimE, PS-mrkA, and PL-mrkA.The expression of type 1 fimbriae was
detected with anti-FimA antibody……….. 88 (B) The purified recombinant PhgS protein resolved on SDS-12.5%
polyacrylamide gel by electrophoresis is shown on the left. The EMSA assessment for the DNA binding activity of the recombinant
PhgS is shown on the right………. 88 (C) Quantitative determination of the mRNA by Limiting-dilution
RT-PCR analysis……… 89
(D) Western blotting analysis of the expression of type 3 fimbriae…… 89 Fig. 13. (A) Locations of the primers used in mapping of the mrkA
transcription start site………. 91
(B) Mapping of the mrkA transcription start site by RT-PCR (upper panel) and Southern blotting hybridization (lower panel).
Abbreviations
Amp Ampcilin
BCIP 5-bromo-4-chloro-3-indolyl phosphate
BSA Bovine serum albumin
Cm Chloramphenicol CRP cAMP Receptor Protein
EMSA Electrophoretic mobility shift assay
GCAA Minimal medium containing 1% glycerol and 0.3% casamino acid HA hemagglutination
IVET In vivo expression technology Km Kanamycin
LD-RT-PCR Limiting-dilution-reverse transcription-PCR LRP Leucine-responsive regulatory protein NBT Nitro blue tetrazolium
PBS Phosphate-buffered saline
RGD Arg-Gly-Asp
RFLP Restriction fragment length polymorphism Sm Streptomycin
STM Signature-tagged mutagenesis
TEM Transmission Electron Microscopy Tc Tetracycline
Chapter I
Klebsiella pneumoniae is an important cause of community-acquired pneumonia
that commonly results in high fatality if untreated (Han, 1995; Podschun and Ulmann,
1998). The vast majority of K. pneumoniae infections are associated with
hospitalization, which has been estimated to cause up to 8% of all nosocomial
bacterial infections in developed countries (Bergogne-Berezin, 1995; Schaberg et al.,
1991). Nasopharynx and intestinal tract have been thought as the primary reservoirs
and colonization of the bacterium in perigenital area is an important phase for
subsequent infection of the urinary tract (Mayhall et al., 1980; Seidler et al., 1975). In
Taiwan, K. pneumoniae has been attributed to be the major cause of liver abscess
especially in diabetes mellitus patients (Chang and Chou, 1995; Wang et al., 1998).
The infections often cause serious complications such as septic endogenous
endophthalmitis, metastatic infections of brain and lung, and necrotizing fasciitis
(Cheng et al., 2003). Several factors are known to participate in the pathogenesis of
Klebsiella spp., including capsular polysaccharides, lipopoly- saccharides,
iron-acquisition systems, and adhesins (Podschun and Ulmann, 1998). Most recently,
in vivo expression technology (IVET) and Signature-tagged mutagenesis (STM)
assays have been applied to identify other factors involved in the bacterial
pathogenesis (Boddicker et al., 2006; Lai et al., 2001; Struve et al., 2003). However,
Adherence to host tissues is an essential early phase in many bacterial infections.
The attachment to host surfaces is thought to increase the infection potential by
providing resistance to the mechanical clearance by the host defense system
(Switalski et al., 1989). In K. pneumoniae, the adhesion process is frequently
mediated by fimbriae and the specific interaction is determined by adhesins, a
component on the tip of fimbriae. Among the five adhesins identified in K.
pneumoniae, type 1 fimbriae are present frequently in pyelonephritis isolates of
Escherichia coli (Iwahi et al., 1983). Type 3 fimbriae, referred to as the
mannose-resistant Klebsiella hemagglutin (Mr/KH), have been shown to be
produced by some uropathogenic isolates of K. pnenumoniae (Old, 1972). The
nonfimbrial adhesin CF29K, which is involved in the adherence to human intestinal
cell line Caco-2 (Di-Martino et al., 1995), and a fimbrial antigen KPF-28 that has
been shown to be a determinant for the colonization on human gut (Di-Martino et al.,
1996), are both encoded by a β-lactamase-producing plasmid. The fifth has recently
been described as a nonfimbrial adhesin to mediate an aggregative adhesion pattern
to intestinal cell lines (Favre-Bonte et al., 1995).
Bacterial adherence determines the tissue tropism during infection and the
process is generally mediated by adhesin (Clegg and Gerlach, 1987;
dramatically reduce the number of fimbriae expressed on the cell surface (Schembri et
al., 2002). The effect of allelic variation of the adhesive molecule on adhesive activity
of type 1 and type P fimbriae also has been reported. For instance, a minor mutation
in fimH, the adhesin encoding gene of type 1 fimbriae, rendered approximately 70%
of the uropathogenic Escherichia coli an increasing ability to recognize
monomannose (Man 1). While 80% of the feces isolates bind only to trimannose
(Man 3) receptors (Firon et al., 1987). The variation of PapG, the adhesin of P
fimbriae, also appeared to alter the fimbrial receptor specificity (Stromberg et al.,
1991).
Type 3 fimbriae is often found on Klebsiella strains as well as in a variety of
enteric bacteria (Old and Adegbola, 1983). Similar to the operon structure coding for
type 1 fimbriae, type 3 fimbriae is encoded by mrk gene cluster (Allen et al., 1991).
The mrk operon includes mrkA, which encodes the major fimbrial subunit; mrkB and
mrkC, respectively coding for chaperone and usher proteins required for assembly and
anchorage of the fimbriae; mrkF, encoding a protein which has been shown to affect
the stability of the fimbriae; and mrkD which encodes the adhesin responsible for the
activity of MR/K hemagglutination. Until recently, only three mrkD variants, a
plasmid-encoded MrkD1p and chromosomally occurred MrkD1C1 and MrkD1C2, each
reported (Sebghati et al., 1998). Although a protein receptor has been suggested
(Hornick et al., 1992), the identity of MrkD receptor remains unknown.
