國 立 交 通 大 學
生 物 科 技 研 究 所
碩 士 論 文
Regulatory roles of MrkH, MrkI, MrkJ and CsgD on
the expression of type 1 and type 3 fimbriae of
Klebsiella pneumoniae CG43
克雷白氏肺炎桿菌 CG43 中 MrkH、 MrkI、MrkJ 和 CsgD
在調控第一型與第三型線毛表現所扮演的角色
研究生: 鄭崴云 (Wei-Yun Cheng)
學號: 9828507
指導教授: 彭慧玲 博士 (Hwei-Ling Peng, Ph. D.)
中華民國一百年六月
i
Abstract in Chinese
第一型和第三型線毛已被證實是克雷白氏肺炎桿菌的重要的致病因子。分析 克雷白氏肺炎桿菌 CG43 基因體,我們發現這兩型線毛基因組相鄰座落,暗示二 者的基因表現有協同調控的可能。本論文中,我們探討 MrkH、MrkI、MrkJ、CsgD 調控蛋白對此兩型線毛表現的影響。首先,分別構築 mrkH、 mrkI 、mrkJ 和 csgD 的基因剔除菌株,再藉由甘露醣競爭酵母菌凝集測試、西方墨點法、生物膜形成 能力分析這些基因缺損的影響。結果顯示,mrkI 基因剔除菌中第一型線毛表現 增加而第三型線毛的表現量減少;mrkH 基因的剔除也減少 MrkA 的表現量及降 低生物膜活性,但不影響 FimA 的表現量;在 mrkJ 基因剔除株中,MrkA 的表現 量有些許增加,但生物膜活性卻略微降低;另一方面,csgD 基因剔除後降低第 三型線毛基因組啟動子活性。而在 mrkI 基因剔除株中,第三型線毛基因組啟動 子的活性降低,但 fimS 翻轉變為正向的趨勢卻增加,此結果進一步證實 MrkI 調 控蛋白在轉錄層次正向調控第三型線毛、負向調控第一型線毛的表現。此外,實 驗顯示 MrkJ 重組蛋白具有磷酸二酯酶活性,這也暗示二級信使 c-di-GMP 在第 三型線毛表現上扮演重要角色。最後,增加 c-di-GMP 結合蛋白 MrkH 表現能提 高第三型線毛表現量,此結果更進一步支持了上述的假說。ii
Abstract
Klebsiella pneumoniae type 1 and type 3 fimbriae have been reported to be important virulence factors. Analysis of the Klebsiella pneumoniae CG43 genome revealed the two fimbriae encoding gene clusters are physically linked, which suggesting a coordinated -regulation for their expression. In the study, regulatory roles of the MrkH, MrkI, and MrkJ and CsgD on the expression of the two fimbriae were investigated. Firstly, the K. pneumoniae CG43S3-derived mutants respectively with a gene-deletion of mrkH, mrkI, mrkJ, and csgD were generated. The deleting effects on the fimbrial activities were then determined using analysis of mannose-sensitive yeast agglutination, western blot hybridization, and biofilm formation. An increased expression of type 1 fimbriae and a decreased expression of type 3 fimbriae were found for the mrkI strain. The deletion of mrkH also decreased the expression of
MrkA and biofilm formation activity but had no apparent effect on FimA expression. On the other hand, the mrkJ deletion appeared to slightly increase the MrkA expression but decrease the biofilm formation activity while the csgD deletion reduced the promoter activity of MrkA. The mrkI deletion reduced the mrkA promoter activity but increased OFF-to-ON inversion of the fimS suggesting MrkI is the transcription regulator for their reciprocal expression. Moreover, a phosphodiesterase activity of the recombinant MrkJ was demonstrated implying the second messenger c-di-GMP (bis-(3′-5′)-cyclic dimeric GMP) plays a regulatory role on the expression of type 3 fimbriae. The possibility was further supported by the notion that overexpression of the c-di-GMP binding protein MrkH apparently increased the type 3 fimbriae activity.
iii
Acknowledgement
碩士班兩年是我人生很重要的轉捩點,開啟了研究生涯第一頁。最重要的是 要感謝我的指導老師- 彭慧玲博士,每當我遇到困難裹足不前時,總是能推我一 把給我勇氣,教導我如何思考,您就像媽媽一樣很溫暖,謝謝您這兩年來辛苦的 教導。請您好好保重身體,要健康唷~ 謝謝幽默的顗峰學長在我剛進實驗室時,耐心的帶領我設計實驗,讓我開始 適應研究的生活;謝謝大胃王丸子太后和享受生活的雅雯學姊,總是讓我們吃到 飽又哈哈大笑,很喜歡這快樂氣氛,真好! 謝謝很有喜感的小新和健誠學長,對 我平日的照顧以及給我實驗上寶貴的意見,也很懷念你們哥倆演內心戲的趣事; 謝謝身材超棒的靜柔學姊,在我情緒低落的時候傾聽並給我鼓勵;謝謝志桓學長 送我一些壓箱寶;謝謝可愛的哲充,你的某些想法真的很有趣,相信你內心應該 還住著一個青春的小男孩,我不會忘記你愛的廣播電台;謝謝剛柔並濟的假花和 親切的佩君學姊,常帶土產和水果填飽我們的五臟廟;謝謝跟我一起奮鬥兩年的 品瑄、宜臻和欣怡,有你們讓我很幸福,永遠都要有 woman’s talk 唷!非常聰明 的豪君,我們一起與為了實驗奮鬥的心路歷程,永難忘懷。活潑大方的小波、很 有安全感的郁勝、心思細密的舉豪和熱心的力成,實驗室的未來就交給你們了! 誠心的感謝實驗室所有成員,有你們的幫忙才有現在的我,非常感謝大家! 最後,我想要謝謝我的家人、小豆和大牙牙,陪我走過人生很重要的階段, 讓我一路走來不孤單,支撐我堅持到最後,我把碩士這份榮耀獻給愛我的人,謝 謝你們。希望在未來的日子裡我們都可以過得幸福快樂,當我們再次相聚的時候 還是有聊不完的趣事。 崴云 謹致於 交通大學生物科技研究所 中華民國 100 年 6 月iv
Contents
Abstract in Chinese...i Abstract………..ii Acknowledgement………...……iii Contents……….…ivList of tables and figures………....vii
Abbreviation………...viii
Introduction 1. Clinical importance of Klebsiella pneumonia………..……1
2. Adhesion properties…….……….2
2.1 Type 1 fimbriae………...2
2.2 Type 3 fimbriae………...3
2.3 Curli fibers and CsgD………...4
3. Cyclic-di-GMP signaling pathway………..….4
4. Specific aims……….5
Materials and methods 1. Plasmids, primers, bacterial strains and growth conditions………7
2. DNA manipulation………..7
3. Preparation of genomic DNA……….7
4. Bioinformatics analysis………..…8
5. Construction of specific gene-deletion in K. pneumoniae CG43…………...8
6. Yeast agglutination activity assay………...9
7. Construction of a complementation strain………..…9
v
9. Constructions of the recombinant His6-tagged proteins………...10
10. Overexpression and purification of the insoluble the His6-tagged FimA….10
11. Western blot analysis of the expression of type 1 and type 3 fimbriae……11 12. Construction of the LacZ reporter gene fusion……….……12 13. β-galactosidase activity assay……….…….….12 14. Motility assay………...……12 15. Overexpression and purification of the His6-tagged MrkJ………….……..13
16. Phosphodiesterase activity of MrkJ………..13 Results
1. BLAST analysis and Pfam searches……….……14 2. Generation of the mrkH, mrkI, mrkJ and csgD deletion mutants………….15 3. Analysis of the deletion effects on the activity of type 1 and type 3 fimbriae………15 4. Analysis of the deletion effects on the expression of FimA pilin of type 1 fimbriae and MrkA pilin of type 3 fimbriae……….16 5. MrkI affected the fimS inversion………..…17 6. MrkA, FimA, FimB and FimE promoter activity analysis………..…17 7. The recombinant MrkJ exhibited a phosphodiesterase activity……...……18 Discussion
1. MrkH is a positive regulator for type 3 fimbriae………19 2. An inverse regulatory role of MrkI on the expression of type 1 and type 3 fimbriae………19 3. MrkJ exerted a PDE activity………..20 4. CsgD is also a positive regulator for the expression of type 3 fimbriae….20 References………22
vi
Tables……….31 Figures………34
vii
List of tables and figures
Table 1. Bacteria strains used in this study………31
Table 2. Plasmids used in this study……….…..32
Table 3. Oligonucleotide primers used in this study………..……33
Fig. 1. Gene organization and domain analysis of mrkH, mrkI, mrkJ and csgD….34 Fig. 2. Comparative sequence alignment of the PilZ domains………35
Fig. 3. Comparative sequence alignment of the EAL domains………36
Fig. 4. Comparative sequence alignment of the CsgDs………...37
Fig. 5. Schematic depiction of the deletion-construct on the left panel and PCR analysis of the deletion mutants on the right panel..………38
Fig. 6. Growth of CG43S3 and the derived mutants in LB and M9 media……….39
Fig. 7. Yeast agglutination activity assay……….40
Fig. 8. Biofilm formation in polystyrene Material at 24hrs or 48hrs...41
Fig. 9. SDS-PAGE analysis of the expression of the recombinant FimA in E. coli BL21(DE3) and purity of the protein………...42
Fig. 10. Western blot hybridization analysis of the deletion mutants………43
Fig. 11. The fimS promoter analysis………..44
Fig. 12. The promoter activity measurement using LacZ as a reporter……….45
Fig. 13. Motility assay………...47
Fig. 14. SDS-PAGE analysis of the expression of the recombinant MrkJ in E. coli BL21(DE3) and purity of the protein………...48
viii
