1535-9778/05/$08.00⫹0 doi:10.1128/EC.4.1.147–155.2005
Copyright © 2005, American Society for Microbiology. All Rights Reserved.
A Homolog of Ste6, the a-Factor Transporter in Saccharomyces
cerevisiae, Is Required for Mating but Not for Monokaryotic
Fruiting in Cryptococcus neoformans
Yen-Ping Hsueh† and Wei-Chiang Shen*
Department of Plant Pathology and Microbiology, National Taiwan University, Taipei, Taiwan Received 16 May 2004/Accepted 13 October 2004
Fungal pheromones function during the initial recognition stage of the mating process. One type of peptide pheromone identified in ascomycetes and basidiomycetes terminates in a conserved CAAX motif and requires extensive posttranslational modifications to become mature and active. A well-studied representative is the a-factor of Saccharomyces cerevisiae. Unlike the typical secretory pathway utilized by most peptides, an alter-native mechanism involving the ATP-binding cassette transporter Ste6 is used for the export of mature a-factor. Cryptococcus neoformans, a bipolar human pathogenic basidiomycete, produces CAAX motif-contain-ing lipopeptide pheromones in both MATa and MAT␣ cells. Virulence studies with a congenic pair of C.
neoformans serotype D strains have shown that MAT␣ cells are more virulent than MATa cells.
Characteriza-tion of the MAT␣ pheromones indicated that an autocrine signaling loop may contribute to the differentiation and virulence of MAT␣ cells. To further address the role of pheromones in the signaling loop, we identified a
STE6 homolog in the C. neoformans genome and determined its function by gene disruption. The ste6 mutants
in either mating-type background showed partially impaired mating functions, and mating was completely abolished in a bilateral mutant cross. Surprisingly, the MAT␣ ste6 mutant does not exhibit a defect in monokaryotic fruiting, suggesting that the activation of the autocrine signaling loop by the pheromone is via a Ste6-independent mechanism. MF␣ pheromone itself is essential for this process and could induce the signaling response intracellularly in MAT␣ cells. Our data demonstrate that Ste6 is evolutionarily conserved for mating and is not required for monokaryotic fruiting in C. neoformans.
Cryptococcus neoformans is a human fungal pathogen which primarily infects individuals with compromised immune func-tions. Unlike most of the frequently encountered human fun-gal pathogens, which are ascomycetes, C. neoformans is a ba-sidiomycete. As a consequence of the increasing prevalence of immunosuppression caused by AIDS, chemotherapy, and high-dose steroid treatment, C. neoformans has emerged as the leading cause of fungal meningitis in the past 2 decades (3).
The bipolar sexual cycle of C. neoformans was first identified in 1975 (21). The mating process is initiated by the fusion of haploid yeast cells of opposite mating types (a and␣) and leads to the production of heterokaryotic hyphae with fused clamp connections. A specialized sporulation structure called the ba-sidium forms at the tip of the hypha, where karyogamy and meiosis occur to produce sessile basidiospores terminally in basipetal chains by repetitious budding. An analysis of single basidiospore isolates revealed a 1:1 segregation of the two mating types, which indicated that a bipolar mating system existed (20). Upon germination, the basidiospores form yeast cells to regenerate the haploid yeast phase.
An alternative route for the vegetative cells to produce fruit-ing-body-like structures with spores is called monokaryotic, or haploid, fruiting. Upon desiccation and nitrogen starvation,
MAT␣ haploid yeast cells can differentiate, in the absence of a mating partner, into monokaryotic filaments with unfused clamp connections, producing four chains of basidiospores on the basidia. This haploid hyphal phase was initially reported to be associated exclusively with the␣ mating type (38), and it has been thought to be one of the factors contributing to the predominance of MAT␣ cells over MATa cells in the environ-ment. A recent study, however, reported the discovery of hap-loid fruiting in some MATa strains (36); as a consequence, whether haploid fruiting accounts for the␣ mating type pre-dominance is still unclear.
Mating specificity in fungi is controlled by the mating-type (MAT) locus. The MAT locus was first characterized, and has been extensively studied, in the budding yeast Saccharomyces cerevisiae (16). In this region, homologous chromosomes con-tain nonhomologous sequences. The term idiomorph has been introduced to specify this variation and to distinguish the MAT loci from classical alleles. Information encoded by the MAT loci determines sexuality. Studies on the well-characterized mating systems of ascomycetous and basidiomycetous fungi present a conserved mechanism utilized by fungal cells (18). S. cerevisiae, which is an ascomycete, harbors a single MAT locus which differs between a and␣ cells. MATa encodes the tran-scriptional regulator a1, and MAT␣ encodes the transcriptional regulators␣1 and ␣2. Basidiomycetous fungi typically have a tetrapolar mating system with two unlinked MAT loci in the genome; one locus encodes homeodomain proteins, and the other encodes the pheromones and pheromone receptors. Un-like most of the basidiomycetes, C. neoformans does not em-ploy a tetrapolar mating system. Instead, it has a bipolar mat-* Corresponding author. Mailing address: Department of Plant
Pa-thology and Microbiology, Room 216, Building Number 1, National Taiwan University, No. 1 Roosevelt Rd., Sec. 4, 106 Taipei, Taiwan. Phone: 886-2-23630231-2733. Fax: 886-2-23636490. E-mail: wcshen @ntu.edu.tw.
† Present address: Department of Molecular Genetics and Microbi-ology, Duke University Medical Center, Durham, NC 27710.
147
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
ing system. The C. neoformans MAT locus is larger than any other known fungal MAT loci (⬎100 kb) and contains multiple genes which have never been previously observed in a mating-type locus. The unusual size and structure of the C. neoformans MAT locus may therefore indicate that it is an evolutionary intermediate between the MAT loci of fungi and the sex chro-mosomes of higher multicellular eukaryotic organisms, such as humans (14, 23).