In addition to the major subunit pilin protein and the adhesin protein on the tip of
a fimbria, there are other proteins, minor subunits, found in the filamentous hair. For
example, type 1 fimbriae contains minor subunits, FimF and FimG, which are
required for integration of FimH adhesin into the fimbirae at the tip location (Krogfelt
and Klemm, 1988). Their roles in determining morphology and assembly of the
fimbrial structure have been demonstrated (Klemm and Christiansen, 1987). The
minor subunits of P pilus, PapF, PapK and PapH have also been reported to function
as adaptor, initiator, and terminator, respectively, for the growth of the filament
(Jacob-Dubuisson et al., 1993; Verger et al., 2006). In addition, PapE was shown to be
responsible for binding to fibronectin indicating another role of the minor subunit
(Westerlund and Korhonen, 1991). MrkF has been reported to affect type 3 fimbrial
activity by stabilizing the structure of the filament (Allen et al., 1991). We have
shown previously by TEM analysis that MrkF was able to serve as an initiator for the
growth of the filament and to control the length of the fimbriae (廖心瑋,民國九十五
年). Furthermore, as determined by the measurements including collagen binding
activity, biofilm formation, and autoaggregation phenotype, MrkF also played a
Bacteria can often express multiple adhesins during infection in order to
establish the attachment to specific niches. In the genome of Salmonella enterica
serovar Typhi CT18, twelve fimbrial gene clusters of the chaperone-usher-dependent
assembly class have been identified. Through prevalence analysis of these fimbrial
gene clusters among serotypes of S. enterica subspecies, S. Typhi was shown to
possess a unique repertoire of fimbrial gene sequences possibly due to specific
selective pressures (Townsend et al., 2001). Using of flow cytometry to detect
expression of eleven S. Typhimurium fimbrial operons also showed that in vivo
growth conditions drastically alter the repertoire of fimbrial antigens expressed in S.
Typhimurium (Humphries et al., 2003). In uropathogenic E. coli, the mutant deficient
in both type 1 and P fimbrial expression produced F1C fimbriae, which was not
produced by wild-type strain during growthin aerated or static culture conditions
(Snyder et al., 2005). This also suggested that the presence of a regulatory network in
controlling differential expression of these fimbriae.
The two most commonly associated fimbriae with E. coli urinary tract infections
are type 1 and type P fimbriae (Holden et al., 2006). The expression of type 1
fimbriae requires two recombinases, FimB, to promote the inversion of the promoter
DNA in off-to-on phase, and FimE to mediate an on-to-off orientation (Gally et al.,
by alternative DNA methylation patterns (Blomfield, 2001). PapB, a positive
regulator for type P fimbriae expression, has been shown to affect the expression of
type 1 fimbriae via inhibition of the FimB-promoted recombination but enhancement
of the expression of fimE (Xia et al., 2000). The DNA microarray analysis also
demonstrated a coordinate expression betweentype 1 fimbriae and P fimbriae (Snyder
et al., 2005). Moreover, a study further determined that the cross-regulation between
P and type 1 fimbriae occurs in E. coli clinical isolates with the level of PapB that are
produced from its natural promoteron the chromosome (Holden et al., 2006).
K. pneumoniae has become an increasingly common pathogen of adult
community-acquiredbacterial meningitis (Lee et al., 2003). In part I of the study, we
intend to investigate the association between variation of MrkD adhesin of type 3
fimbriae and the meningitis-associated K. pneumoniae isolates. Since the expression
of type 3 fimbriae is generally low in K. pneumoniae clinical isolates, a type 3
fimbriae display system carrying respectively each of the MrkD variants was
constructed and the influences of the mrkD allelic variation on the fimbrial activity
investigated.
Type 1 and type 3 fimbriae are commonly reported in K. pneumoniae. While the
regulationof type 1 fimbriae has been clear, thatof type 3 fimbriae remains to be
of type 3 fimbriae in K. pneumoniae CG43, a highly virulent clinical isolate of K2
serotype (Chang et al., 1996). A lamda phage clone containing mrkABCDF gene
cluster has previously been isolated from a genomic library (Peng et al., 1992) and the
sequences determined. The fim gene cluster coding for type 1 fimbriae and
divergently transcribed phgS and phgM, homologues of Erwinia chrysanthemi
virulence regulator encoding genes pecS and pecM (Reverchon et al., 1994), were
identified upstream of the mrk gene cluster. In the study, a coordinate expression
between type 1 and type 3 fimbriae, and PhgS/PhgM-dependent PhgS/PhgM
Chapter II
2.1Bacterial strains, plasmids and media
Bacterial strains and plasmids used in this study are listed in Table 1. The growth media were LB (Luria Broth) and GCAA (minimal medium containing 1% glycerol and 0.3% casamino acid) (Gerlach et al., 1989) supplemented with appropriate antibiotics. The bacteria were grown at 37°C unless otherwise indicated.
2.2 PCR-RFLP analysis of the mrkD genes
Genomic DNA of the K. pneumoniae isolates were prepared as the template and the
primers used are corresponding respectively to the 5’ and the 3’-end of mrkD1p coding
region (Gerlach and Clegg, 1988). The PCR products were then digested with Sau3AI
and the restriction fragments resolved on a 2% agarose by gel electrophoresis.
2.3 Cell adhesion assay
Three epithelial cell lines including human laryngeal carcinoma cell line Hep-2,
ileocecal epithelial cell line HCT-8, and embryonic intestinal epithelial cell line
Int-407 were used. According to the cellular adherence assay (Oelschlaeger and Tall,
1997), the cells were seeded into 24 well plate (TPP, Trasadingen, Switzerland) and
incubated to confluent growth in 5% CO2 for 48 h. Approximately 107 bacteria were
then added to each well containing about 105 cells, and the incubation continued for 1 h. To determine if the RGD motif contained in MrkDV3 plays a role in cell adhesion,
the hexapetides GRGDSP (Calbiochem 03340035) and GRADSP (Calbiochem
03340052), and anti-integrin monoclonal antibody α5β1 (Chemicon JBS5) were
added. Finally, the plates were washed three times with phosphate-buffered saline
(PBS), and the cells were lysed with 0.1% Triton X-100. The cell-adhesive bacteria
were measured by recovery of the bacteria from the lysates.
2.4 Preparation of the recombinant proteins FimA, MrkA and PhgS
The genes fimA and mrkA, respectively encoding the major pilin subunit of type 1
fimbriae and type 3 fimbriae, and phgS were isolated by PCR cloning from K.
pneumoniae CG43S3 (Lai et al., 2003) and ligated into pET30 expression vector. The
recombinant plasmid was then transformed into E. coli NovaBlue(DE3) and
overexpression of the recombinant protein was induced by addition of 0.5 mM IPTG
(isopropyl-β-D-thiogalactopyranoside). Finally, the recombinant proteins FimA,
MrkA, and PhgS, N-terminal fusion with His-tag respectively, were purified using the
affinity column charged with nickel resins (Novagen, Madison, WI, USA).
2.5 Antisera preparation
Five-week-old female BALB/c mice, purchased from the animal center of National
or MrkA. Ten days later, the mice were immunized again with 5 μg of the protein and
the antisera obtained by intracardic puncture.
2.6 Western blot analysis of the expression of type 1 and type 3 fimbriae
Total cellular lysates from the bacteria grown overnight in either LB or GCAA broth
were resolved by SDS-PAGE to determine the expression of type 1 and type 3
fimbriae in K. pneumoniae CG43. The proteins were then electrophoretically
transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica,
MA, USA). After incubation with 5﹪skim milk at room temperature for 1 h, the
membrane was washed 3 times with 1X PBS. Subsequently, the membrane was
incubated at room temperature for 1 h with diluted anti-FimA or anti-MrkA serum.