Abbreviation
APS ammonium persulfate bp Base pair (s)
c-di-GMP Bis-(3’-5’)-cyclic dimeric guanosine monophosphate DGC di-guanylate cyclase
EDTA N’N’N’N’-ethylenediaminetetraacetate ESBL extended-spectrum β-lactamase
IPTG isopropyl-β-D-thio-galactopyranoside kDa kiloDalton (s) LB Luria-Bertani broth μg microgram μl microliter MR Mannose resistance
NBT nitro blue tetrazolium chloride OD optical density
ONPG ο-nitrophenyl-β-D-galactopyranoside PAGE polyacrylamide gel electrophoresis PDE phosphodiesterase
PBS phosphate buffer saline PCR polymerase chain reaction PVDF polyvinylidene fluoride rpm revolutions per minutes
SDS-PAGE sodium dodecyl sulfate-polyacrylamid gel electrophoresis X-gal 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside TEMED N, N, N’, N’- tetramethylethyl –enediamide
1
Introduction
1. Clinical importance of Klebsiella pneumoniae
Klebsiella pneumonia, a member of the the Enterobacteriaceae family is a
Gram-negative opportunistic pathogen that infects immunocompromised patients (16, 41). Since 1980s, K. pneumoniae is emerging as an important pathogen both in the community and the hospital setting. In the community the emergence of virulent strains with predominantly of K1/K2 capsular serotypes has been observed (41). K.
pneumoniae causes a wide spectrum of infections, including septicemia, pneumonia,
urinary tract infection, meningitis, and purulent abscess at various sites. Especially in Taiwan, K. pneumoniae has been attributed to be the major cause of liver abscess in diabetes mellitus patients (14, 92). In particular, a distinctive clinical syndrome, which is characterized by community-acquired K. pneumoniae bacteremia with primary liver abscess, metastatic meningitis, and endophthalmitis, has been recognized (47, 50, 89, 93).
In the hospital environment with the extensive use of antibiotics, multiple drug resistance has been increasingly observed in K. pneumonia isolated, especially the extended-spectrum β-lactamase (ESBL)-producing strains (24, 25, 41, 51, 53). The prevalence of ESBL is 25.0% for Klebsiella spp., 12.3% for Escherichia coli and less than 5% for other Enterobacteriaceae. Carbapenems has been considered to be the best option for the treatment of serious infections caused with ESBL-producing K.
pneumoniae (18). The recent report of NDM-1 K. pneumonia which is a
carbapenems-resistant strain and also produces a novel metallo-β-lactamase from a patient in New Delhi hospital has demanded a potent drug for effective clinical treatment (33, 44, 56, 94).
2
2. Adhesion properties
In addition to its antibiotic resistance feature (77), there are five major virulence factors identified to participate in K. pneumonia infections, which include capsular polysaccharides (32), lipopolysaccharides (88), iron-acquisition systems (60), and adhesion property (83). Adherence to host tissues is an essential early phase in many bacterial infections. Fimbriae, one of the adherence factors, are long, thread-like appendages on bacterial surface. They are found in as many as 500 copies per cell (43). Each fimbrial fiber is a polymer composed of hundreds of structural subunits called pilin. The adhesin which is located on the tip of fimbriae determines the specific binding to the host cell. The attachment to host surfaces is thought to increase the infection potentially by providing resistance to the mechanical clearance of the host defense system (84). Most clinical K. pneumoniae isolates are known to express two types of fimbrial adhesins, type 1 and type 3 fimbriae (27).
2.1 Type 1 fimbriae
Type 1 fimbriae, which facilitate colonization of uroepithelial cells (79), are heteropolymeric fibers produced by all members of the Enterobacteriaceae family and expressed by fimACDFGHIK gene cluster (42). They are right-handed, 6.9 nm wide pilus rod containing 500-3000 copies of the major structural subunit FimA, and of a linear tip fibrillum. The fibrillum is formed by the D-manose-specific adhesin FimH at the tip and by several copies of the subunits FimG and FimF (29, 40). In addition to mediating fimbriae attachment to the bladder epithelium, the adhesion FimH also enables bacteria to get internalized into bladder cells (13). The fimC and
fimD genes respectively encode a fimbrial chaperone and usher protein. The fimK
gene located directly downstream of fimH is only present in K. pneumoniae but not in
3
fimbria expression (70). The function of fimI gene product is unknown, but this product has been found to be essential for type 1 fimbriae biosynthesis in E. coli (91). Regulation of the type 1 fimbriae expression in E. coli is very complex, and several regulatory factors that act by altering the expression of fimB and fimE have been described. The fimB and fimE genes located upstream to the fim operon encode DNA recombinases that mediate the expression of type 1 fimbriae (1, 8, 72, 80). The recombinases FimB and FimE regulate the phase switch of type 1 fimbriae. FimB facilitates inversion from phase-OFF to phase-ON, as well as inversion from phase-ON to phase-OFF. FimE, on the other hand, causes only inversion from phase-ON to phase-OFF of type 1 fimbriae plays a significant role in the ability of E.
coli infection of the urinary tract (19). Recently, type 1 fimbriae were also found to be
essential for the ability of K. pneumoniae to cause UTI (urinary tract infection) (74).
2.2 Type 3 fimbriae
Type 3 fimbriae are 2 to 4 nm wide and 0.5 to 2 μm long organelles that are characterised by their ability to mediate mannose-resistant agglutination of tannic acid-treated human RBC (MR/K agglutination) (23). The fimbriae are similar to type 1 fimbriae, both are produced by the chaperone-usher assembly pathway (37). Type 3 fimbriae are encoded by mrkABCDF gene cluster (2, 23): MrkA and MrkF, the major fimbrial subunit protein and minor subunit, respectively (36), MrkD, the fimbrial adhesin of which the N terminal domain is responsible for receptor binding (86). Specifically, MrkD adhesin has also been shown to mediate adhesion to type IV and type V collagen (75); MrkB, a periplasmic chaperon; MrkC, an outer membrane usher protein which anchors the fimbriae to the bacterial cell surface. Most pathogenic K.
pneumoniae strains produce type 3 fimbriae, which are essential for bacterial biofilm
4
of bacteria attached to surfaces, composed of polysaccharides, nucleic acids and proteins known as extracellular polymeric substances (EPS) (11). In addition, type 3 fimbriae have been demonstrated to mediate bacterial attachments to several cell types including tracheal epithelial cells, renal tubular cells, extracellular matrix proteins, and components of basement membranes of human lung tissue (35, 86, 87) .