An intriguing correlation between virulence and the MAT␣ locus has been noticed. Over 95% of all C. neoformans isolates are MAT␣ (3, 22). Congenic serotype D ␣ cells have been shown to be more virulent than a cells in a murine model of systemic cryptococcosis (22). Additionally, previous studies showed that a MAT␣ pheromone triple-deletion mutant was greatly impaired for monokaryotic fruiting, and overexpression of the ␣ pheromone enhanced this process. Pheromoneless MAT␣ mutants were modestly attenuated for virulence. All the data suggested that an autocrine signaling loop may function and contribute to the differentiation and virulence of MAT␣ cells (33). To further address how this signaling loop is acti-vated by the pheromone, we have identified a homolog of the S. cerevisiae STE6 gene in the C. neoformans genome and determined its function.
It has been shown that fungal pheromones function in the initial recognition stage of the mating process (2, 28). Fungal pheromones can be divided into two broad categories based on their hydrophobicities. First, pheromones can be hydrophilic, like␣-factor in S. cerevisiae. Second, pheromones can be hy-drophobic, like a-factor in S. cerevisiae. The mating pheromone a-factor is a specific, diffusible signaling molecule expressed only by a cells and is similar to peptide hormones secreted by higher eukaryotes. It is initially synthesized as a larger precur-sor that undergoes posttranslational modification. Pheromone maturation occurs by sequential events involving a carboxyl-terminal CAAX modification (in which C is cysteine, A is an aliphatic amino acid, and X is cysteine, serine, methionine, glutamine, or alanine) and amino-terminal processing. The mature pheromone is finally exported from the cell via an alternative (nonclassical) mechanism that involves the ATP-binding cassette (ABC) transporter Ste6 (5, 27). Many phero-mone precursors terminating in a conserved CAAX motif have been identified in ascomycetes and basidiomycetes. Unlike those in S. cerevisiae, the MF␣ and MFa pheromones in C. neoformans are both CAAX motif-containing lipopeptide pheromones (25, 29, 33). Characterization of the MF␣ phero-mone showed that only the mature form is capable of trigger-ing morphological responses in MATa cells (9), and the mattrigger-ing efficiency of the pheromoneless MAT␣ mutant is dramatically decreased to 1% of that of wild-type cells (33).
STE6 encodes the a-factor transporter, which is essential for mating in S. cerevisiae (19, 26). Ste6 is a member of the ABC transporter superfamily composed of two homologous halves, each with six membrane-spanning segments and an ATP nu-cleotide binding domain (NBD). The only observable pheno-type of the ste6 null mutant in S. cerevisiae is the inability to export a-factor and, consequently, to mate; thus, it appears that Ste6 has a defined role in S. cerevisiae as a transporter dedicated to a-factor (11). In addition, biochemical evidence has shown that Ste6 couples ATP hydrolysis with pheromone
export, so Ste6 is one of a very few ABC transporters in which the presumed ATPase activity has been proven (17).
In this study, we report the identification of a C. neoformans STE6 homolog that is not linked to the mating-type locus. Mutants lacking STE6 exhibit a bilateral mating defect, al-though discrepancies are noticed in different mating-type back-grounds. Monokaryotic fruiting in the MAT␣ ste6 mutants is surprisingly unaffected. Our results indicate that STE6 func-tions in both mating types and is required for mating but not for haploid fruiting in C. neoformans.
MATERIALS AND METHODS
Strains and media.The C. neoformans strains used in this study are listed in Table 1. Congenic serotype D strains JEC20 (MATa) and JEC21 (MAT␣) and their auxotrophic derivatives were used throughout the study (10, 29). All strains were handled by use of standard techniques and media as previously described (1, 12). Yeast extract-peptone-dextrose (YPD), yeast nitrogen base (YNB), V8 mating, and synthetic low ammonia dextrose (SLAD) media and filament agar were prepared as previously described (13, 38).
Identification of a STE6 homolog in C. neoformans.C. neoformans primers
WC6 (5⬘-GTCAGGAGAGATTACTATGGA-3⬘) and WC7 (5⬘-CTTCACCTC CTCTCTTGCA-3⬘) were designed based on the regions homologous to the S.
cerevisiae STE6 gene and were used for PCR amplification of a partial sequence
of the C. neoformans STE6 gene. This 1.8-kb PCR product was then used as a probe for subsequent identification of a C. neoformans genomic clone from a JEC21 bacterial artificial chromosome library (Research Genetics). A 7.9-kb ClaI fragment from this clone was subcloned into pBluescript SK(⫹) (Strat-agene) to generate plasmid pYPE3. Reverse transcription-PCR and sequencing with primers WC156 (5⬘-GCCCACGTCCGACGGCTCGCCTTTCCA-3⬘), WC185 (5⬘-CGTCGGAAACGATCTGTGAAAGGTCGT-3⬘), WC179 (5⬘-CG TGGTGAGCTTGCCCGACGGTTAT-3⬘), and WC180 (5⬘-GTACAACATTA ACGCAAGAGCAACC-3⬘) allowed the coding regions to be recognized.