After 3 washes with 1X PBS, a 3000-fold diluted alkaline phosphatase-conjugated
anti-mouse immunoglobulin G was added and the incubation continued for one more
hour. The blot was again washed and the bound antibodies were detected by using the
chromogenic reagents BCIP (5-bromo-4-chloro-3-indolyl phosphate) and NBT (Nitro
blue tetrazolium).
2.6 Construction of the type 3 fimbriae expression plasmid
pneumoniae CG43. Each of the mrkD variants was then subcloned respectively into
pmrkABC, which resulted in the plasmids pmrkABCDV1, pmrkABCDV2,
pmrkABCDV3 and pmrkABCDV4 (陳美甄,民國九十一年). The gene coding for
MrkF, which helps to stabilize type 3 fimbriae (Allen et al., 1991), was then inserted
downstream to each of the mrkD alleles. The plasmids were named pmrkABCDV1F,
pmrkABCDV2F, pmrkABCDV3F, and pmrkABCDV4F, respectively.
2.7 Transmission electron microscopy (TEM)
Twenty microliters of bacterial suspension (108 cfu/ml) were added to collodion-
coated copper grids (300 mesh) and negatively stained by 2% phosphotungstic acid,
pH 7.2. For immunogold TEM analysis, the bacteria coated on the grids (10-nm
diameter) were reacted with the raised anti-MrkA polyclonal antibody (1:50 dilution)
and the anti-mouse IgG -gold conjugate of 1:65 dilution before staining with 2%
phosphotungstic acid. The grids were examined under a JEOL JEM 2000EXII trans
-mission electron microscope at an operating voltage of 100 kV (Hornick et al.,
1995).
2.8 RT-PCR analysis to assess an operon structure of mrkABCDF.
Total RNA was isolated from K. pneumoniae cells by extraction with the TRI reagent
(Molecular Research Center, Cincinnati, Oh, USA) and the residual DNA was
eliminated with RQ1 RNase-free DNase (Promega, Madison,WI, USA). The cDNAs
used for PCR amplification were each synthesized from 1μg of total RNA using the
random hexamer primer from a RT-PCR kit (Stratagene, La Jolla, CA, USA). The
primers used to amplify the major pilin MrkA of type 3 fimbriae are MrkA-RT-1
(5’-CTC TGA CAA GGA AAT GGC AAT G-3’) and MrkA-RT-2 (5’-GGT AAG TAA
TTT CGT AAG TCG CGT-3’). The primers for MrkD adhesin are MrkD-RT-1
(5’-ATG TCG CTG AGG AAA TTA CTA ACG-3’) and MrkD-RT-2 (5’-GCT GAA
ACG CAT GCC GAT-3’). The primers for MrkF are MrkF-RT-1 (5’-ATG AAG GGA
TTG CCG AAA AA-3’) and MrkF-RT-2 (5’-GCT CCA TCC GGC AAG GTA-3’).
The primers specific for intergenic region between mrkD and mrkF are MrkDF-RT-1
(5’-AGG AGA CCC GCT ACA TCA CC-3’) and MrkF-RT-2. The PCRwas carried
out with initial denaturation at 95°C for 10 min, 35 cycles of denaturation at 95°C for
1 min, annealing at 46°C for 1 min, and elongation at 72°C for 1 min, and finally 10
min of elongation at 72°C. Finally, the amplified productswere resolved on 1%
agarose gels.
The hemagglutination assay was performed as described (Gerlach and Clegg, 1988).
Briefly, overnight grown bacteria were collected and suspended in PBS to
approximately 109 CFU/ml. Human erythrocytes (group A) were treated with 0.01%
tannic acid for 15 min at 37°C and subsequently washed twice with PBS. A series of
four-fold dilution of the bacterial suspension with 2% D-mannose were mixed with an
equal volume of 3% (vol/vol) tanned-erythrocytes in PBS. The mixture was incubated
at room temperature for 30 min to allow erythrocytes settle to the bottom of the glass
tube.
2.10 Binding to type IV- and type V-collagen
The binding assay was carried out as described (Sebghati et al., 1998). Essentially, the
wells of flatbottom microtiter plate (Nunc, Rochester, NY, USA) were coated
following incubation overnight at 4°C with optimal concentrations of type IV
collagen (Sigma C7521) or type V collagen (Sigma C3657). The non-specific
bindings were prevented by incubation for 2 h at 22°C with a 1% (wt/vol) solution of
bovine serum albumin (BSA). Subsequently, each well was added with 100 μl bacteria
(108 cfu/ml) and the incubation continued for 2 h at 22°C with gentle shaking. The
unattached bacteria were removed by washing three times with 0.05% Tween-20 in of
adhesion was determined by the recovery of the bacteria.
2.11 Preparation of collagen-coated beads
Polystyrene beads (Polysciences, Warrington, PA, USA) of 1 lm in diameter were
added into PBS containing 10 μg collagen and then incubated at 4°C for 16 h. The
beads were then blocked with the blocking reagent (2% BSA in PBS) at room
temperature for 1 h. After blocking, the beads were washed and re-suspended in 100
ml PBS. BSA-coated beads were also prepared similarly, with the replacement of the
collagen to BSA solution.
2.12 Biofilm formation
The ability of bacteria to form biofilm was analyzed as described with a minor
modification (O'Toole and Kolter, 1998). 100 μl of the overnight grown bacteria
diluted 1/100 in GCAA medium were inoculated into each well of a 96-well
microtiter dish and incubated at 37°C for 48 h. After washing, 150 μl of crystal violet
(1%) was added to each well and incubated for 30 min at room temperature. The plate
was then washed, the dye was solubilized in 1% SDS, and the absorbance at 595 nm
was determined. The mean of three separate experiments represents the biofilm
2.13 Construction of mrkA, mrkF, phgS, and phgM deletion mutants
DNA fragments of 1 kb in length flanking both ends of the target genes mrkA, phgS,
and phgM were amplified by PCR with the primer sets (Table 2) and the amplified
DNA fragments were cloned into the suicide vector pKAS46(Lai et al., 2003). The
plasmids were transformed respectively into E. coli S17-1pir (de Lorenzo and Timmis,
1994) and then mobilized to the streptomycin-resistant strain K. pneumoniae CG43S3
(Lai et al., 2003) by conjugation. A kanamycin resistant transconjugant was selected
and propagated in 2 ml LB overnight, and a small aliquot of the culture was plated on
LB agar containing 500 μg/ml streptomycin. The streptomycin resistant colonies were
screened further for their susceptibility to ampicillin and kanamycin, a property
reflecting the loss of vector sequence. To construct mrkF deletion mutant, the
amplified DNA fragments were cloned into the suicide vector pwA01 (Table 1). The
resulting plasmid was transformed to the K. pneumoniae NTUH-K2044 by
conjugation. A kanamycin resistant transconjugant was selected and propagated in 2
ml LB overnight, and a small aliquot of the culture was plated on LB agar containing
20% sucrose. The colonies resistant to sucrose were screened further for their
susceptibility to kanamycin, a property reflecting the loss of vector sequence.
flanking mrkA, mrkF, phgS, or phgM, and confirmed with Southern hybridization.