2.3 Curli fibers and CsgD
Curli fibers are a major adhesin factor to surfaces, and also affect cell aggregation and biofilm formation in many enterobacteria, such as Salmonella and pathogenic E. coli strains (20, 58, 62, 68). Expression of both curli fibers and cellulose depends on the CsgD protein, a response regulator of the LuxR family. The CsgD activates transcription of the csgBAC operon (5), which encodes curli structural subunits, and transcription of the adrA gene, a positive effector of cellulose biosynthesis (66). The AdrA is a member of the GGDEF protein family (26, 85) which can catalyze the synthesis of bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) and in turn stimulates the enzymes responsible for expression of cellulose and adhesive curli fibers (76). The expression of curli is also affected by environmental factors, such as a low growth temperature (<32°C), low osmolarity, and slow growth or starvation conditions (28, 30, 58, 69).
3. Cyclic-di-GMP signaling pathway
Almost 20 years after its discovery, bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) has come to be recognized as a ubiquitous second messenger in bacteria. The second messenger has been shown to control a variety of bacterial cellular processes, including motility, fimbriae expression, biofilm formation and cell cycle progression (31). The intracellular levels of cyclic-di-GMP is regulated by the GGDEF domain
5
proteins with diguanylate cyclase (DGC) activity and the EAL domain or HD-GYP domain proteins with cyclic di-GMP specific phosphodiesterase (PDE) activity (73). The synthesis of c-di-GMP from two GTP molecules is catalysed by the cooperative action of two GGDEF domains that each binds one GTP substrate. On the other hand, degradation of c-di GMP is catalysed by the highly specific HD-GYP or EAL-containing PDEs. Recent investigation has demonstrated that glutamate at position 352, conserved motif DDFG(T/A)GYSS ( loop 6 domain) and Mg2+ or Mn2+ is required for PDE activity (67).
The recently study of BlrP1 (also known as KPN 01598) crystal structure from K.
pneumonia shows that BlrP1 consist of, in addition to a phosphodiesterase EAL
output domain, a BLUF photoreceptor domain which is able to senses blue light using a FAD chromophore (6). In 2006, PilZ- domain proteins were identified as a cellular c-di-GMP receptor using a bioinformatic approach (3). A nuclear magnetic resonance (NMR) structure of the PilZ-containing protein PA4608 from P. aeruginosa (63) and a crystal structure of the Vibrio cholera PlzD (7) have also been resolved and analyzed. Moreover, the residues in the RxxxR and D/NxSxxG which are conserved in the PilZ-domain proteins were shown to be able to bind c-di-GMP at the level of sub-micromolar affinity (17, 52, 71). However, it is still unclear how the binding of c-di-GMP to different PilZ-containing proteins affects the expression of their downstream target genes.
4. Specific aims : To investigate the functional role of MrkH, MrkI, MrkJ and CsgD on the expression of type 1 fimbriae and type 3 fimbriae in K. pneumoniae CG43
In K. pneumoniae CG43, a highly virulent liver abscess isolate of K2 serotype (15), the type 1 fimbriae encoding genes are physically linked with the type 3
6
fimbriae encoding genes mrkABCDF, which suggesting a coordinated regulation is involved in controlling their expression. Next to mrkF, three regulatory genes mrkJ,
mrkI, and mrkH are found (Fig. 1A). The mrkH gene product is annotated to be a
PilZ-domain protein, mrkI gene product is a transcriptional regulator containing an uncharacterized N-terminal region and a C-terminal LuxR-like DNA-binding domain, and MrkJ is an EAL-containing phosphodiesterase. Deletion of mrkJ was found to be able to increase the type 3 fimbriae production and biofilm formation activity, which resulted from the accumulation of intracellular c-di-GMP (38). Although no curli fimbriae genes csgBAC (12) could be identified in the published K. pneumoniae genomes, csgD was found to be clustered with csgEFG (65) in the genome of K.
pneumoniae CG43 (Fig. 1B). In this study, functional roles of MrkH, MrkI, MrkJ and
CsgD on the expression of type 3 fimbriae or and type 1 fimbriae in K. pneumoniae CG43 are investigated. We anticipate that a better understanding of the adhesion property allow identification of a potent target for the development of antibacterial agents.
7
Materials and methods
1. Plasmids, primers, bacterial strains and growth conditions
Bacterial strains, plasmids and primers used in this study are respectively listed in Table 1, Table 2 and Table 3. K. pneumonia CG43, a clinical isolate of serotype K2 recovered from Chang Gung Memorial Hospital, Linkou, Taiwan and is highly virulent to mice (15). K. pneumonia CG43S3, which is a derivative of CG43, is a streptomycin-resistant mutant (46). K. pneumoniae and E. coli strains were generally propagated at 37°C in Luria-Bertani (LB) broth and M9 minimal medium. Bacterial growth was assessed by measuring the optical density at 600 nm (OD600). The
antibiotics used include ampicillin (100 g/ml), chloramphenicol (20 g/ml), kanamycin (25 g/ml), tetracycline (12.5 g/ml), chlorhexidine (15 g/ml) and streptomycin (500 g/ml).
2. DNA manipulation
Plasmids were purified by using the High-Speed Plasmid Mini kit (Geneaid, Taiwan). All DNA-modifying and -restriction enzymes were used as recommended by the manufacturer (Fermentas, USA). PCR amplifications were performed with Taq DNA polymerase (MDBio, Inc, Taiwan), Blend Taq DNA polymerase (TOYOBO, Japan) or High-Fidelity DNA Polymerase (Finnzymes, New England). PCR products and DNA fragments were purified using the Gel/PCR DNA Fragments Extraction kit (Geneaid, Taiwan). The primers used in this study were synthesized by MDBio, Inc, Taiwan. Transformation of E. coli cells was performed following the method of Dower (22).
8
Bacteria cultured at 37°C in LB medium overnight were collected by centrifugation for 3 min (8,000 rpm). The pellet was resuspended in 800 l lysis buffer (5 mM DTT, 100 g/ml lysozyme, 200 mM NaCl, 20 mM EDTA, 40 mM Tris-HCl pH 8.0, 0.2% Triton X-100) and heated at 37°C for 1 hr. Proteinase K solution 100 g/ml was added and the mixture incubated at 50°C overnight. After sitting on ice for 10 min, 250 l of saturated NaCl was added, mixed by gently shaking for 10 min, and then precipitated by centrifugation for 10 min (13000 rpm). Finally, 1000 l 99% alcohol was added to the collected supernatant (500 l) and the mixture subjected to centrifugation for 10 min (13000 rpm). After gently rinsed with 75% alcohol, the pellet was dried and resuspended in sterile water.
4. Bioinformatics analysis
Homology search analysis and gene annotation were performed with the BLAST program provided by NCBI (http://www.ncbi.nlm.nih.gov) or VectorNTI (Invitrogen Vector NTI™ Advance). Functional domains of proteins were predicted using Pfam (http://pfam.sanger.ac.uk/) and promoter prediction was carried out by (http://www.softberry.com/all.htm).
5. Construction of specific gene-deletion in K. pneumoniae CG43
DNA fragments of 1 kb in length flanking both ends of the target genes mrkH,
mrkI, mrkJ, or csgD were amplified by PCR with the respective primer sets (Table 3)
and the amplified DNA fragments were cloned into the suicide vector pKAS46 (78), a suicide vector containing rpsL, which allows counter-selection with streptomycin for loss of the vector plasmid. The plasmids were transformed respectively into E. coli S17-1 λpir (78) and then mobilized to the streptomycin-resistant strain K. pneumoniae CG43S3 (46) by conjugation. Several kanamycin resistant transconjugant were
9
selected and propagated in 4 ml LB overnight, and a small aliquot of the culture was plated on LB agar containing 500 g/ml of streptomycin. The streptomycin resistant colonies were analyzed further for their susceptibility to ampicillin and kanamycin, a property reflecting the loss of the vector sequence. The streptomycin-resistant and kanamycin sensitive colonies were isolated and the deletion of mrkH, mrkI, mrkJ, or
csgD was confirmed by PCR with the gene specific primers (Table 3). The resulting
bacteria with mutation in mrkH, mrkI, mrkJ, or csgD were respectively named WYC09, WYC42, WYC12, and WYC45 (Table 1).
6. Yeast agglutination activity assay
The agglutination analysis of yeast Saccharomyces cerevisiae AH109 was carried out as described (9). Briefly, bacteria (~108 c.f.u./ml) were suspended in PBS with or without 2% mannose and then mixed with 10 mg/ml of yeast (Sigma, YSC2) on a glass slide. After 5 min incubation at room temperature on an orbital shaker, agglutination of yeast caused by bacteria could be assessed.