Disruption and reintroduction of the STE6 gene.The C. neoformans STE6 gene was disrupted by replacing a 3.7-kb region within the open reading frame with a 1.9-kb fragment containing the URA5 gene. Primer pair WC46 (5⬘-GGA CGGAGAATTCTGTGATCTTGATCTTGAGGCCTGA-3⬘) and WC47 (5⬘-C ATCATCGGGATCCCTGCGGTGGTACGG TCA-3⬘) and primer pair WC48 (5⬘-CGCATTGGATCCCGACAACGCTAGGGCTGTA-3⬘) and WC49 (5⬘-GT GGATGATCTAGATGTTGATATCGGTGCGA-3⬘) were used to amplify the 5⬘- and 3⬘-end homologous flanking regions of the STE6 open reading frame, respectively. The URA5 selectable marker (10) was released from pRCD69 (8) and cloned into the BamHI site to generate a ste6::URA5 deletion construct containing the left and right portions of STE6. The resulting ste6::URA5 deletion construct, pYPE1, was introduced into the MAT␣ ura5 strain JEC43 by biolistic transformation as described previously (35). Uracil prototrophic transformants were picked and screened by PCR and Southern analyses. To isolate MATa ste6 strains, the MAT␣ ste6 strain YPC4 was crossed with JEC34 (MATa ura5), the progeny were isolated on synthetic dextrose medium lacking uracil, and MATa
TABLE 1. Strains used in this study
Strain Description Reference or source
JEC20 MATa 21
JEC21 MAT␣ 21
JEC34 MATa ura5 28
JEC43 MAT␣ ura5 28
JEC170 MAT␣ ade2 lys2 J. Edman
JEC171 MATa ade2 lys2 J. Edman
WSC18 MAT␣mf␣1::ADE2 mf␣2,3::URA5 ade2 ura5 32
YPC4 MAT␣ ste6::URA5 This study
YPC7 MATa ste6::URA5 This study
YPC13 MAT␣ ura5 ste6 (5-FOAr) This study
YPC15 MATa ura5 ste6 (5-FOAr) This study
YPC17 MAT␣ ura5 ste6 STE6-URA5 This study YPC18 MATa ura5 ste6 STE6-URA5 This study
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
strains containing the disruption construct were identified by mating and PCR and Southern analyses. The ura5 versions of ste6 mutant strains used for recon-stitution were generated by selecting the original mutants on 5-fluoroorotic acid (5-FOA) medium, which is toxic to URA5 cells. The STE6 reconstitution clone pYPE4 was constructed by introducing the 1.9-kb URA5 gene fragment into pYPE3 by blunt-end ligation and transformed into ura5 ste6 mutant strains.
Southern blot analysis.Genomic DNA used for Southern analysis was pre-pared by a large-scale genomic DNA isolation method described previously (30). DNA was digested with ClaI and electrophoresed in a 0.8% 1⫻ Tris-acetate-EDTA agarose gel. A 1.5-kb probe of the STE6 flanking region was generated by PCR amplification with primers WC48 (5⬘-CGCATTGGATCCCGACAACGC TAGGGCTGTA-3⬘) and WC49 (5⬘-GTGGATGATCTAGATGTTGATATCG GTGCGA-3⬘) and labeled by use of a Prime-It II random primer labeling kit (Stratagene) with [␣-32
P]dCTP (NEN Life Science Products). Blotting and au-toradiography were carried out by standard procedures.
Northern blot analysis.RNA used in the Northern analysis was isolated from yeast cells by using TRIzol total RNA isolation reagent according to the man-ufacturer’s instructions (Invitrogen). Twenty micrograms of total RNA from each sample was separated by electrophoresis in a 1.2% agarose–formaldehyde gel. RNA was transferred by capillary action to a nylon membrane (Immobilon-Ny⫹; Millipore) and was hybridized in hybridization buffer (0.12 M Na2HPO4 [pH 7.2], 0.25 M NaCl, 1 mM EDTA [pH 8], 7% sodium dodecyl sulfate, 50% formamide). A 0.5-kb STE6 probe was amplified with WC9 (5⬘-TCTGGTCAT TCTTCCTTTCCAA-3⬘) and WC7 (5⬘-TCTTCCACCTCCTCTCTTGCA-3⬘). Probes used for the detection of MFa1 and MF␣1 were amplified with primer pair WC121 (5⬘-CGCGGATCCAATGGACGCCTTCACTGCTATCT-3⬘) and WC122 (5⬘-CGGGGTACCCGACTAGATATATTATGCATTCT-3⬘) and primer pair WC83 (5⬘-CTCGAGGCTTTCCCCCTTTTTCT-3⬘) and WC84 (5⬘-ATTTG AAAAAGAGATCACAGTG-3⬘), respectively. All probes were labeled as de-scribed for Southern blot analysis.
Mating, haploid fruiting, and confrontation assays.Strains for mating, hap-loid fruiting, and confrontation assays were first grown on YPD at 30°C for 2 days. Mating reactions were performed by coincubating the cells with desired partners on V8 or SLAD medium at 26°C in the dark for 1 to 5 days. The mating tester strains used were JEC20 (MATa) and JEC21 (MAT␣). Filamentation was evaluated by observing the periphery of the mating reaction under a microscope. Pictures were taken 1 day postincubation. For haploid fruiting assays, cells were resuspended in sterile water, spotted onto filament agar or SLAD medium, and incubated at 26°C for up to 4 weeks in the dark. For confrontation assays, cells were streaked in parallel lines onto filament agar roughly 3 to 4 mm apart. Plates were incubated at 26°C in the dark and observed after 2 days. Pictures were taken 4 days postincubation.
Fusion assays.To test the fusion efficiencies, equal amounts (107cells) of the wild-type and ste6 strains carrying ura5 mutations (JEC43, JEC34, YPC13, and YPC15) were mixed with strains containing complementing auxotrophic markers
ade2 and lys2 (JEC170 and JEC171). The cell suspensions were spotted onto V8
medium and incubated for 24 or 48 h. The portions of media showing mating reactions were then cut out and resuspended in 2 ml of water. Two microliters of the suspension was plated onto minimal YNB medium to select for prototrophic fusion products. Colonies on each YNB plate were counted after 3 days.
Nucleotide sequence accession number.The sequence for STE6 has been assigned GenBank accession number AY587551.