2.14 Yeast-cell agglutination
Agglutination of yeast Saccharomyces cerevisiae AH109 was carried out as described
(Blumer et al., 2005). Briefly, the bacteria were suspended in PBS or PBS with 2 %
mannose (2 x 109/ml) and then mixed with 10 mg/ml of yeast on a glass slide. The
degree of clumping was assessed by eyes and the agglutination count was expressed
as the highest dilution of the bacteria causing visible agglutination of yeast.
2.15 Promoter activity measurements
The putative promoters of fimB, fimE and mrkA were PCR amplified using the
specific primers (Table 2) and the PCR products subcloned in front of the
promoterless lacZ gene on placZ15 (Lin et al., 2006). The bacteria carrying each of
the reporter plasmids were grown statically overnight in LB medium, and the
β-galactosidase activities were measured essentially as described (Miller, 1972). The
data presented were derived from a single experiment, which is representative of at
last three independent experiments. Every sample was assayed in triplicate, and the
2.16 Limiting-dilution RT-PCR (LD-RT-PCR) analyses
LD-RT-PCR was performed as described (Schwan et al., 2005; Schwan et al., 2002).
The cDNA was prepared as described in RT-PCR analysis to assess an operon
structure of mrkABCDF and then subjected to PCR amplification. Each of the cDNA
was diluted in four-fold graded dose and then subjected to PCR amplification. The
primers used to amplify the major pilin MrkA of type 3 fimbriae are MrkA-RT-1
(5’-CTC TGA CAA GGA AAT GGC AAT G-3’) and MrkA-RT-2 (5’-GGT AAG
TAA TTT CGT AAG TCG CGT-3’). The primers specific for 23S-rRNA are
Kp-23S-rRNA-1 (5’-CCC CCG AAG ATG AGT TCA CG-3’) and Kp-23S-rRNA-2
(5’-GGC GAT GTC CGA ATG GGG AA-3’). The PCRwas carried out with initial
denaturation at 95°C for 10 min, 35 cycles of denaturation at 95°C for 1 min,
annealing at 46°C for 1 min, and elongation at 72°C for 1 min, and finally 10 min of
elongation at 72°C. Finally, the amplification productswere resolved on 1.5% agarose
gels.
2.17 Electrophoretic mobility shift assay (EMSA)
Since the overexpressed PhgS protein formed insoluble inclusion bodies, 6 M urea
was used to dissolve the insoluble protein and then the denatured protein purified. The
The DNA fragment comprising the putative promoter PS-mrkA was obtained by PCR
amplification and then labeled with [γ-32P]ATP using T4 polynucleotide kinase as
described (Lai et al., 2003). The purified PhgS was then incubated with the
radioactively labeled DNA in 20 μl containing 20 mM Tris-HCl (pH 8.0), 4 mM
MgCl2, 50 mM KCl, 1 mM CaCl2 and 1 mM dithiothreitol at 37°C for 20 min. To
demonstrate the binding specificity, excess amount (approximately 100 times more
than the labeled DNA) of the unlabeled DNA was used. The samples were then
loaded onto a running gel of 5% nondenaturing polyacrylamide in 0.5× TBE (45 mM
Tris-HCl, pH 8.0, 45 mM boric acid, 1 mM EDTA) and the gel electrophoresed with a
20-mA current at 4°C. Finally, the gel was analyzed by InstantImagerTM (Packard
Chapter III
Allelic variation of type 3 fimbrial adhesin MrkD affects
the fimbriation and adhesion activity
3.1 Abstract
Four novel mrkD alleles namely mrkDV1, mrkDV2, mrkDV3 and mrkDV4, were
identified in seventeen Klebsiella pneumoniae meningitis strains using PCR-RFLP
and sequence determination. Comparative analysis revealed a highly variable region
containing an RGD motif in the receptor domain of MrkDV3. In order to determine if
the sequence confers the K. pneumoniae mrkDV3 the highest level of the fimbrial
activity, a type 3 fimbriae display system was constructed in Escherichia coli. The E.
coli JM109[pmrkABCDV3F] displaying meshwork-like fimbriae also had the most
fimbrial activity, supporting a possible role of the varied sequences. In a
dose-dependent manner, the GRGDSP hexapeptide appeared to inhibit the adhesion of
the E. coli JM109[pmrkABCDV3F] to HCT-8, an ileocecal epithelial cell line. In
addition, the adhesion activity was reduced by the addition of anti-α5β1 integrin
monoclonal antibody, indicating that the RGD containing region in MrkDV3 is
3.2 Results and discussion
3.2.1 Identification of four novel mrkD alleles
Recently, the incidence of K. pneumoniae meningitis in newborns and adult
patients have been reported worldwide (Lee et al., 2003). Since the role of type 3
fimbriae in determining the tissue tropism has been suggested (Hornick et al., 1992;
Schurtz et al., 1994), the presence of a specific type 3 fimbrial adhesin mrkD allele in
meningitis isolates was investigated. Using the primers specific to mrkD1p, PCR
analysis showed that all the 17 meningitis isolates carry mrkD gene, and four different
mrkD RFLP types were obtained. Each of the PCR products was then cloned and their
sequences determined. The BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/) analysis revealed 4 novel mrkD alleles, designated mrkDV1, mrkDV2, mrkDV3, and
mrkDV4 (under the GenBank accession number AY225462, AY225463, AY225464,
and AY225465). Notably, fourteen of the isolates carry mrkDV1 RFLP and others
include one each of the variants mrkDV2 (VHm2), mrkDV3 (VHm5) and mrkDV4
(VHm10). This suggests that the K. pneumoniae carrying mrkDV1 RFLP is a prevalent
strain. All the isolates carry mrkD gene implying a possible correlation of type 3
fimbriae with the disease. Nevertheless, more isolates are needed to establish the
3.2.2 Amino acid sequence analysis
Comparative analysis with sequence of K. pneumoniae MGH78578
(http://genome.wustl.edu/) revealed an identical mrkD except that a G deletion was found in mrkDV1 at the position 355. The nucleotide deletion caused a frame shift and
resulted in a truncated protein, of which the pilin domain was replaced with a garbled
sequence of 57 amino acid residues at the C-terminus. It is hence the name MrkDV1T
for the truncated form of the adhesin. As shown in Fig. 1, the conserved receptor
binding and pilin domains, and the cysteine residues could be identified in each of the
MrkD variants. The comparison indicated that MrkDV2 and MrkDV4 share the most
identity, which is 88.1%. Less were found between MrkDV2 and MrkDV3 with 79.3%
identity, and MrkDV3 and MrkDV4 with 80.2%. In the receptor domain of MrkDV3, a
varied sequence from residues 120 to 140, and an RGD motif of integrin recognition
site (Ruoslahti and Pierschbacher, 1987) were identified (Fig. 1). In addition, the
residues which have been proposed to facilitate the interaction of MrkD with other
fimbrial component (Sebghati and Clegg, 1999), were unique in MrkDV3 (C102 and
R200). The D-R-N (residues 68 to 70) of MrkD1P that has been shown to affect the
fimbrial activity (Sebghati and Clegg, 1999) appeared to be replaced by different
varied sequences for the fimbriae activity.