7. Construction of a complementation strain
The complementation plasmids which carrying respectively mrkHJ and mrkHIJ coding sequences were constructed by PCR cloning using the specific primers into yT&A (Yeastern Biotech, Taiwan), and named pWY05 and The SacI/ XbaI fragment from pWY05 and pWY24 was then subcloned to pRKAS46 and the resulting plasmids were individually transformed into E. coli S17-1 λpir, and the transformants namedmrkI[pKAS46-mrkHIJ] and mrkI[pKAS46-mrkHJ] (Table 1) and then
mobilized to the streptomycin-resistant strain K. pneumoniae CG43S3 by conjugation.
10
The measurement of biofilm formation was performed according to the method described (57). Overnight grown bacteria were diluted (1:100) in LB medium, and 150 l diluted bacteria were inoculated into each well of a 96-well microtiter dish (Orange Scientific, Belgium, cat #5530100 or TPP Scientific, America, cat #92096) and the plate incubated at 37°C for 24 hr or 48 hr to allow the biofilm formation. Each well was then washed with water and 150 l of 1% crystal violet was added and the incubation at room temperature continued for 30 min. After washing with water, 150 l of 1% SDS was subsequently added to each well and the microtiter dish was shaken to dissolve the dye. The capability of biofilm formation was quantified by determining the absorbance at 595 nm (ELx800, BIO-TEK). The biofilm formation activity result represented the mean of three separate experiments.
9. Constructions of the recombinant His6-tagged proteins
The DNA fragments which respectively contains the major pilin of type 1 fimbriae, MrkI-HTH domain and MrkJ-EAL domain were PCR amplified from the genomic DNA of K. pneumoniae CG43S3 with primers wc27 /wc12 and wc21/wc08 (Table 3). The amplified PCR products were cloned into the cloning vector yT&A (Yeastern Biotech, Taiwan), and then subcloned using proper restriction enzymes specific enzyme and then ligated into pET30 expression vector. The recombinant plasmid was then transformed into E. coli NovaBlue(DE3) or E. coli BL21(DE3).
10. Overexpression and purification of insoluble the His6-tagged FimA
The bacterial cells were grown in 100 ml of LB medium at 37°C with shaking until OD600 reached 0.6. Isopropyl-1-thio-β-D-galactopyranoside (IPTG) was then
added to a final concentration of 0.5 mM and the growth was continued for 4 hrs at 37°C. Subsequently, the cells were harvested by centrifugation at 8000 rpm for 10
11
min, resuspended in lysis buffer (50mM Tris-HCl [pH8.0], 1mM EDTA and 100 mM NaCl), and the cell suspension disrupted by sonication and then the cell debris removed by centrifugation at 13000 rpm for 10 min. The recombinant FimA was insoluble. Finally, the His6-tagged proteins were purified from the pellet via affinity
chromatography using His-Bind resin (Novagen), and the elution was carried out with elution buffer (20 mM Tris-HCl, 0.5 M NaCl, 250 mM imidazole, 6N urea, [pH 7.9]). Aliquots of the collected fractions were analyzed by SDS-PAGE and the fractions containing most of the purified His6-tagged protein were dialyzed against the 1 mL of
1X PBS buffer (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4, [pH 7.4])
containing 6N urea. The 5.26mg/ ml anti-rabbit FimA was generated by Kelowna International Scientific Inc.
11. Western blot analysis of the expression of type 1 and type 3 fimbriae
Total cellular lysates from the bacteria grown overnight in LB medium were resolved by 12% SDS-PAGE to determine the expression of type 1 and type 3 fimbriae in K. pneumoniae CG43. The proteins were then electrophoretically transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After incubation with 5% skim milk at room temperature for 1 hr, the membrane was washed 3 times with 1X PBS. Subsequently, the membrane was incubated at room temperature for 2 hrs with diluted anti-FimA or anti-MrkA serum. After 3 washes with 1X PBS, a 5000-fold diluted alkaline phosphatase-conjugated anti-rabbit immunoglobulin G was added and the incubation continued for 1 hr. The blot was again washed and the bound antibodies were detected using the chromogenic reagents BCIP (5-bromo-4-chloro-3-indolyl phosphate), NBT (Nitro blue tetrazolium) and alkaline phosphatase buffer (10 mM, 5 mM and 100 mM Tris-HCl pH 9.5).
12
12. Construction of the LacZ reporter gene fusion
The putative promoters of fimA, fimB, fimE and mrkA were PCR amplified using the specific primers (Table 3) and the PCR products subcloned in front of the promoterless lacZ gene on placZ15 (48). The bacteria carrying each of the reporter plasmids were grown sharking overnight in LB medium, and the β-galactosidase activities were measured essentially as described(55). The data is representative of at last three independent experiments. Every sample was assayed in triplicate, and the average activity and standard deviation were presented.
13. β-galactosidase activity assay
β-galactosidase was assayed according to the method of Miller (54). The bacteria in the early or late logarithmic growth phase (optical density at 600 nm 0.5 or 0.8) were taken 100 l, and mixed with 900 l Z buffer (60 mM Na2HPO4 , 40 mM
NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol), 17 l of 0.1%
SDS and 35 l chloroform and incubated for 15 min at 28°C. Subsequently, 200 l of 4 mg/ml ο-nitrophenyl-β-D-galactopyranoside (ONPG) was added and the mixture vortexed for 10 sec, then incubated at 28°C until yellow color was apparent. Finally, the reaction was stopped by adding 500 l of stop solution (1 M Na2CO3) and the
absorbance of the supernatant was measured OD420. One unit of β-galactosidase is
defined as the hydrolysis of 1 nmol ONPG per min per mg protein.
14. Motility assay
Essentially as described (49), 3 l overnight-grown bacteria was inoculated onto trypton swimming plate (0.3% Bacto Agar, 0.5% NaCl, and 1% tryptone) and the plate incubated at 30°C for 9 hrs. The diameter of the zone created by the swimming bacteria was measured.
13
15. Overexpression and purification of the His6-tagged MrkJ
The bacterial cells were incubated in 100 ml of LB medium at 37°C with shaking until OD600 reached 0.6. Isopropyl-1-thio-β-D-galactopyranoside (IPTG) was then
added to a final concentration of 0.5 mM and the growth was continued for 4 hrs at 37°C. Subsequently, the cells were harvested by centrifugation at 8000 rpm for 10 min, resuspended in lysis buffer (50mM Tris-HCl [pH8.0], 1mM EDTA and 100mM NaCl), and the cell suspension disrupted by sonication and then the cell debris removed by centrifugation at 13000 rpm for 10 min. Finally, the His6-tagged proteins
were purified from the supernatant via affinity chromatography using His-Bind resin (Novagen), and the elution was carried out with buffer A (20 mM Tris-HCl, 500 mM NaCl, 250 mM imidazole, [pH 7.9]). Aliquots of the collected fractions were analyzed by SDS-PAGE and the fractions containing most of the purified His6-tagged protein
were dialyzed against the buffer containing 20 mM Tris-HCl [pH 8.5], 200 mM NaCl, and 10% glycerol.
16. Phosphodiesterase activity of MrkJ
Phosphodiesterase activity of the recombinant MrkJ was performed as previously described (39). In the assay buffer (50 mM Tris-HCl, 1 mM MnCl2 [pH 8.5])
supplemented with 5 mM bis(p-nitrophenol) phosphate (bis-pNPP), 20g of the purified MrkJ was added and the mixture incubated for 3 hrs at 37°C. Reactions were incubated for 3 hrs at 37°C, and the release of p-nitrophenol was measured at 410 nm.
14
Results
1. BLAST analysis and Pfam searches
As shown in Fig. 1A, the MrkH, MrkI, and MrkJ encoding genes are located downstream of the type 3 fimbriae operon mrkABCDF (Fig. 1A) and the intergenic region of mrkH-mrkI and mrkI-mrkJ are respectively 5 and 143 bp. The gene organization has been found to be conserved in the published K. pneumoniae genomes (97). As previously demonstrated by reverse transcription PCR (RT-PCR), the three ORFs mrkH, mrkI, and mrkJ could be transcribed in a transcriptional unit (96). These suggested a possibility of a coordinated expression of the physically linked genes
mrkABCDF and mrkHIJ. The gene coding for csgD, which has been associated with
bacterial virulence (31), is located within yggR, a putative ATPase, and yqgF which is an essential protein for Holliday junction resolvase (HJR) (4) (Fig. 1B).