RESULTS
Identification and characterization of the C. neoformans
STE6 homolog.Based on the typical CAAX motif in the car-boxyl terminus of C. neoformans MF␣ and MFa, we hypothe-sized that a homolog of the S. cerevisiae Ste6 pheromone trans-porter may exist and be responsible for pheromone secretion in C. neoformans. Therefore, we used a reverse genetics ap-proach to identify a STE6 homolog candidate in the C. neo-formans genome. BLAST searches with S. cerevisiae Ste6 were performed with the Stanford C. neoformans genome database (see http://www-sequence.stanford.edu/group/C.neoformans /index.html). A partial sequence which was found to have sim-ilarity with S. cerevisiae Ste6 (29% identity and 52% simsim-ilarity) has since been designated CNBA7570 in the database and has been submitted to GenBank under accession number
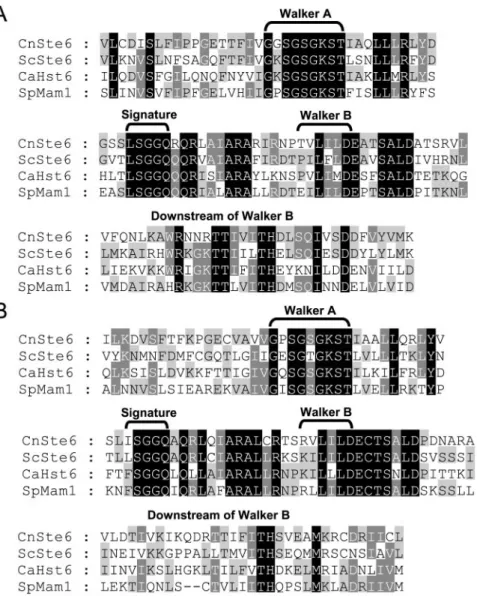
AY587551. BLAST searches of the putative C. neoformans STE6 homolog in GenBank revealed several members of the ABC transporter superfamily in various organisms. Genomic sequence analysis revealed a predicted coding region of 5.3 kb, and the gene has the expected modular architecture with two homologous halves. Each half contains one membrane-spanning domain (MSD) and one NBD that are distinct among the ABC transporter superfamily. The four core domains are in a single polypeptide with a forward order (MSD1-NBD1-MSD2-NBD2). Analysis of the cDNA sequence presented six introns within the coding sequence, and the predicted number of amino acid residues is 1,656. PCR and Southern blot anal-yses revealed that this gene is in strains with either mating type (see Fig. 2). The NBDs are the most conserved regions in ABC transporters, and several conserved motifs are located within this region. The results of the amino acid sequence alignment of Ste6 fungal homologs for NBD1 and NBD2 are shown in Fig. 1. Protein alignment showed that C. neoformans Ste6 shares 45% similarity with Candida albicans Hst6 (31) and 50% similarity with Schizosaccharomyces pombe Mam1 (6).
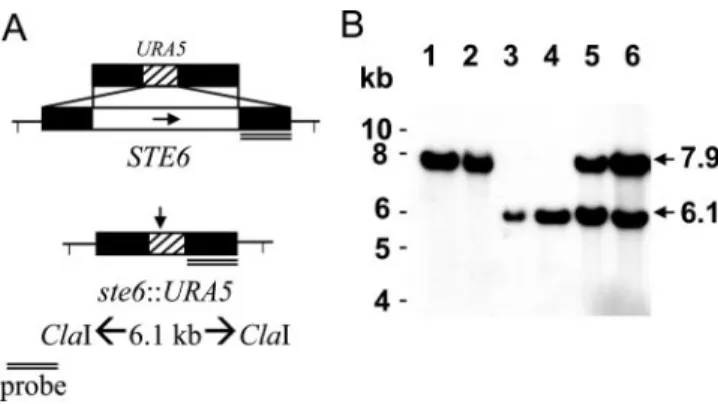
Disruption of the C. neoformans STE6 gene.To determine the function of the putative STE6 homolog, the C. neoformans STE6 gene was disrupted by homologous recombination. The ste6::URA5 disruption allele (Fig. 2A) was introduced by bi-olistic transformation into the ura5 strain JEC43 (MAT␣). Transformants were selected on synthetic medium lacking ura-cil and containing 1 M sorbitol and were then screened by the presumptive, impaired mating phenotype on V8 medium in crosses with the MATa tester strain JEC20. Four isolates with apparent reductions of filamentation were obtained among the 64 transformants selected. Genomic DNA from the four pu-tative deletion strains was extracted, and a PCR analysis was conducted to confirm that the isolates impaired in mating all lacked wild-type STE6 and contained only the ste6::URA5 dis-rupted allele. Southern blot analysis with a flanking 1.5-kb fragment of the ste6::URA5 construct confirmed the gene re-placement by the reduction of the hybridization signal from a 7.9- to a 6.1-kb fragment among all four isolates upon ClaI digestion (Fig. 2B and data not shown). To generate a MATa
ste6 mutant, the MAT␣ ste6 strain was crossed with the ura5
strain JEC34 (MATa) on V8 medium and incubated for more than 1 month until substantial filaments and basidiospores had formed. Progeny were isolated by sectioning the agar block with filaments into sterile water, and the suspension was spread onto synthetic medium lacking uracil. Single colonies were picked and screened for mating type to identify MATa strains and analyzed by PCR to identify the ste6::URA5 disrupted allele. Approximately half of the progeny were MATa strains with the ste6::URA5 allele. Two strains were confirmed by Southern blot analysis to be ste6::URA5 disruption strains and were selected for subsequent analysis. The MAT␣ ste6 and MATa ste6 strains with the reconstitution of the wild-type frag-ment of the STE6 gene were also confirmed by Southern blot analysis (Fig. 2B).
ste6 mutant strains are impaired in mating.For C.