3.2.3 Type 3 fimbriae activity of the meningitis isolates
It has been reported that type 3 fimbriae of K. pneumoniae mediate a specific
adherence to different kinds of human epithelial cells (Schurtz et al., 1994; Tarkkanen
et al., 1997). To examine influences of the mrkD allelic variation on the fimbrial
adhesive activity, three epithelial cell lines Hep-2, HCT-8, and Int-407 were used. As
shown in Fig. 2A, the bacteria VHm5 of mrkDV3 allele exerted a relatively higher
level of the cell adhesion activity. While, 14 of the mrkDV1 strains revealed different
levels of activity. The subsequent analysis using western blotting hybridization with
the anti-MrkA antiserum indicated that the expression of type 3 fimbriae could only
be observed in VHm2 (mrkDV2), VHm5 (mrkDV3), and 6 of the mrkDV1 strains (Fig.
2B). These implied that, besides type 3 fimbriae, other factor(s) such as capsular
polysaccharide, which has been reported to impede the bacterial adherence to cells
(Sahly et al., 2000), is/are involved in determining the cellular adherence activity.
3.2.4 Expression of the recombinant type 3 fimbriae
To rule out the possibility that other factors resided in K. pneumoniae interfere
established. The production of type 3 fimbriae on the surface of the recombinant
bacteria was confirmed by western blot analysis (Fig. 3). The TEM analysis (Fig. 4)
revealed that no fimbriae on the surface of JM109[pGEMT-easy] could be observed.
Only in a small portion, approximately one tenth of the bacteria JM109[pmrkABC],
some short and erect fimbriae were found. In addition, several long fimbriae were
found on the surface of E. coli JM109[pmrkABCF], suggesting that MrkF, as a minor
fimbrial subunit, is able to function as an initiator for the growth of the filament. In
the absence of MrkD adhesin, however, the growth of filament could not be properly
terminated and hence appeared lengthy. The speculation is supported by the
appearance of extremely long and bundle fimbriae on the surface of E. coli
JM109[pmrkABCDV1TF], which could be caused by an interaction of the truncated
MrkDV1T with the usher protein leading to uncontrollable length of the fimbriae.
Different from the uniform fimbrial pattern observed on JM109[pmrkABCDV2F]
and JM109[pmrkABCDV4F], the fimbriae on the surface of JM109[pmrkABCDV3F]
are entangled and give rise to a meshwork-like morphology. The sequence
comparison in Fig. 1 indicated that unique residues of MrkD V3 are probably the
determinants in facilitating MrkD interaction with other fimbrial protein for the
3.2.5 The mrkF gene is a component of mrkABCDF operon
The common structural characteristics of major pilin subunit are (i) two cysteine
residues; (ii) a conserved pattern of alternating hydrophobic residues at position 4, 6,
and 7 from the COOH terminus; (iii) a penultimate tyrosine; and (iv) a Gly at position
14 from COOH terminus (Girardeau et al., 2000). As shown in Fig. 5, sequence
analysis of MrkF revealed a signal peptide using LipoP in ExPASy proteomic tools
and all the characteristics of major subunit. In addition, MrkF also possessed
sequence motifs that are conserved among fimbrial subunits (Girardeau and Bertin,
1995). The intergenic sequence of 16 bp between mrkD and mrkF also implied mrkF
is a component of the mrk operon structure of mrkABCDF. The RT-PCR analysis of
mrk expression in K. pnemoniae NTUH-K2044 revealed a co-expression of mrkA,
mrkD, mrkF, and the intergenic region between mrkD and mrkF (Fig. 6). This
indicated mrkF belongs to mrk operon. The previous study using E. coli display
system has shown MrkF played a role in regulating the length of the filament and the
fimbrial activity (廖心瑋,民國九十五年). A mrkF deletion mutant is being
constructed, and the fimbrial morphology and activity will be compared to that of
3.2.6 Activity assessments of the recombinant fimbriae
As shown in Table 3, the bacteria JM109[pmrkABCDV3F] and
JM109[pmrkABCDV4F] expressed approximately 16 HA units, and
JM109[pmrkABCDV2F] had less of the activity. Whereas, JM109[pmrkABCDV1TF] as
well as the bacteria carrying pGEMT-easy, pmrkABC, or pmrkABCF exhibited no
hemagglutination. This suggested that the MrkDV1T truncation alters conformation of
the MrkD receptor binding domain and hence no hemagglutination activity could be
detected. As shown in Fig. 7A, JM109[pmrkABCDV3F] expressed the highest level of
adhesive activity to either of the three cell lines. Allelic variation of MrkD has been
shown to affect the binding activity and specificity to collagen (Schurtz et al., 1994).
The Fig. 7B showed that JM109[pmrkABCDV3F] also revealed the strongest binding
activity to collagen IV and V, and JM109[pmrkABCDV4F] had a medium level
activity. Moreover, the biofilm formation analysis revealed that JM109
[pmrkABCDV3F] retained the highest activity (Fig. 7C). JM109[pmrkABCDV2F] and
JM109[pmrkABCDV4F] also exhibited a comparable activity of biofilm formation.
These support the finding that type 3 fimbriae is a major determinant for K.
pneumoniae biofilm formation (Jagnow and Clegg, 2003). As shown in Fig. 1, the
sequence comparison indicated that the D-R-N (residues 68 to 70) of MrkD1P and the
the fimbrial activity. Interestingly, an autoaggregation phenotype was observed only
for JM109[pmrkABCDV3F] (Fig. 7D) suggesting the meshwork like fimbriae
increased the interaction of the bacteria. The alteration of receptor-binding domain of
FimH has been shown to affect the autoaggregation (Schembri et al., 2001). It is also
likely that the varied sequence in the receptor domain of MrkDV3 also confers the
bacteria an autoaggregation property.