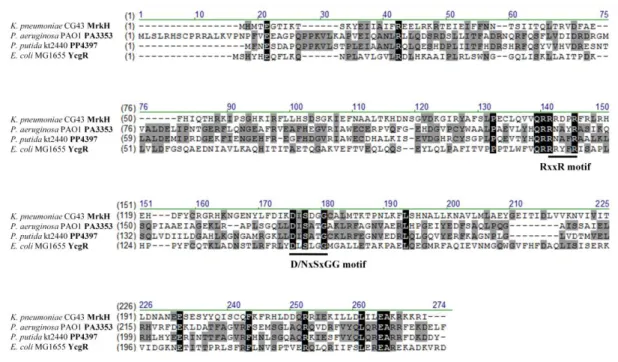
Analysis using BLAST (Basic Local Alignment Search Tool) (38) and Pfam database (protein family database) revealed that mrkH, mrkI, mrkJ and csgD respectively encode c-di-GMP binding protein (PilZ domain protein), LuxR-type transcription regulator, c-di-GMP phosphodiesterase (EAL domain protein) and LuxR-type transcription regulator (Fig.1C). As shown in Fig. 2, the conserved RxxxR motif and D/NxSxGG motif, which play essential roles for c-di-GMP binding in many PilZ domain proteins (3, 10, 31, 52, 61, 71), is found in MrkH. The conserved DDGF(T/A)GYSS motif and glutamate residue critical for phosphodiesterase activity of many EAL domain protein, is also present in MrkJ (Fig. 3). In K. pneumoniae, several EAL domain proteins including BlrP1 (6), YjcC (45), FimK (80), MrkJ (38) have been reported. As shown in Fig.4, the sequence alignment revealed K.
pneumoniae CsgD had 36% identity shared with E. coli CsgD and Salmonella enterica.
15
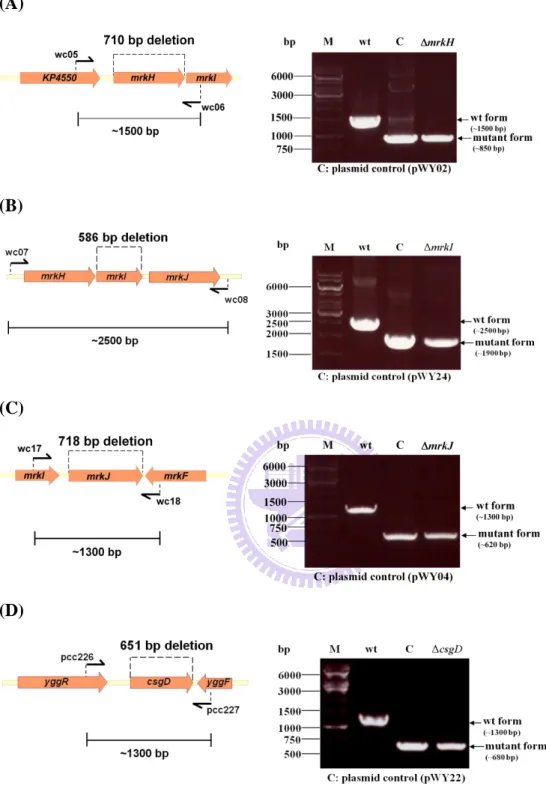
2. Generation of the mrkH, mrkI, mrkJ and csgD deletion mutants
The respective gene deletion was assessed using PCR analysis with the specific primer pairs, wc05/wc06 formrkH, wc07/wc08 formrkI, wc17/wc18 for mrkJ,
and pcc226/pcc227 for csgD. As shown in Fig. 5, the amplicons of 1500-bp and
850-bp were obtained for wild type strain andmrkH strain; 2500-bp and 1900-bp for
wild type strain andmrkI strain; 1300-bp and 620-bp for wild type strain andmrkJ
strain; 1300-bp and 680-bp for wild type strain andcsgD strain, respectivrly which
confirmed the individual deletion for each of the mutants. The selection percentage for mrkH, mrkI, mrkJ and csgD were respectivrly 57%, 64%, 10% and 21%.
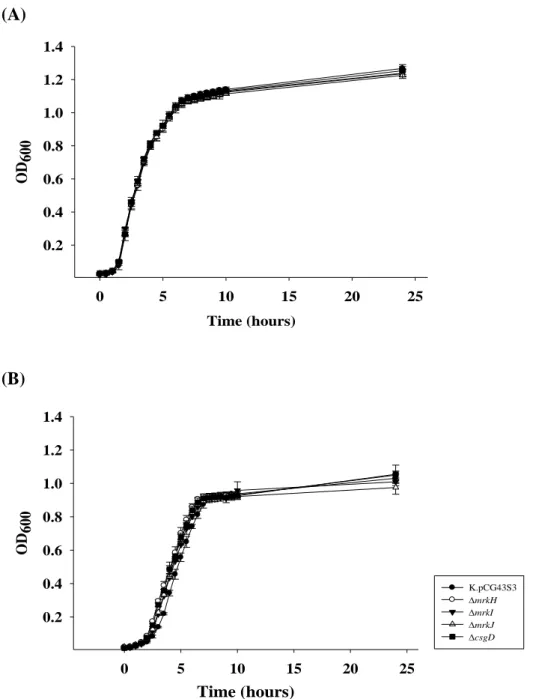
To determine if the gene deletion effects the bacterial growth, growth curve of the mrkH, mrkI, mrkJ and csgD in LB or M9 medium were determined. The
four mutant strains appeared to show similar growth curve as the wild type strain in LB medium (Fig. 6A) or M9 medium (Fig. 6B).
3. Analysis of the deletion effects on the activity of type 1 and type 3 fimbriae
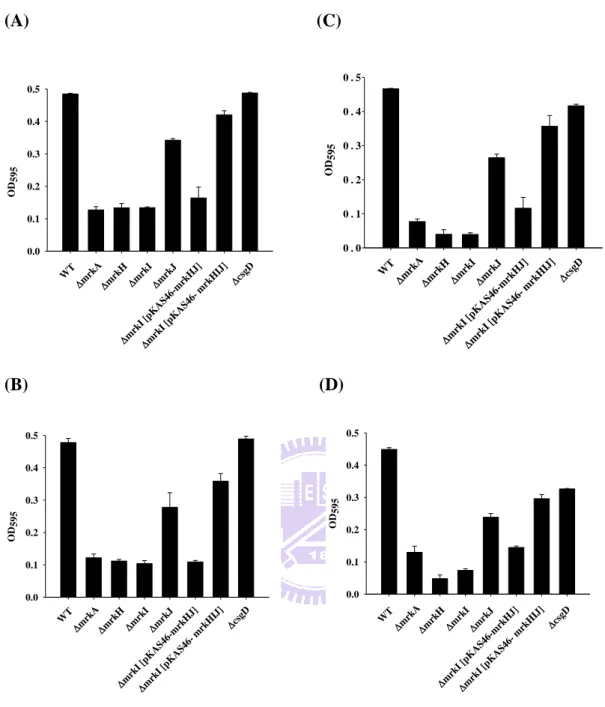
The activity of type 1 fimbriae was assessed using yeast agglutination analysis. As shown in Fig. 7A, agglutination could be observed for mrkAmrkH, mrkI and
mrkI[pKAS46-mrkHJ] strains. The mrkI deletion effect was able to be
complemented by introducing pKAS46-mrkHIJ into mrkI strain. Addition of 2%
mannose could inhibit the agglutination activity of mrkA, mrkI or
mrkI[pKAS46-mrkHJ] indicating a mannose-sensitive agglutination activity (Fig.
7B).
Expression of K. pneumonia type 3 fimbriae has been reported to be able to promote the biofilm formation (21, 36). Compared to wild type, the biofilm formation activity of mrkA, mrkH or mrkI was apparently reduced while the activity slightly
16
and in 24 hrs or 48 hrs incubation (Fig. 8A and B or Fig. 8C and D). The deletion effect of mrkI was able to be complemented by introduction pKAS46-mrkHIJ into mrkI strain. However, deletion of csgD did not affect biofilm formation activity
except a reduced biofilm formation was observed in TPP-microtiter dish for 48 hrs.