neofor-mans, mating occurs when MAT␣ and MATa strains are
cocul-tured on V8 or SLAD medium and is characterized morpho-logically by cell fusion, filamentation, basidium formation, nuclear fusion, meiosis, and sporulation. To determine the role of the C. neoformans STE6 gene in mating, the ste6 mutant
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
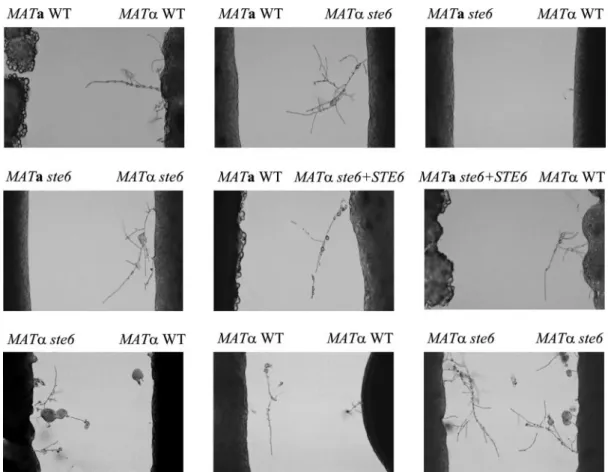
strains were crossed with the wild-type tester strains JEC20 (MATa) and JEC21 (MAT␣) on V8 or SLAD medium (Fig. 3 and data not shown). As predicted, on the basis of the sterile phenotype of the S. cerevisiae MATa ste6 deletion mutant, the MAT␣ ste6 and MATa ste6 mutant strains displayed a dramatic reduction in filament formation when crossed with a tester strain of the opposite mating type. Interestingly, different de-grees of filamentation were observed in the mutants with dif-ferent mating-type backgrounds. Filamentation in MAT␣ ste6 mutants was better than that in MATa ste6 mutants. Reconsti-tution with the wild-type STE6 gene in the mutant strains restored the mating efficiency to the wild-type level (Fig. 3). The mating phenotype of the C. neoformans ste6 mutants is not identical to that of the S. cerevisiae MATa ste6 mutant, which is sterile. Instead, it is similar to that of the C. neoformans mf␣1 mf␣2 mf␣3 triple deletion mutant, in which mating filaments, basidia, and basidiospores were still produced (33). In the
bilateral crosses (MAT␣ ste6 strain ⫻ MATa ste6 strain), mating behavior was completely abolished (Fig. 3). These findings show that the STE6 gene plays an important role in mating for both a and␣ cells. It functions bilaterally and is required but not essential for mating in C. neoformans. Furthermore, since we think that ste6 mutants might be impaired in the courtship stage of the mating process, fusion assays were performed to test this hypothesis. Compared to that of the wild-type cells, the fusion efficiency of the MATa
ste6 or MAT␣ ste6 mutant was reduced to less than 1% after
24 h of incubation. The MATa ste6 mutant exhibited de-creased fusion efficiency, to about 1.5%, after a 48-h period, and the MAT␣ ste6 mutant retained about 40.7% of the fusion efficiency (data not shown). This result is consistent with the filamentation phenotypes observed with V8 mating medium and suggests that ste6 mutants are partially im-paired in the fusion step.
FIG. 1. Amino acid sequence alignment of NBD1 (A) and NBD2 (B) of Ste6 fungal homologs. Amino acid sequences from C. neoformans (Cn) Ste6 (accession number AY587551), S. cerevisiae (Sc) Ste6 (accession number NP_012713), C. albicans (Ca) Hst6p (accession number P53706), and S. pombe (Sp) Mam1 (accession number P78966) are compared. Walker A, Signature, and Walker B are conserved motifs in the NBDs of ABC transporters. Amino acids identical among all four proteins are shaded black, and amino acids identical among two or three proteins are shaded light grey or dark grey, respectively.
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
ste6 mutant strains fail to secrete pheromone molecules in
confrontation assays.Confrontation assays have been estab-lished to examine the capabilities of the cells to secrete and sense pheromone (33, 37). MAT␣ and MATa cells cultured in close proximity but without contact on filament agar induce morphological changes in response to the opposite mating type. Filamentation at the edges of MAT␣ cells and the pres-ence of swollen MATa cells are typically observed. Cells re-spond to pheromones secreted from cells with the opposite mating type by undergoing these morphological changes. Therefore, this assay not only determines the ability to respond to pheromones but also analyzes the pheromone secretion of the cells. As shown in Fig. 4, filamentation and swollen cell production of the MAT␣ and MATa cells were observed in the confronting wild-type pairs. When a MAT␣ ste6 strain was
confronted with the MATa type strain, the MATa wild-type cells did not respond to the opposite mating-wild-type cells, suggesting that the MAT␣ ste6 cells failed to secrete MF␣ pheromone. Similar results were observed when the MATa ste6 strain was confronted with the MAT␣ wild-type strain. On the other hand, both MAT␣ STE6 and MATa STE6 reconstitution strains can fully restore the ability to trigger morphogenesis in response to cells of the opposite mating type. These results indicated that the ste6 mutants are unable to secrete phero-mones to induce morphological changes in opposite-mating-type cells.
STE6 is dispensable in haploid fruiting.The MF␣ phero-mone has been shown to regulate haploid fruiting of MAT␣ cells, and the pheromone mf␣1 mf␣2 mf␣3 triple deletion mutant was found to have a significant defect in haploid fruit-ing when grown on a nitrogen-limitfruit-ing medium. Overexpres-sion of the MF␣1 pheromone gene enhanced haploid fruiting in the wild-type cells (33). To our surprise, the MAT␣ ste6 mutant was fully capable of undergoing haploid filamentation (Fig. 5). Under nitrogen limitation and desiccation conditions, the MAT␣ ste6 mutant produced monokaryotic filaments and blastospores to at least the same degree as, if not more pro-lifically than, the wild-type MAT␣ cells. Similar observations were also obtained with the confrontation assay, in which the MAT␣ ste6 mutant cells produced filaments to an extent similar to that of the wild-type MAT␣ cells while confronting the MAT␣ or MATa wild-type or MAT␣ ste6 or MATa ste6 mutant cells (Fig. 5). These results indicated that STE6 is not required for haploid filamentation in the MAT␣ C. neoformans cells and suggested that the autocrine signaling loop may be triggered intracellularly.