Since the number of fimbrial filaments varies among bacterial cells, the
above-mentioned activity measurements could only determine the average binding
activity between a given bacterial population and the target molecules. To determine
precisely the direct interacting force between a fimbria and its target molecules, the
optical tweezers was also used to investigate whether the MrkD really plays a role in
presenting the highest activity of highest JM109[pmrkABCDV3F]. Therefore, a typical
record of the bead’s displacement during the measurement is illustrated in Fig. 8. The
adhesive force of a single fimbria, which expressed with each of mrkD alleles, to
collagen IV measured using optical tweezers were 2.03 ± 0.03 pN, 3.79 ± 0.12 pN,
and 2.87 ± 0.15 pN for mrkDV2, mrkDV3, and mrkDV4, respectively. This further
supported the result of collagen binding analysis. It has been reported that interacting
force between a single type 1 fimbriae and an α-C-mannoside ligand (Liang et al.,
between type 3 fimbriae and collagen IV, suggesting that type 1 and type 3 fimbriae
may be expression in the part of body with same fluid flow rate. According to
Stokes’s Law (F=6(pi)RnVc, R is the radius of the sphere, n is the viscosity, and Vc is
the velocity through a continuous fluid), it is calculated that 1 pN could support 1 μ
m bead to resistant 50 μm/sec flow rate. In addition, the calculated flow rate is
about 40 mm/sec in the bladder and about 300 μm/sec in blood capillary. In addition,
the data of the adhesive force of a single fimbria of type 3 fimbriae will give more
information in the bacterial infection research.
3.2.7 RGD peptide inhibits the adhesion of JM109[pmrkABCDV3F] to HCT-8
It has been reported that the RGD sequence in FHA (Filamentous hemagglutinin)
of Bordetella pertussis is involved in the interaction of the bacteria with macrophage
(Relman et al., 1990). To determine if the RGD motif in MrkDV3 affects the bacterial
adherence to cells, the peptide GRGDSP was added as a competitor in the cell
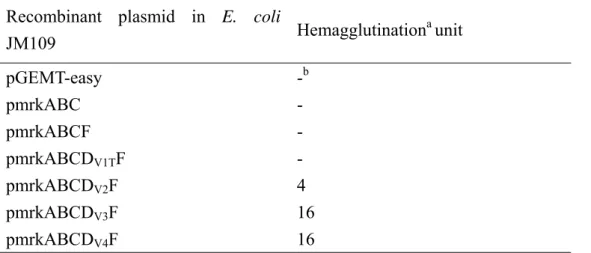
adherence assay. As shown in Table 2, the adhesion of JM109[pmrkABCDV3F] to
HCT-8 cell was reduced by the addition of GRGDSP and the inhibition was in a
dose-dependent manner. In contrast, no inhibition was observed when GRADSP
peptide was added. This supported a role of the RGD sequence in affecting the
ECM and cell surface proteins, is recognized by integrins on the cell surface
(Ruoslahti, 1996). The RGD sequence of B. pertussis FHA has been demonstrated to
interact specifically with α5β1 integrin (Ishibashi et al., 2001). As shown in Table 4,
the anti-α5β1 integrin monoclonal antibody was able to inhibit the adhesion of
JM109[pmrkABCDV3F] to HCT-8, indicating the presence of an interaction of MrkD V3 with α5β1 integrin.
Taken together, we have shown in the study that MrkF is able to serve as an
initiator for the growth of type 3 fimbriae and mrkF is a component of mrkABCDF
operon. In addition, the proper growth of the filament and fimbrial morphology
appeared to be MrkD adhesin dependent. Moreover, MrkDV3 may promote the
bacterial adhesion to HCT-8 cells through the interaction of its RGD sequence with
Chapter IV
Regulation of expression of type 3 fimbriae in Klebsiella
pneumoniae CG43
4.1 Abstract
A lamda phage clone containing type 1- and type 3-fimbriae gene clusters was
isolated from a genomic library of Klebsiella pneumoniae CG43, a K2 isolate with a
high virulence. In-between the two gene clusters, homologues of Erwinia
chrysanthemi virulence regulatory genes pecS and pecM were identified and named
phgS and phgM respectively. The expression of type 3 fimbriae in LB or GCAA
medium could be readily demonstrated in the bacteria. Whereas, the expression of
type 1 fimbriae was evident only in the mrkA deletion mutant or in the overexpression
mutant of fimB, a recombinase-encoding gene. Interestingly, overexpression of fimB
in turn diminished the expression of type 3 fimbirae. The promoter activity
measurement using LacZ as reporter revealed that the deletion of phgS or phgM
reduced the activity of the putative promoter PS-mrk, which suggesting a
PhgS/PhgM-dependent expression of type 3 fimbriae. The binding of the recombinant
PhgS to PS-mrk demonstrated by EMSA also supported a transcriptional regulation of
PhgS on the expression of type 3 fimbriae. Nevertheless, a PhgS/PhgM independent
promoter PL-mrk was also identified indicating a complicated regulation on the
4.2 Results and discussion
4.2.1 Type 1 and 3 fimbrial gene clusters are physically linked in K. pneumoniae
CG43
The entire gene cluster was isolated from a λDASH genomic library, which was
constructed previously in the laboratory (Peng et al., 1992), and the nucleotide
sequence determined. Comparative analysis with the corresponding sequences in the
genome of K. pneumoniae MGH78578 (http://genome.wustl.edu/) revealed 98% nucleotide sequence homology. The mrkE and mrkG that were reported upstream of
the type 3 fimbrial gene cluster mrkABCDF (Sebghati et al., 1998) could not be found.
Instead, four open reading frames namely orf1, phgS, phgM, and orf2 were identified
(Fig. 9). Orf1 encodes a protein of 35% amino acid sequence identity with a
hypothetical membrane protein of E. coli (Dobrindt et al., 2002) and orf2 shares 42%
sequence identity with an ABC-type transporter of Yersinia frederiksenii. PhgS and
PhgM reveal 36% and 44% amino acid sequence identities respectively with the
proteins PecS and PecM of E. chrysanthemi. PecS, a transcription regulator with a
MarR type of DNA binding domain, and PecM, a putative membrane protein, have
been shown to work cooperatively to regulate the virulence of E. chrysanthemi
segments and PhgS carries also a MarR type of DNA binding domain. The finding
implies that the regulatory genes, the phgM and phgS, were recruited to the
mrkABCDF gene cluster after the gene cluster was organized.
Interestingly, a type 1 fimbriae encoding gene cluster was found next to orf1 in a
divergent transcription orientation (Fig. 9). The close association of the fim and mrk
fimbrial gene clusters is intriguing and has not been reported in any other bacteria.