4. Analysis of the deletion effects on the expression of FimA pilin of type 1 fimbriae and MrkA pilin of type 3 fimbriae
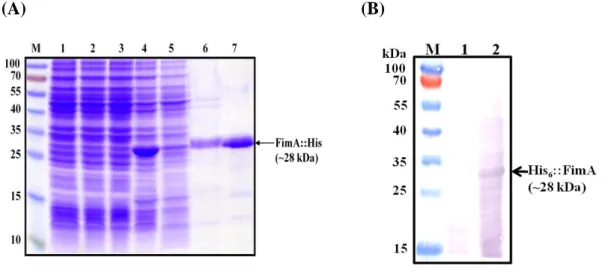
In order to obtain a good amount of the recombinant FimA protein, the recombinant plasmid pETfimA-23 (Table 2) was used to transform E. coli Novablue (DE3) and expression of the recombinant FimA was analyzed. As shown in Fig. 9A, an IPTG-induced overexpression of the His6-FimA could be observed, however, most
of the recombinant proteins were in the pellet-fraction. Urea (6N) was employed to unfold the aggregated protein. The purified His6-FimA of approximately 28 kDa (Fig.
9A) was used to immunize rabbit to raise anti-FimA antibody. The specific of anti-FimA antibody was tested by a 10000-fold diluted anti-FimA at room temperature for 1 hr. As shown in Fig. 9B, the anti-FimA antibody could specifically bind to the recombinant FimA.
Western blot analysis using the prepared FimA antiserum and the MrkA antiserum obtained from Dr. HY Chang’s lab (NTHU, College of Life Science (96)) was then performed to determine the expression of type 1 and type 3 fimbriae. As shown in Fig. 10, approximately same amount of the bacterial total proteins were applied to gel stained by coomassie brilliant blue. Compared to wild type, expression of MrkA was abolished in mrkH, mrkI, mrkA while slightly decreased in csgD.
MrkA expression could be restored in mrkI[pKAS46-mrkHIJ] strain. By contrast, a
slight increase on the expression of MrkA was observed inmrkJ strain. On the other
17
expression of FimA was observed in mrkI as well as in mrkA strains further
suggesting a reciprocal expression of the two fimbriae.
5. MrkI affected the fimS inversion
As shown in Fig. 11A, two primer pairs pcc248/ pcc249 and pcc247/ pcc249
were designed to respectively assess ON-phase or OFF-phase of fimS. Compare to wild type, the level of ON-phase of fimS represented by a 478-bp amplicon in mrkI
and mrkI[pKAS46-mrkHJ]strain increased (Fig. 11B). The mrkI deletion effect
could be complemented by introduction of pKAS46-mrkHIJ into mrkI strain,
suggesting MrkI negatively regulates the fimS promoter activity. By contrast, a slight decrease of ON-phase of fimS level was found for mrkJ and csgD strains. However,
no apparent change of ON-phase or OFF-phase of fimS (599-bp amplicon) level in mrkH was observed (Fig. 11B).
6. MrkA, FimA, FimB and FimE promoter activity analysis
As shown in Fig. 12A, the putative promoters of 551-bp, 358-bp, 400-bp and 270-bp noncoding DNA respectively located upstream of mrkA, fimA, fimB and fimE were individually fused with the promoterless lacZ gene of the reporter plasmid placZ15 (Table 2). Thus, the activity of PmrkA, PfimA, PfimB and PfimE could be determined by the activity of LacZ. The promoter reporter plasmids were then individually transformed into the parental strain lacZ (CG43S3Z01), or each of the
specific gene deletion strains lacZmrkH (CG43S3Z01mrkH), lacZmrkI
(CG43S3Z01mrkI) and lacZcsgD (CG43S3Z01csgD). The deletion of mrkH
had no apparent effect on the activity of PfimA, PfimB or PfimE (Fig. 12A left panel) which is consistent with the result of fimS analysis (Fig. 11). On the contrary, the mrkH deletion dramatically decreased PmrkA activity (Fig. 12A right panel). As shown in Fig.
18
12B, similar deletion effect on the promoter activity was observed for mrkI compared to mrkH. However, mrkJ deletion had no apparent effect on either of the promoter activity (Fig. 12C). Although no apparent change of the PfimA, PfimB or PfimE activity by the csgD deletion (Fig. 12D left panel), reduced activity of PmrkA in lacZcsgD was observed (Fig. 12D right panel).
7. The recombinant MrkJ exhibited a phosphodiesterase activity
To examine whether mrkJ encodes a functional phosphodiesterase, the MrkJ expression plasmid was transformed into E.coli MG1655 for motility analysis. As shown in Fig.13, E. coli MG1655[pRK415-MrkJ] exhibited the highest level of motile activity, while E. coli MG1655[pRK415-Ydeh] which expresses c-di-GMP cyclase activity (95), had the lowest level of motility activity. This implied that MrkJ encodes a functional phosphodiesterase activity to reduce the cellular c-di-GMP leading to increase the bacterial swimming activity.
The in vitro analysis was performed using the purified recombinant MrkJ and phosphodiesterase-specific substrate bis(pNPP). As shown in Fig. 14, an IPTG-induced overexpression of the His6-MrkJ could be observed in E. coli BL21
(DE3), however, the recombinant proteins were found mostly in pellet-fraction but some in supernatant fractions. The recombinant MrkJ was then purified from the supernatant fraction for the assay of phosphodiesterase activity. The purified His6-MrkJ of approximately 34 kDa was found to exhibit a 40-fold increase of the p-nitrophenol release compared to the reaction with BSA (Fig. 15). This further
supported that MrkJ is a phosphodiesterase playing a role in modulation of the level of the secondary messenger c-di-GMP.
19
Discussion
Type 1 and type 3 fimbriae are important factors for bacterial invasion, biofilm formation, cell motility and persistence in specific cell surface. Regulation of type 1 fimbriae is well known which is mediated by the DNA recombinase FimB and FimE to control the inversion DNA sequence of fimS (34). By contrast, the regulation of type 3 fimbriae is poorly understood. Downstream to the type 3 fimbrial gene clusters,
mrkH, mrkI, and mrkJ have recently been demonstrated to be transcribed in a
transcription unit (96). This also implies that mrkHIJ acts as a regulatory operon for the expression of type 3 fimbriae. If mrkHIJ operon is also involved in the type 1 fimbriae expression is hence investigated.
1. MrkH is a positive regulator for type 3 fimbriae
The deletion of mrkH from K. pneumoniae CG43S3 caused an increased of mannose-resistant yeast agglutination implying that MrkH controls an unknown type of sugar-mediated binding adhesion activity. On the other hand, a reduced level of biofilm formation was found for the mrkH deletion strain which suggesting a positive regulatory role on the expression of type 3 fimbriae. A decreased expression of MrkA and PmrkA activity in the mrkH strain further supports that MrkH play a positive role on the expression of type 3 fimbriae through influencing PmrkA activity. Science MrkH is a putative c-di-GMP binding protein, how the second messenger-mediated regulation carried out remains be investigated.
2. An inverse regulatory role of MrkI on the expression of type 1 and type 3 fimbriae
20
mannose-sensitive yeast agglutination, ON-phase fimS inversion, and FimA expression. This implies MrkI plays an inhibitory role for the expression of type 1 fimbriae via altering of the fimS direction. However, promoter activity measurement (Fig. 12B) revealed that none of the promoters PfimA, PfimB, or PfimE were affected by the deletion of mrkI. If MrkI indirectly affects the expression of type 1 fimbriae remains to be clarified. On the other hand, a reduced level of biofilm formation, PmrkA activity, promoter activity and MrkA pilin expression were found for the mrkI deletion strain suggesting MrkI plays as an activator at the transcription level for the expression of type 3 fimbriae. RT-qPCR analysis of the mrkI deletion effect and an electrophoresis mobility shift assay (EMSA) of MrkI binding to PmrkA performed by Dr. Ching-Ting Lin (School of Chinese Medicine, China Medical University) indicated that the recombinant MrkI was able to bind PmrkA further supporting that
MrkI reciprocally regulates the expression of the fimbriae at the transcription level.