STE6 expression is induced during the mating process.To examine the expression pattern of STE6, 6⫻ 107cells of the
MATa JEC20 or MAT␣ JEC21 overnight YPD culture/ml were
inoculated onto solid V8 medium or mixed in a 1:1 ratio, respectively. Cells were harvested at 2, 6, 12, 24, and 48 h FIG. 2. Construction of the ste6::URA5 allele and Southern
hybrid-ization analysis of wild-type, ste6, and STE6 reconstitution strains. (A) The ste6 deletion allele was created by replacing STE6 with the
URA5 selectable marker. (B) The genomic DNA of each strain was
digested with ClaI, electrophoresed, blotted, and hybridized with the
32P-labeled STE6 fragment indicated. Lane 1, MAT␣ wild type; lane 2, MATa wild type; lane 3, MAT␣ ste6 mutant; lane 4, MATa ste6 mutant;
lane 5, MAT␣ ste6 mutant plus STE6; lane 6, MATa ste6 mutant plus
STE6.
FIG. 3. STE6 is required but not essential for mating in both mating types of C. neoformans. Wild-type (WT) and ste6 mutant strains were coincubated with mating partners on SLAD plates in the dark for 30 h at 26°C. The edges of the mating mixtures were photographed at a magnification of⫻100.
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
postincubation. RNA was extracted, and transcription of STE6, MF␣, and MFa was examined by Northern blot analysis. Hybridization results revealed that STE6 was expressed at a basal level in response to nutrient limitation in both MATa and MAT␣ cells (Fig. 6 and data not shown). Previous studies showed that a coculture of cells of opposite mating types dra-matically induces the transcription of MF␣ and MFa genes (9, 33); therefore, we further examined the transcription of STE6 during mating. As shown in Fig. 6, a coculture of cells of opposite mating types significantly induced STE6 transcription at 6 h postincubation, and the expression of STE6 returned to the basal level at later time points. Hybridizations with probes for the MF␣ and MFa transcripts similarly demonstrated that
the highest expression level of the pheromone also occurred at 6 h postinoculation (Fig. 6). Thus, the hybridization results suggest that the transcription of pheromones and pheromone transporter genes is highly coordinated and that the expression of STE6 might also be under the control of the pheromone response pathway in C. neoformans.
DISCUSSION
STE6 functions bilaterally in C. neoformans. By disrupting the gene and analyzing the phenotypes of mutants in different mating-type backgrounds, we found that Ste6 is responsible for pheromone secretion in both MAT␣ and MATa cells and is the FIG. 4. ste6 mutant strains fail to secrete pheromone molecules in confrontation assays. Congenic wild-type (WT) and ste6 mutant strains of opposite mating types were streaked in parallel on filament agar. Pictures were taken 4 days postincubation at a magnification of⫻100.
FIG. 5. STE6 is dispensable for haploid fruiting. Suspensions of the MAT␣ wild-type (WT), MAT␣ ste6 mutant, and MAT␣ ste6 reconstitution cells were spotted onto the filament agar and incubated in the dark at 26°C. The edges of the spots were photographed at a magnification of⫻100 after 12 days.
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
first pheromone transporter to be characterized in a basidio-mycete. In S. cerevisiae, STE6 is expressed only in a cells, in which it functions as a transporter to secrete the mating pher-omone a-factor. Studies of S. cerevisiae have shown that the C-terminal methyl moiety of a-factor is critical for recognition by Ste6 and secretion (32). Contributions of particular amino acids in this dodecapeptide have been assessed. Interestingly, most of the mutations on the a-factor do not affect the export of a-factor but impede interaction with the pheromone recep-tor (26). It seems that the interaction of a-facrecep-tor with its trans-porter is more permissive than the interaction of a-factor with its receptor, for which high specificity is required (26). Studies of the C. neoformans MF␣ and MFa pheromone genes have shown that the structure of these pheromones is conserved and that 3 out of 10 or 13 amino acids are identical in the mature MF␣1 and MFa1 peptides, respectively (9, 25). Therefore, it is plausible that in C. neoformans, the same transporter functions bilaterally in MAT␣ and MATa cells through different substrate affinities.
STE6 is involved in, but not essential for, mating in C. neoformans.The mating ability of the MF␣ pheromone triple deletion mutant is not completely abolished (33). As predicted, similar results have been observed for the pheromone trans-porter mutant. However, this mutant is in stark contrast to the ste6 deletion mutant in S. cerevisiae, which exhibits a sterile phenotype. The early cell-cell interaction during mating in S. cerevisiae has been inspected extensively. High levels of pher-omone molecules have been proven to be required as signals for prezygotes to initiate cell fusion (2, 15). However, unlike
the case with ascomycetes, such as S. cerevisiae, in which cell fusion is strictly controlled, cell fusion in basidiomycetes is more promiscuous. This fact may account for the leaky, non-sterile phenotypes observed in the pheromoneless, pheromone receptor, and pheromone transporter mutant strains of C. neo-formans (7, 33).
MAT␣ ste6 cells have a higher mating efficiency. Interest-ingly, the mating efficiency of the ste6 mutant in the MAT␣ background is higher than the mating efficiency of the ste6 mutant in the MATa background. One possible explanation may be that there is a second nonspecific pheromone trans-porter in the C. neoformans genome and that it is capable of secreting the MF␣ pheromone but does not have affinity for the MFa pheromone. However, when we examined the MAT␣ ste6 and wild-type MATa cells, no morphological changes in the MATa cells were discerned. This result poses a problem for the aforementioned hypothesis. However, studies of other ba-sidiomycetous systems may provide some hints. In Schizophyl-lum commune, a homobasidiomycete, numerous pheromone genes encoding the lipopeptide pheromones have been iden-tified in the B mating-type locus. In a study, Fowler et al. (12) reported that heterologous expression of the Schizophyllum sex pheromones and receptors in S. cerevisiae can substitute for the original yeast pheromones and receptors to induce the phero-mone response pathway and lead to cell cycle arrest. Because the S. commune lipopeptide pheromones are predicted to have structural similarity to the S. cerevisiae a-factor precursor, it was hypothesized that the same machinery used for the pro-cessing and secretion of the a-factor in S. cerevisiae is also used FIG. 6. STE6 expression is induced during mating. Total RNA was prepared from cells grown on V8 plates for 0, 2, 6, 12, 24, and 48 h. A Northern blot was hybridized in succession with probes for STE6, MF␣, and MFa. RNA loading is demonstrated by the ethidium bromide-stained RNA gel.