Comparative analysis of the available sequences of the partly completed genome of K.
pneumoniae MGH78578 also revealed a similar structure, indicating that the
physically linked gene organization is not uncommon in K. pneumoniae. It has been
demonstrated that multiple fimbrial adhesins are required for the establishment of a
systemic infection caused by some bacteria and expression of the individual adhesin is
known to be conditionally regulated (Schwan et al., 2002). It is therefore reasonable
to propose that a coordinate regulation like that between type 1 and the type P
fimbriae in E. coli is also present in K. pneumoniae, between the type 1 and type 3
fimbriae.
In addition, comparing to the type 1 fimbriae operon in E. coli (Blattner et al.,
1997), the inverted repeats contained in fim promoter recognized by the recombinase
FimB or FimE were found to be preserved. However, the intergenic regions of
sequence homologies, respectively. The transcription factors NagC and NanR binding
elements contained upstream of E. coli fimB (Sohanpal et al., 2004) could not be
found, suggesting a different regulation on the expression of type 1 fimbirae in K.
pneumoniae.
4.2.2 Expression of type 1 and type 3 fimbriae in K. pneumoniae CG43S3
As shown in Fig. 10A, non-specific protein bands were found to cross react with
the generated anti-MrkA antiserum in the western blotting analysis. Nevertheless, the
detection of specific band with the molecular weight corresponding to MrkA
suggested expression of the major pilin of type 3 fimbriae. The expression of type 3
fimbriae in K. pneumonaie CG43S3 was further confirmed using TEM detection with
an immunogold-labeled anti-MrkA antibody (Fig. 10B). Expression of type 3 fimbriae
could be readily detected while K. pneumoniae CG43 grown in LB or GCAA medium,
suggesting a primary role of type 3 fimbriae in attachment to solid surface. This is
consistent with the report that type 3 fimbriae played as a major adherence factor for
K. pneumoniae to establish infections (Tarkkanen et al., 1997).
Although some of the DNA resulted from incomplete digestion was observed,
the DNA pattern shown in Fig. 10C type 1 indicated an “OFF” phase promoter of the
has been commonly shown to be required for a stable cell-to-surface attachment (Pratt
and Kolter, 1998). On the other hand, the expression of type 1 fimbriae were rarely
observed in K. pnemoniae (Di-Martino et al., 2003; Matatov et al., 1999). Therefore,
it is not surprising to find that the promoter of type 1 fimbriae in K. pneumoniae
CG43 is in OFF phase.
The mutant with a specific deletion of mrkA gene was subsequently generated to
investigate if a cross-talk regulation is involved in the expression of the two fimbriae.
The specific band corresponding to MrkA, which was not detected in the mrkA mutant,
reappeared in the bacteria transformed with the mrkA expression plasmid pYJA (Fig.
10A). As assesses by TEM analysis, the mrkA mutant exhibited fimbriated phenotype
suggesting a coordinate regulation for the expression of type 3 fimbriae and an
unknown type of fimbriae. The expression of FimA protein, the major pilin of type 1
fimbriae, became evident in the mrkA deletion mutant as determined using anti-FimA
antibody (Fig. 10D) suggesting the unknown type of fimbriae appeared on the surface
of mrkA mutant is type 1 fimbriae. As shown in Fig. 10E, an increase of yeast
agglutination activity which could be diminished with mannose in the mrkA deletion
mutant also supported the expression of type 1 fimbriae. Nevertheless, a certain level
wild type bacteria or the mrkA mutant carrying with pYJA, suggesting a
co-expression of type 1 and type 3 fimbriae in the bacteria.
4.2.3 Overexpression of fimB turned on the expression of type 1 fimbriae but
repressed type 3 fimbrial expressio
Multiple fimbrial adhesins have been demonstrated to be required for bacteria to
establish a systemic infection (Schwan et al., 2002). It is generally anticipated that
expression of the individual adhesin is cooperatively regulated. In order to
demonstrate further a coordinate control of the expression between type 1 and type 3
fimbriae, a construct to overexpress FimB recombinase in K. pneumoniae CG43S3
was generated. The analysis of fim promoter as shown in Fig. 11A revealed
“ON”-phase of type 1 fimbriae in the bacteria carrying the expression plasmid
pETQ33-fimB. The expression of type 1 fimbriae could be switched on by
overproduction of FimB recombinase indicating the fim gene cluster is functional in
the bacteria. An apparent increase of mannose-sensitive yeast agglutination activity
further supported the expression of type 1 fimbriae (Fig. 11B). The overexpression of
FimB recombinase was evident by the analysis of protein pattern (Fig. 11C-1).
Notably, the fimB overexpression turned on the expression of FimA (Fig. 11C-2) but
The deletion of mrkA increased the expression of type 1 fimbriae, while the
overproduction of FimB recombinase repressed MrkA expression. Together, both
indicated that the expression of type 3 fimbriae and type 1 fimbriae are regulated in a
coordinate manner in K. pneumoniae. It has been reported that planktonic cells of
nonfimbriated and fimbriated cell differ in their OMP patterns (Otto et al., 2001), and
ompA deletion decreased the expression of type 1 fimbriae in E. coli K1 (Teng et al.,
2006). Furthermore, subunit misfolding of P pili was sensed by two parallel pathways:
the Cpx two component signaling system and the sigma E modulatory pathway (Jones
et al., 1997). A report also indicated that CpxR-P competes with Lrp for binding to
both promoter proximal and distal pap DNA binding sites, inhibiting pap transcription
in vitro and pili expression in vivo (Hernday et al., 2004). Thus, we proposed that the
deletion of mrkA or overexpression of fimB may result in the envelope stress and
activation of the sensing system in the inner-membrane leading to cross-talk
regulation.
4.2.4 PhgS/PhgM-dependent or PhgS/PhgM-independent expression of type 3
fimbriae
In E. chrysanthemi, PecS has been reported to regulate the expression of
1994). In addition, PecS could positively regulate the expression of polygalacturonase
enzyme (Nasser et al., 1999). The phgS and phgM deletion mutants were generated to
investigate whether PhgS and PhgM proteins play regulatory role in the expression of
the physically linked fim and mrk gene clusters. The putative promoters of PfimB and
PfimE (Fig. 9), containing respectively the intergenic regions of fimB-orf1and
fimE-fimB, and the putative promoters of mrk genes, PL-mrkA and PS-mrkA (Fig. 9),
containing intergenic region of orf2-mrkA and approximately 500-bp non-coding
region upstream of mrkA start codon were also isolated by PCR cloning. Each of the
putative promoters was then cloned in front of the promoterless lacZ on pLacZ15. The
resulting plasmids were transformed into wild type, phgS deletion mutant, or phgM
deletion mutant, respectively, and the β-galactosidase activities measured and
compared. Since the promoter activities appeared to be much higher in static cultures
in either LB or GCAA, the following measurements were carried out while the
bacteria grown in LB in static cultures.