3. MrkJ exerted a PDE activity
MrkJ has been reported in K. pneumoniae IApc35 as a functional c-di-GMP phosphodiesterase (38). Here, overexpression of MrkJ in E. coli appeared to increase the motility further supporting that MrkJ function as a PDE to decrease the cellular c-di GMP level. In the mrkJ-deletion mutant, a slightly increased of MrkA product was found whereas no obvious effect on the expression of FimA. This implied MrkJ plays a negative role in regulating the expression of type 3 fimbriae. Nevertheless, biofilm forming activity was decreased by the deletion of mrkJ indicated that MrkJ may play an indirect role to affect type 3 fimbriae activity.
4. CsgD is also a positive regulator for the expression of type 3 fimbriae
21
reduced the MrkA expression and PmrkA activity, suggesting CsgD is a positive regulator at the transcription level for the expression of type 3 fimbriae. However, no apparent effect of the csgD deletion on type 1 fimbriae was observed. Different regulatory role from the CsgD of E. coli or Salmonella is speculated because K.
pneumoniae is non-flagellated bacteria.
In summary, this study indicated that MrkH, MrkI and CsgD play as activators whereas MrkJ plays as an inhibitor for the expression of type 3 fimbriae. In addition, MrkI played a negative role for the expression of type 1 fimbriae. The deletion of
mrkA has been shown to cause an increase of type 1 fimbriae expression, however, by
an unknown mechanism (82). It is concluded that MrkI is probably the regulator determining the reciprocal expression between the two types of fimbriae.
22
References
1. Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985.
An invertible element of DNA controls phase variation of type 1 fimbriae of
Escherichia coli. Proc Natl Acad Sci U S A 82:5724-7.
2. Allen, B. L., G. F. Gerlach, and S. Clegg. 1991. Nucleotide sequence and
functions of mrk determinants necessary for expression of type 3 fimbriae in
Klebsiella pneumoniae. J Bacteriol 173:916-20.
3. Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial
c-di-GMP binding protein. Bioinformatics 22:3-6.
4. Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. SURVEY AND
SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res 28:3417-32.
5. Arnqvist, A., A. Olsen, and S. Normark. 1994. Sigma S-dependent
growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol 13:1021-32.
6. Barends, T. R., E. Hartmann, J. J. Griese, T. Beitlich, N. V. Kirienko, D. A. Ryjenkov, J. Reinstein, R. L. Shoeman, M. Gomelsky, and I. Schlichting. 2009. Structure and mechanism of a bacterial light-regulated
cyclic nucleotide phosphodiesterase. Nature 459:1015-8.
7. Benach, J., S. S. Swaminathan, R. Tamayo, S. K. Handelman, E. Folta-Stogniew, J. E. Ramos, F. Forouhar, H. Neely, J. Seetharaman, A. Camilli, and J. F. Hunt. 2007. The structural basis of cyclic diguanylate
signal transduction by PilZ domains. EMBO J 26:5153-66.
8. Blomfield, I. C. 2001. The regulation of pap and type 1 fimbriation in
Escherichia coli. Adv Microb Physiol 45:1-49.
9. Blumer, C., A. Kleefeld, D. Lehnen, M. Heintz, U. Dobrindt, G. Nagy, K. Michaelis, L. Emody, T. Polen, R. Rachel, V. F. Wendisch, and G. Unden.
2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287-98. 10. Boehm, A., M. Kaiser, H. Li, C. Spangler, C. A. Kasper, M. Ackermann, V. Kaever, V. Sourjik, V. Roth, and U. Jenal. 2010. Second
messenger-mediated adjustment of bacterial swimming velocity. Cell
141:107-16.
11. Braxton, E. E., Jr., G. D. Ehrlich, L. Hall-Stoodley, P. Stoodley, R. Veeh, C. Fux, F. Z. Hu, M. Quigley, and J. C. Post. 2005. Role of biofilms in
23
12. Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene
expression regulation by the Curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol 188:2027-37.
13. Capitani, G., O. Eidam, R. Glockshuber, and M. G. Grutter. 2006.
Structural and functional insights into the assembly of type 1 pili from
Escherichia coli. Microbes Infect 8:2284-90.
14. Chang, F. Y., and M. Y. Chou. 1995. Comparison of pyogenic liver
abscesses caused by Klebsiella pneumoniae and non-K. pneumoniae pathogens. J Formos Med Assoc 94:232-7.
15. Chang, H. Y., J. H. Lee, W. L. Deng, T. F. Fu, and H. L. Peng. 1996.
Virulence and outer membrane properties of a galU mutant of Klebsiella
pneumoniae CG43. Microb Pathog 20:255-61.
16. Chen, K. Y., P. R. Hsueh, Y. S. Liaw, P. C. Yang, and K. T. Luh. 2000. A
10-year experience with bacteriology of acute thoracic empyema: emphasis on
Klebsiella pneumoniae in patients with diabetes mellitus. Chest 117:1685-9.
17. Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine
monophosphate receptors and controls flagellar motor function in Caulobacter
crescentus. Proc Natl Acad Sci U S A 104:4112-7.
18. Colodner, R., R. Raz, B. Chazan, and W. Sakran. 2004. Susceptibility
pattern of extended-spectrum beta-lactamase producing bacteria isolated from inpatients to five antimicrobial drugs in a community hospital in Northern Israel. Int J Antimicrob Agents 24:409-10.
19. Connell, I., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli
virulence for the urinary tract. Proc Natl Acad Sci U S A 93:9827-32.
20. Cookson, A. L., W. A. Cooley, and M. J. Woodward. 2002. The role of type
1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol 292:195-205.
21. Di Martino, P., N. Cafferini, B. Joly, and A. Darfeuille-Michaud. 2003.
Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation
on abiotic surfaces. Res Microbiol 154:9-16.
22. Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency
transformation of E. coli by high voltage electroporation. Nucleic Acids Res
16:6127-45.
23. Duguid, J. P. 1959. Fimbriae and adhesive properties in Klebsiella strains. J
24
24. Flidel-Rimon, O., E. Leibovitz, A. Juster-Reicher, M. Amitay, A. Miskin, Y. Barak, and B. Mogilner. 1996. An outbreak of antibiotic multiresistant
Klebsiella at the Neonatal Intensive Care Unit, Kaplan Hospital, Rehovot,
Israel, November 1991 to April 1992. Am J Perinatol 13:99-102.
25. French, G. L., K. P. Shannon, and N. Simmons. 1996. Hospital outbreak of
Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and
beta-lactam-beta-lactamase inhibitor combinations by hyperproduction of SHV-5 beta-lactamase. J Clin Microbiol 34:358-63.
26. Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains
of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11-21.
27. Gerlach GF, C. S., and Allen BL. 1989. Identification and characterization
of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella
pneumoniae. J Bacteriol 171:1262-70.
28. Gerstel, U., and U. Romling. 2001. Oxygen tension and nutrient starvation
are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ Microbiol
3:638-48.
29. Hahn, E., P. Wild, U. Hermanns, P. Sebbel, R. Glockshuber, M. Haner, N. Taschner, P. Burkhard, U. Aebi, and S. A. Muller. 2002. Exploring the 3D
molecular architecture of Escherichia coli type 1 pili. J Mol Biol 323:845-57. 30. Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995.
Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol
18:661-70.
31. Hengge, R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev
Microbiol 7:263-73.
32. Highsmith, A. K., and W. R. Jarvis. 1985. Klebsiella pneumoniae: selected
virulence factors that contribute to pathogenicity. Infect Control 6:75-7.
33. Hirsch, E. B., and V. H. Tam. Detection and treatment options for Klebsiella
pneumoniae carbapenemases (KPCs): an emerging cause of
multidrug-resistant infection. J Antimicrob Chemother 65:1119-25.
34. Holden, N., I. C. Blomfield, B. E. Uhlin, M. Totsika, D. H. Kulasekara, and D. L. Gally. 2007. Comparative analysis of FimB and FimE recombinase
activity. Microbiology 153:4138-49.
35. Hornick, D. B., B. L. Allen, M. A. Horn, and S. Clegg. 1992. Adherence to
respiratory epithelia by recombinant Escherichia coli expressing Klebsiella
25
36. Huang, Y. J., H. W. Liao, C. C. Wu, and H. L. Peng. 2009. MrkF is a
component of type 3 fimbriae in Klebsiella pneumoniae. Res Microbiol
160:71-9.
