Molecular Cloning, Characterization, and Transgenic Expression of CCFMADS: a MADS-Box Gene
from Calocedrus formosana
Chia-Chen Wu,1,3) Jia-Bin Tsai,2) Fang-Hua Chu3,4)
【Summary】
In this research, a MADS-box gene, CCFMADS, was cloned from Calocedrus formosana.
Through phylogenetic analyses and similarities of specific C-terminal motifs, this gene was cat- egorized into C-function lineages belonging to the AGAMOUS family. The CCFMADS-specific C-terminal motif shared high similarity with C and D-functional types of genes, and showed little divergence with other gymnosperms. In addition, transgenic Arabidopsis thaliana was used to evaluate the function of the CCFMADS gene. All sense transgenic plants displayed abnormal veg- etative growth, such as smaller and curling leaves. According to a Northern blot analysis of CCF- MADS expression in different months in C. formosana, we assumed that the CCFMADS gene may play an important role at the stage when vegetative buds convert to reproductive buds.
Key words: Calocedrus formosana, MADS-box gene, reproductive period, transgenic, Arabidopsis thaliana.
Wu CC, Tsai JB, Chu FH. 2018. Molecular cloning, characterization, and transgenic expression of CCFMADS: a MADS-box gene from alocedrus formosana. Taiwan J For Sci 33(1):1-15.
1)Silviculture Division, Taiwan Forestry Research Institute, 53 Nanhai Rd., Zhongzheng District, Taipei 10066, Taiwan. 林業試驗所育林組,10066台北市中正區南海路53號。
2)Liouguei Research Center, Taiwan Forestry Research Institute, 198 Chunghing Village, Liouguei District, Kaohsiung City 84443, Taiwan. 林業試驗所六龜研究中心,84443高雄市六龜區中興村198 號。
3)School of Forestry and Resource Conservation, National Taiwan University, 1 Roosevelt Road, Sec.
4, Taipei 106, Taiwan. 國立台灣大學森林環境暨資源學系,106台北市大安區羅斯福路4段1號。
4)Corresponding author, e-mail: fhchu@ntu.edu.tw 通訊作者。
Received November 2016, Accepted February 2017. 2016年11月送審 2017年2月通過。
研究報告
台灣肖楠MADS-box基因CCFMADS之選殖、
轉基因表現與特性分析
吳家禎1,3) 蔡佳彬2) 曲芳華3,4)
摘 要
本研究從台灣特有種台灣肖楠(Calocedrus formosana)中選殖MADS-box基因。透過親緣關係 圖 以 及 胺 基 酸 序 列C端區塊之相似性分析,將此基因歸類於C功能型之家族(lineages),並命名為 CCFMADS。透過序列分析結果得知,CCFMADS基因在序列上面和C功能型與D功能型之基因的 C-terminal之區塊相似,並與其他裸子植物無太大的差異性。透過轉殖阿拉伯芥觀察CCFMADS基因功 能的結果發現,轉殖CCFMADS會影響阿拉伯芥營養生長之狀況,呈現出葉小,或是葉片捲曲等表現
性狀。在台灣肖楠生殖週期CCFMADS基因的表現偵測中,發現CCFMADS基因在營養芽轉變成為生殖
芽的時期出現表現量,推測CCFMADS基因於營養生長轉變到繁殖生長的階段扮演重要角色。
關鍵詞:台灣肖楠,MADS-Box基因,生殖週期,阿拉伯芥基因轉殖。
吳家禎、蔡佳彬、曲芳華。2018。台灣肖楠MADS-box基因CCFMADS之選殖、轉基因表現與特性分 析。台灣林業科學33(1):1-15。
INTRODUCTION
In plants, flower and seed production are very important for reproduction and disper- sal. The development of floral organs of the flower is one of the most important processes in a plant’s life-cycle (Skipper et al. 2006). In the past decade, there has been much research on flowering control genes, such as those that modify floral organs, early flowering, and seed sterility. The mechanism of plant flow- ering, the ABC model, was examined in the past 2 decades. According to the ABC model, most flowering control genes can be classified into 3 functions of homeotic genes (termed A, B, and C) (Theißen et al. 2001, Causier et al. 2010). In this model, B-function genes control the formation of floral-meristems and floral-organs by interacting with Aand-C- function genes. In other words, A-function genes alone specify sepal formation, the com- bination of Aand-B-function genes specifies
the formation of petals, and B together with C function genes specifies stamen formation. C- function genes alone determine carpel forma- tion. The model also assumes an antagonistic relationship between A and C-function genes.