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
for the Schizophyllum pheromones. Interestingly, research has shown that one of the pheromones is secreted in a Ste6-inde-pendent manner (12).
In S. cerevisiae, the secretion of an AFRP (a-factor-related peptide) has also been found to be Ste6 independent. AFRP corresponds to the C-terminal 7 amino acids of mature a-fac-tor, including both farnesyl- and carboxymethylcysteine. The AFRP does not have pheromone activity, and its biological function is still unknown (4). There are over 30 ABC trans-porters with similarity to Ste6 in S. cerevisiae, and it is possible that the secretion of the AFRP is through one of these trans-porters (34). In order to determine whether residual amounts of pheromone could still be secreted in the C. neoformans MAT␣ or MATa ste6 mutants, an immunochemical assay, such as immunoprecipitation of the pheromones, is required.
Another explanation for why the MAT␣ ste6 mutant mates better is the intrinsic nature of filamentation of the C.
neofor-mans MAT␣ cells. It is known that some of the wild-type MAT␣
strains undergo monokaryotic filamentation, while very few wild-type MATa strains demonstrate this capability (36). Therefore, it is possible that this intrinsic nature of filamenta-tion contributes to the better mating efficiency of the MAT␣ ste6 mutant.
The expression of STE6 is coordinated with nutritional sta-tus and pheromone sensing.The expression pattern of C. neo-formans STE6 is somewhat different from the pattern of the homolog in S. cerevisiae. First, C. neoformans STE6 expression is not mating type specific; cells of both mating types express the gene at comparable levels (Fig. 6 and data not shown), in contrast to S. cerevisiae, in which the expression of STE6 is restricted to a cells. Second, the C. neoformans STE6 gene is not transcribed at a detectable level under nutrient-rich con-ditions, and the expression is induced by nutrient limitation. Despite these differences, the expression levels of both the C. neoformans and S. cerevisiae STE6 genes are elevated in re-sponse to pheromone signaling. The STE6 homolog from an-other pathogenic yeast, C. albicans (HST6), has a very different expression pattern. The HST6 gene is constitutively expressed in different cell types of diploid strains at similar levels. HST6 was originally isolated by complementation of the S. cerevisiae ste6 mutant (31). In a recent study, it was demonstrated that the HST6 gene is required for mating in MTLa but not in MTL␣ cells in C. albicans (24), indicating that the mating processes in S. cerevisiae and C. albicans are highly conserved. Evolutionarily, C. albicans is much closer to S. cerevisiae than C. neoformans, and currently we are addressing the question of whether C. neoformans STE6 can also functionally comple-ment the mating defect of an S. cerevisiae ste6 mutant.
The autocrine signaling response is regulated via a Ste6-independent manner.The finding that the MAT␣ ste6 mutant does not exhibit a defect in monokaryotic fruiting is intriguing. The MAT␣ ste6 mutant appears to undergo haploid fruiting to a greater extent than the wild-type MAT␣ cells do. Addition-ally, the MAT␣ ste6 mutant strain reconstituted with the wild-type copy of STE6 produces fewer haploid filaments (Fig. 5). This observation suggests that the reconstitution strain may have an elevated level of STE6 expression and this, in turn, results in the hypersecretion of the pheromone molecules. Using real-time PCR analysis, we confirmed that the expres-sion level of STE6 in the reconstitution strain is 1.5-fold higher
than that of the wild-type MAT␣ cell (data not shown). Taken together, these results imply that monokaryotic fruiting is reg-ulated by the intracellular level of MF␣ pheromone and that Ste6 pheromone transporter is not required for the MF␣ pher-omone-mediated autocrine signaling response.
To further address whether the autocrine signaling loop operates intracellularly or extracellularly in MAT␣ cells, we also created and analyzed mutants defective in the pheromone receptor gene. The capacity for monokaryotic filamentation in the pheromone receptor cpr␣ mutant has been shown to be largely intact (7), and we have confirmed this finding in our experiment. Additionally, we have found that the MAT␣ cpr␣ ste6 double mutant is also capable of undergoing haploid fila-mentation and that no significant difference in filafila-mentation was discernible when it was compared to the cpr␣ single mu-tant (data not shown). Based on all these results, our present hypothesis is that the autocrine signaling loop is triggered intracellularly by the MF␣ pheromone. If this is the case, it is a novel phenomenon that has never been reported. Research to identify the corresponding intracellular targets is now under way to further characterize MF␣ pheromone signaling in C.
neoformans MAT␣ cells.
ACKNOWLEDGMENTS
We thank Joseph Heitman for his enthusiastic support and encour-agement. We also thank Cristl Arndt and Deborah S. Fox for assis-tance and Kirsten Nielsen, James A. Fraser, and Alexander Idnurm for their comments on the manuscript. We finally thank the C. neoformans Genome Project at Stanford Genome Technology Center for provid-ing the C. neoformans sequence information.
This work was supported by National Science Council grants NSC91-2311-B-002-047 91C2801 and NSC90-2311-B-002-062 to Wei-Chiang Shen.
REFERENCES
1. Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus
neofor-mans mating and virulence are regulated by the G-protein␣ subunit GPA1 and cAMP. Genes Dev. 11:3206–3217.
2. Brizzio, V., A. E. Gammie, G. Nijbroek, S. Michaelis, and M. D. Rose. 1996. Cell fusion during yeast mating requires high levels of a-factor mating pher-omone. J. Cell Biol. 135:1727–1739.
3. Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
4. Chen, P., J. D. Choi, R. Wang, R. J. Cotter, and S. Michaelis. 1997. A novel a-factor-related peptide of Saccharomyces cerevisiae that exits the cell by a Ste6-independent mechanism. Mol. Biol. Cell 8:1273–1291.
5. Chen, P., S. K. Sapperstein, J. D. Choi, and S. Michaelis. 1997. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol.
136:251–269.
6. Christensen, P. U., J. Davey, and O. Nielsen. 1997. The Schizosaccharomyces
pombe mam1 gene encodes an ABC transporter mediating secretion of
M-factor. Mol. Gen. Genet. 255:226–236.
7. Chung, S., M. Karos, Y. C. Chang, J. Lukszo, B. L. Wickes, and K. J.
Kwon-Chung.2002. Molecular analysis of CPR␣, a MAT␣-specific phero-mone receptor gene of Cryptococcus neoformans. Eukaryot. Cell 1:432–439. 8. Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M.
Hull, C. D’Souza, P. Wang, and J. Heitman.2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Micro-biology 148:2607–2615.
9. Davidson, R. C., T. D. E. Moore, A. R. Odom, and J. Heitman. 2000. Characterization of the MF␣ pheromone of the human fungal pathogen
Cryptococcus neoformans. Mol. Microbiol. 39:1–12.
10. Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from
Cryptococcus neoformans var. neoformans and its use as a selective marker
for transformation. Mol. Cell. Biol. 10:4538–4544.
11. Elia, L., and L. Marsh. 1996. Role of the ABC transporter Ste6 in cell fusion during yeast conjugation. J. Cell Biol. 135:741–751.
12. Fowler, T. J., S. M. DeSimone, M. F. Mitton, J. Kurjan, and C. A. Raper. 1999. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol. Biol. Cell 10:2559–2572.
13. Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif.
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org
14. Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557–615.
15. Jackson, C. L., and L. H. Hartwell. 1990. Courtship in Saccharomyces
cer-evisiae: an early cell-cell interaction during mating. Mol. Cell. Biol. 10:2202–
2213.
16. Johnson, A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552–558.
17. Ketchum, C. J., W. K. Schmidt, G. V. Rajendrakumar, S. Michaelis, and
P. C. Maloney.2001. The yeast a-factor transporter Ste6, a member of the ABC superfamily, couples ATP hydrolysis to pheromone export. J. Biol. Chem. 276:29007–29011.
18. Kronstad, J. W., and C. Staben. 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31:245–276.
19. Kuchler, K., R. E. Sterne, and J. Thorner. 1989. Saccharomyces cerevisiae
STE6 gene product: a novel pathway for protein export in eukaryotic cells.
EMBO J. 8:3973–3984.
20. Kwon-Chung, K. J. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821–833.
21. Kwon-Chung, K. J. 1975. A new genus, Filobasidiella, the perfect state of
Cryptococcus neoformans. Mycologia 67:1197–1200.
22. Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic associa-tion of mating types and virulence in Cryptococcus neoformans. Infect. Im-mun. 60:602–605.
23. Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich,
and J. Heitman.2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704–718.
24. Magee, B. B., M. Legrand, A. M. Alarco, M. Raymond, and P. T. Magee. 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 46:1345–1351. 25. McClelland, C. M., J. Fu, G. L. Woodlee, T. S. Seymour, and B. L. Wickes. 2002. Isolation and characterization of the Cryptococcus neoformans MATa pheromone gene. Genetics 160:935–947.
26. Michaelis, S. 1993. STE6, the yeast a-factor transporter. Semin. Cell Biol.
4:17–27.
27. Michaelis, S., P. Chen, C. Berkower, S. Sapperstein, and A. Kistler. 1992.
Biogenesis of yeast a-factor involves prenylation, methylation and a novel export mechanism. Antonie Leeuwenhoek 61:115–117.
28. Michaelis, S., and I. Herskowitz. 1988. The a-factor pheromone of
Saccha-romyces cerevisiae is essential for mating. Mol. Cell. Biol. 8:1309–1318.
29. Moore, T. D. E., and J. C. Edman. 1993. The␣-mating type locus of
Cryp-tococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol.
13:1962–1970.
30. Pitkin, J. W., D. G. Panaccione, and J. D. Walton. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557–1565.
31. Raymond, M., D. Dignard, A. M. Alarco, N. Mainville, B. B. Magee, and
D. Y. Thomas.1998. A Ste6/P-glycoprotein homologue from the asexual yeast Candida albicans transports the a-factor mating pheromone in
Saccha-romyces cerevisiae. Mol. Microbiol. 27:587–598.
32. Sapperstein, S., C. Berkower, and S. Michaelis. 1994. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyl-transferase, and demonstration of its essential role in a-factor export. Mol. Cell. Biol. 14:1438–1449.
33. Shen, W.-C., R. C. Davidson, G. M. Cox, and J. Heitman. 2002. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in
Cryptococcus neoformans. Eukaryot. Cell 1:366–377.
34. Taglicht, D., and S. Michaelis. 1998. Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease. Methods Enzymol. 292: 130–162.
35. Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405–1411.
36. Tscharke, R. L., M. Lazera, Y. C. Chang, B. L. Wickes, and K. J.
Kwon-Chung.2003. Haploid fruiting in Cryptococcus neoformans is not mating type ␣-specific. Fungal Genet. Biol. 39:230–237.
37. Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein subunit GPB1 is required for mating and haploid fruiting in Cryptococcus
neofor-mans. Mol. Cell. Biol. 20:352–362.
38. Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimor-phism and haploid fruiting in Cryptococcus neoformans: association with the ␣-mating type. Proc. Natl. Acad. Sci. USA 93:7327–7331.
at NATIONAL TAIWAN UNIV MED LIB on February 2, 2010
ec.asm.org