37. Hultgren, S. J., S. Normark, and S. N. Abraham. 1991. Chaperone-assisted
assembly and molecular architecture of adhesive pili. Annu Rev Microbiol
45:383-415.
38. Johnson, J. G., and S. Clegg. 2010. Role of MrkJ, a phosphodiesterase, in
type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J Bacteriol 192:3944-50.
39. Johnson, J. G., and S. Clegg. Role of MrkJ, a phosphodiesterase, in type 3
fimbrial expression and biofilm formation in Klebsiella pneumoniae. J Bacteriol 192:3944-50.
40. Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is
assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci U S A 92:2081-5.
41. Keynan, Y., and E. Rubinstein. 2007. The changing face of Klebsiella
pneumoniae infections in the community. Int J Antimicrob Agents 30:385-9.
42. Klemm, P., and K. A. Krogfelt. 1994. Type 1 fimbriae of Escherichia coli,
pp.9–26. In Fimbriae, Adhesion, Genetics, Biogenesis and Vaccines. Klemm, P. (ed.). Boca Raton, FL: CRC Press.
43. Klemm, P., and M. A. Schembri. 2000. Fimbrial surface display systems in
bacteria: from vaccines to random libraries. Microbiology 146 Pt 12:3025-32. 44. Kumarasamy, K. K., M. A. Toleman, T. R. Walsh, J. Bagaria, F. Butt, R.
Balakrishnan, U. Chaudhary, M. Doumith, C. G. Giske, S. Irfan, P. Krishnan, A. V. Kumar, S. Maharjan, S. Mushtaq, T. Noorie, D. L. Paterson, A. Pearson, C. Perry, R. Pike, B. Rao, U. Ray, J. B. Sarma, M. Sharma, E. Sheridan, M. A. Thirunarayan, J. Turton, S. Upadhyay, M. Warner, W. Welfare, D. M. Livermore, and N. Woodford. 2010.
Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis
10:597-602.
45. Lai, Y. C., H. L. Peng, and H. Y. Chang. 2001. Identification of genes
induced in vivo during Klebsiella pneumoniae CG43 infection. Infect Immun
69:7140-5.
46. Lai, Y. C., H. L. Peng, and H. Y. Chang. 2003. RmpA2, an activator of
capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 185:788-800.
26
47. Lee, C. C., C. Y. Chen, F. H. Chen, R. A. Zimmerman, and H. S. Hsiao.
1998. Septic metastatic endophthalmitis from Klebsiella pneumoniae liver abscess: CT and MR imaging characteristics--report of three cases. Radiology
207:411-6.
48. Lin, C. T., T. Y. Huang, W. C. Liang, and H. L. Peng. 2006. Homologous
response regulators KvgA, KvhA and KvhR regulate the synthesis of capsular polysaccharide in Klebsiella pneumoniae CG43 in a coordinated manner. J Biochem 140:429-38.
49. Lin, C. T., Y. J. Huang, P. H. Chu, J. L. Hsu, C. H. Huang, and H. L. Peng. 2006. Identification of an HptB-mediated multi-step phosphorelay in
Pseudomonas aeruginosa PAO1. Res Microbiol 157:169-75.
50. Lin, J. C., F. Y. Chang, C. P. Fung, J. Z. Xu, H. P. Cheng, J. J. Wang, L. Y. Huang, and L. K. Siu. 2004. High prevalence of phagocytic-resistant
capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect
6:1191-8.
51. Medeiros, A. A. 1993. Nosocomial outbreaks of multiresistant bacteria:
extended-spectrum beta-lactamases have arrived in North America. Ann Intern Med 119:428-30.
52. Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The
second messenger bis-(3'-5')-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas
aeruginosa. Mol Microbiol 65:876-95.
53. Meyer, K. S., C. Urban, J. A. Eagan, B. J. Berger, and J. J. Rahal. 1993.
Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Ann Intern Med 119:353-8.
54. Miller, J. H. 1972. Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
55. Miller, J. H. 1972. Experiments in molecular genetics. . New York: Cold Spring Harbor. .
56. Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella
pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228-36.
57. O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in
Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449-61.
58. Olsen, A., A. Arnqvist, M. Hammar, and S. Normark. 1993.
Environmental regulation of curli production in Escherichia coli. Infect Agents Dis 2:272-4.
27
Monaghan, G. R. Nimmo, D. F. Looke, A. G. McEwan, and M. A. Schembri. 2008. Identification of type 3 fimbriae in uropathogenic
Escherichia coli reveals a role in biofilm formation. J Bacteriol 190:1054-63.
60. Podschun, R., A. Fischer, and U. Ullmann. 1992. Siderophore production of
Klebsiella species isolated from different sources. Zentralbl Bakteriol
276:481-6.
61. Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain
proteins bind cyclic diguanylate and regulate diverse processes in Vibrio
cholerae. J Biol Chem 282:12860-70.
62. Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial
adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol 183:7213-23.
63. Ramelot, T. A., A. Yee, J. R. Cort, A. Semesi, C. H. Arrowsmith, and M. A. Kennedy. 2007. NMR structure and binding studies confirm that PA4608
from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66:266-71.
64. Rao, F., Y. Yang, Y. Qi, and Z. X. Liang. 2008. Catalytic mechanism of
cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol
190:3622-31.
65. Romling, U. 2005. Characterization of the rdar morphotype, a multicellular
behaviour in Enterobacteriaceae. Cell Mol Life Sci 62:1234-46.
66. Romling, U. 2005. Characterization of the rdar morphotype, a multicellular
behaviour in enterobacteriaceae. Cmls-Cellular and Molecular Life Sciences
62:1234-1246.
67. Romling, U. 2009. Rationalizing the evolution of EAL domain-based cyclic
di-GMP-specific phosphodiesterases. J Bacteriol 191:4697-700.
68. Romling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998.
Curli fibers are highly conserved between Salmonella typhimurium and
Escherichia coli with respect to operon structure and regulation. J Bacteriol
180:722-31.
69. Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998.
Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28:249-64. 70. Rosen, D. A., J. S. Pinkner, J. M. Jones, J. N. Walker, S. Clegg, and S. J.
Hultgren. 2008. Utilization of an intracellular bacterial community pathway
28
type 1 pilus expression. Infect Immun 76:3337-45.
71. Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ
domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310-4. 72. Schembri, M. A., J. Blom, K. A. Krogfelt, and P. Klemm. 2005. Capsule
and fimbria interaction in Klebsiella pneumoniae. Infect Immun 73:4626-33. 73. Schirmer, T., and U. Jenal. 2009. Structural and mechanistic determinants of
c-di-GMP signalling. Nat Rev Microbiol 7:724-35.
74. Schroll, C., K. B. Barken, K. A. Krogfelt, and C. Struve. 2010. Role of
type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol 10:179.
75. Sebghati, T. A., T. K. Korhonen, D. B. Hornick, and S. Clegg. 1998.
Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun 66:2887-94.
76. Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF
and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123-34.
77. Sirot, D. 1995. Extended-spectrum plasmid-mediated beta-lactamases. J
Antimicrob Chemother 36 Suppl A:19-34.
78. Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic
exchange. Gene 169:47-52.
79. Snyder, J. A., B. J. Haugen, C. V. Lockatell, N. Maroncle, E. C. Hagan, D. E. Johnson, R. A. Welch, and H. L. Mobley. 2005. Coordinate expression of
fimbriae in uropathogenic Escherichia coli. Infect Immun 73:7588-96.
80. Struve, C., M. Bojer, and K. A. Krogfelt. 2008. Characterization of
Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during
colonization and infection and impact on virulence. Infect Immun 76:4055-65. 81. Struve, C., M. Bojer, and K. A. Krogfelt. 2009. Identification of a conserved
chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun
77:5016-24.
82. Struve, C., C. Schroll, K. B. Barken, and K. A. Krogfelt. 2010. Role of
type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. Bmc Microbiology 10.
83. Svanborg Eden, C., and P. de Man. 1987. Bacterial virulence in urinary tract
infection. Infect Dis Clin North Am 1:731-50.
84. Switalski, L. M., M. Hook, and E. H. Beachey. 1989. Molecular mechanisms of microbial adhesion. . New York: Springer-Verlag.