Some research shows the “quartet model” or
“ABCDE model”. D-function genes control the formation of ovules. E-function genes may interact with B and C function genes to control development of petals, stamens, and carpels (Pelaz et al. 2000). Besides the ABC and ABCDE models, because of different flo- ral mechanisms in different species, there is a newly applicable (A) BC model (Causier et al. 2010).
MADS-box genes (named after MCM1 (minichromosome maintenance) from yeast, AGAMOUS from Arabidopsis thaliana, DE- FICIENS from Antirrhinum majus, and SRF (serum response factor) from Homo sapiens)
encode putative transcription factors sharing a highly conserved domain with about 58 amino acid sequences (De Bodt et al. 2003, Irish 2003). This domain is the MADS-box, which is involved in DNA-binding, and pro- tein-protein interactions (Brent and Ptashne 1985). MADS-box genes can be divided into 2 types: type I and type II. Type II MADS- box proteins in plants, referred to as MIKC- type proteins, contain an “I” domain (inter- vening domain), a low conserved domain, K-box (keratin domain), a weak conserved domain, and the variable C-terminal domain, which appears to promote higher-order pro- tein interactions (De Bodt et al. 2003, Kramer et al. 2004). In the model plant, Ara. thaliana, the AP2 gene is an exception, and all ABC- function genes belong to the MADS-box tran- scription factor family (Immink et al. 2010).
In developmental biology, flowering con- trol is studied to understand how vegetative growth converts to reproductive growth, and phylogenetic analyses of the plant MADS-box gene family illustrate distinct functional roles at the molecular level (Skipper et al. 2006). In angiosperms, gymnosperms, and even in ferns and moss, studying floral organs and flower- ing periods has provided important indications of plant evolution (Ng and Yanofsky 2001).
Previous studies indicated that flower development and differentiation are con- trolled by many MADS-box genes (De Bodt et al. 2003). In the model plant, Arabidopsis, many closely related flower development genes have been cloned, these include: AP1, AP2, AP3, PI, AG, SHP1, SHP2, AGL11, AGL13, AGL3, and SEP. In plant develop- ment, A-function genes (AP1 and AP2), B- function genes (AP3 and PI), C-function genes (AG), D-function genes (SHP1, SHP2, AGL11, and AGL13), and E-function genes (AGL3 and SEP) all have different functions in the development of flower whorls. Various
combinations of these 5 kinds of genes con- trol the development of floral organs. But, in gymnosperms, there are only functional com- binations of B and C-function genes, which A-function genes are lacking (Theißen et al.
2000, Nilsson et al. 2007).
In gymnosperms, most reports regarding reproductive growth focused on Picea abies, Pic. mariana, and Pinus radiata. For example, the transgenic Arabidopsis plant harboring SAG1 cloned from Pic. mariana showed homeotic conversion of sepals to carpels and petals to stamens. Therefore, SAG1 is assumed to be related to control of the development of flowers and cones (Rutledge et al. 1998).
Calocedrus formosana is an endemic species in Taiwan. Florin (1956) recognized that there are only 3 species (C. decurrens, C. macrolepis, and C. formosana) in the ge- nus Calocedrus. Because of its afforestation potential and wood utility, C. formosana is widely planted in northern and central Taiwan.
In this study, we report on the isola- tion and characterization of an AG homolog, CCFMADS, from differentiated leaves of C.
formosana. We observed the phenotype of transgenic Arabidopsis with overexpressed sense and antisense CCFMADS, and ana- lyzed the periodic expression of CCFMADS using Northern blotting in C. formosana.
We also constructed a phylogenetic tree of CCFMADS with other MADS-box amino ac- ids by the Neighbor-Joining (NJ) method. To our knowledge, this is the first report of the isolation and characterization of AG homo- logs from C. formosana.
MATERIALS AND METHODS Plant materials
Plant materials were collected from one mature C. formosana tree located in the seed orchard of the Liouguei Research Center,
southern Taiwan. Young leaves, and male and female cones were sampled with steril- ized scissors, biweekly from April 5, 2005 to February 23, 2006; and sampled weekly from February 24, 2006 to March 23, 2006.
Samples were frozen in liquid nitrogen and immediately stored at -80℃.
Transgenic plant samples (A. thaliana var. Columbia) in this research were collected by sterilized scissors, frozen in liquid nitro- gen, and stored at -80℃.