PecM is an inner membrane protein involved in transduction of an extracellular
signal (Reverchon et al., 1994). Several stimuli including changes of pH, osmolarity,
and temperature that have been reported to affect the expression of type 1 fimbriae
(Schwan et al., 2002) were also applied to evaluate the role of PhgM on expression of
PS-mrkA activity under any of the conditions. The effect of phgM deletion was only
evident while the bacteria grown in static culture, suggesting PhgM senses the low
level of oxygen to trigger the expression of type 3 fimbriae.
As shown in Fig. 12A, the deletion of phgS or phgM appeared to diminish the
activity of PS-mrkA suggesting a PhgS- and PhgM-dependent expression of the
promoter. On the other hand, either deletion exerted no effect on the activity of PL-mrkA,
PfimB, or PfimE. In addition, activity of PL-mrkA that carries extra 247 bp at 5’end of
PS-mrkA (Fig. 9), appeared to be lower than the activity of PS-mrkA implying the presence
of a negative regulatory element in the extended region. As shown in the left panel of
Fig. 12B, a small amount of soluble PhgS could be obtained after the treatment of the
inclusion body with 6M urea and the mixture dialyzed against PBS buffer. The
purified PhgS was shown to be able to bind PS-mrkA DNA and retard the DNA mobility
(on the right panel of Fig. 12B). In addition, the formation of binding complex could
be inhibited by adding excess amount of unlabelled PS-mrkA DNA. However, addition
of pUC19 DNA also appeared to be able to inhibit the formation of binding complex
indicating a non-specific binding of PhgS to the promoter PS-mrkA. It has been reported
that a palindromic sequence (CGANWTCGTA)TAT(TACGANNNCG) recognized by
PecS could be identified (Rouanet et al., 2004). As well as PecS, PhgS has a MarR
found on PS-mrkA. It has been suggested that PecS interacts with PecM to exert its
regulatory activity (Praillet et al., 1997). In analogy with the regulatory mechanism,
the lack of binding specificity of the recombinant PhgS to PS-mrkA may be
compensated by its binding to PhgM. The possibility is currently being investigated.
As shown in Fig. 12A, the PL-mrkA activity was independent to the control by
either PhgS or PhgM. The subsequent RT-PCR analysis revealed that the deletion of
either phgS or phgM exerted no negative effect on the levels of mrkA transcript and
the amount of MrkA transcript in either mutant appeared to be even higher than that
synthesized in wild type bacteria (Fig. 12C). Western blotting analysis shown in Fig.
10D also indicated a PhgS/PhgM-independent expression of MrkA protein.
4.2.5 CRP and LRP probably play a role for the PhgS/PhgM-independent
expression of type 3 fimbriae
The expression of a certain genes promoted by two separate condition-controlled
promoters is not uncommon in bacteria (Heroven et al., 2004). The identification of
PhgS/PhgM-dependent and PhgS/PhgM-independent promoters implies different
transcripts are produced. In order to map the transcription start site of mrkA, RT-PCR
analysis using different pairs of primers were carried out. As shown in Fig. 13, the
Southern blotting hybridization using the PCR product amplified with primer pair
pA2 and pA3 as a probe. No transcript was obtained while the primer pA5 replaced
with pA4 indicating that the transcription start site of mrkA could be assigned to the
sequence between pA4 and pA5. Analysis of the sequences upstream of pA4 revealed
a conserved LRP and CRP binding elements, 5’-YAGHAWATTWTDCTR-3’ (Y=C
or T, H≠G, W=A or T, D≠C, R=A or G) (Cui et al., 1995) and 5’-TGTGA
-N6-TCATC-3’ (Kolb et al., 1993), respectively. We speculate that other than the
regulation of PhgS/PhgM, LRP or CRP also plays a role on the expression of type 3
fimbriae.
CRP, cAMP receptor protein, has been reported to function both aspositive and
negative effector to influence many gene expression (Botsford and Harman, 1992).
LRP, leucine-responsive regulatory protein, regulates the expression of more than 40 genes and proteins in E. coli (Calvo and Matthews, 1994). To determine if CRP and
LRP play a regulatory role on the expression of type 3 fimbriae, RT-PCR is being
employed to analyze the effect of addition of glucose or leucine on the expression of
type 1 and type 3 fimbriae. It is probably that the PhgS/PhgM-independent promoter
PL-mrkA is under control by CRP or/ and LRP, which had been shown to affect the
expression of type 1 fimbriae or P pili (Baga et al., 1985; Kelly et al., 2006) The
determines the cross-regulation of the expression of type 1 and type 3 fimbriae. While
in the presence of a lot of PhgS and PhgM regulatory proteins, PL-mrkA is under a
negative control and PhgS and PhgM play a major role to positively regulate the
expression of type 3 fimbriae. However, the possibility remains to be verified.
Overall, a cross-talk regulation was demonstrated between the expression of type
1 fimbriae and type 3 fimbriae. In addition, PhgS/PhgM appeared to be able to
regulate positively the activity of PS-mrkA, which is probably leading to the expression
of type 3 fimbriae. On the other hand, the deletion of either phgS or phgM had no
effect on the activity of PL-mrkA, PfimB orB PfimE. In addition, a PhgS/PhgM-independent
regulation was also observed indicating a complicated regulatory mechanism is
Chapter V
The incidence of Klebsiella infection is found to increase in the recent years.
Antibiotics are usually used to cure K. pneumoniae infections. However, misuse of
antibiotics caused the nosocomial Klebsiella infections even more severe because of
the emergence of muti-drug resistance strains. Therefore, to find new targets for drug
intervention to the muti-drug resistance strains is important.
Attachment of pathogens to their host is a prerequisite step of infection and
adhesin is one of the major virulence factors involved in K. pneumoniae infections.
Expression of type 3 fimbriae has been correlated with several Klebsiella infections.
However, little is known about the roles of the fimbriae in K. pneumoniae
pathogenesis. In part I, to rule out the possibility that other factors resided in K.
pneumoniae interfere with the activity of type 3 fimbriae, an E. coli type 3 fimbriae
display system was established. By using this display system, we have demonstrated
that the proper growth of the filament and fimbrial morphology were MrkD adhesin
dependent. The changes of fimbrial morphology in turn affected the fimbrial activity
as assessed by collagen binding, cell adherence, and biofilm formation. In addition, E.
coli JM109[pmrkABCDV3F] was found to have the highest level of fimbrial activity
and the adhesion to HCT-8 cells was probably due to the interaction of RGD sequence
on MrkDV3 with integrin. Overall, MrkD adhesin appeared to play a major role in

![Table 4. Effect of GRGDSP or anti-α5β1 integrin antibody on binding of JM109[pmrkABCD V3 F]](https://thumb-ap.123doks.com/thumbv2/9libinfo/8213412.170159/78.892.140.756.176.367/table-effect-grgdsp-anti-integrin-antibody-binding-pmrkabcd.webp)