Isolation and sequencing of the full- length CCFMADS gene
Total RNA was extracted from a flower bud using the CTAB method described by Chang et al. (1993). Reverse-transcription was performed by following the kit manual (Superscript II reverse-transcriptase, Invi- trogen, Waltham, MA, USA). A MADS- box gene (accession no.: U69482) from Pic.
mariana was used to perform a BLASTn search in NCBI. Specific primer pairs were designed according to conserved coding se- quences of the top 8 sequences obtained from the BLASTn alignment results (forward:
CCFMADSf:5’-ATgggCCgTgggAAgATT gAg-3’; reverse: CCFMADS r: 5’- TCAgC CAAgCTgAAgCgTTg-3’). A full-length CCFMADS (NCBI accession no.: KY094491) gene was amplified from complementary (c) DNA with the above specific primers us- ing a Pfu DNA Polymerase kit (Promega, Fitchburg, WI, USA). A polymerase chain reaction (PCR) was carried out for 1 min at 94℃, followed by 30 cycles of 30 s at 94℃, 30 s at 56℃, and 1 min at 72℃, with a 6 min incubation at 72℃ and finally maintained at 4℃ using a GeneAmp™ PCR System Model 3730 (PE Applied Biosystems, Waltham, MA, USA). PCR-amplified DNA fragments were extracted using a DNA Clean/Extraction kit (GMbiolab, Taichung, Taiwan) and ligated
with an adenine tail on the blunt end using a Taq polymerase kit (GenetBio, Daejeon, Korea). The purified PCR product with the adenine tail was fused into the pGEM-T Easy vector (Promega), then was the vector trans- formed into Escherichia coli (DH5α stain) for growth on an LB plate with blue-white selec- tion. After plasmid DNA extraction, plasmid DNA was sequenced using an ABI 3730 DNA analyzer (Thermo Fisher Scientific, Waltham, MA, USA) in the Institute of Plant and Microbial Biology, Academia Sinica (Tai- pei, Taiwan).
Phylogenetic analysis
We retrieved 37 different functional-type MADS-box protein sequences of plant spe- cies from the GenBank database for use in the phylogenetic analysis including Ara. thali- ana: AP1 (CAA78909), CAL (AAA64789), FUL (AAL66878), AGAMOUS (CAA37642), AGL6 (AAA79328),SEP1 (AAA32732), SEP2 (AAA32734), SEP3 (AAB67832), AP3 (AAD51903), PI (BAA06465), and AGL11 (AAC49080); Petunia hybrid: FBP26 (AAF19164), FBP6 (CAA48635), pMADS3 (CAA51417), FBP7 (CAA57311), and FBP11 (CAA57445); Malus domestica: MdMADS2 (AAC83170), MdMADS5 (CAA04321), Md- MADS3 (AAD51422), MdPI (CAC28021), and MdMADS10 (CAA04324); Lyco- persicon esculentum: LeMADS-MC (AAM15774), TDR5 (CAA43170), and TAG11 (AAM33102.2); Antirrhinum majus:
PLENA (AAB25101), DEFH72 (CAA64742), GLO (CAA48725), and DEF (CAA36268);
Populus trichocarpa: PTAG1 (AAC06237);
Pic. abies: DAL2 (CAA55867), DAL12 (AAF18374), and DAL13 (AAP34376);
Pic. mariana: SAG-c (AAC97158); Ginkgo biloba: GBM5 (AAM76208); Pin. radiata:
PrDGL (AAF28863); Gnetum gnemon:
GGM13 (CAB44459); and Cryptomeria
japonica: CjMADS1 (AAL05440). Mul- tiple sequence alignments of these protein sequences were performed using ClustalW 1.82 software downloaded from the EMBL (European Molecular Biology Laboratory;
https://www.embl.de/). A phylogenetic tree was generated by the NJ methodology us- ing MEGA 4.1 software based on p-distance amino acid substitutions with 1000 bootstrap replicates. Multiple sequence alignments of gymnosperms C and D-functional type protein sequences of DAL2 (CAA55867), CyAG (AAM74074), GBM5 (AAM76208), and GGM3 (CAB44449) were analyzed with ClustalW for sequence motif prediction.
Transformation and selection of Ara.
thaliana
In order to observe the function of CCFMADS, we overexpressed and repressed CCFMADS under the control of the cauliflow- er mosaic virus (CaMV) 35S promoter. SacI- BamHI fragments with sense or antisense CCFMADS cDNA were generated by PCR amplification and used to replace the GUS coding region within pBI121 that contains a single 35S promoter. These 2 kinds of pBI121 vectors (sense CCFMADS and antisense CCFMADS) were transformed into Agrobac- terium tumefaciens strain GV3101 follow- ing the MicroPulser electroporation manual (Biorad, Hercules, CA, USA). The transgenic A. thaliana (Columbia) was produced at The Transgenic Plant Core lab at Academia Sinica by following an online protocol (http://trans- plant.sinica.edu.tw/english/protocol/trans/
arabidopsis-eg.doc). The putative transformed plants were selected on media plates with kanamycin. Plants were placed at 22℃, with 16 h of light/8 hr darkness for growth. To validate the presence of 35S-CCFMADS in the transgenic plants, transgenic plants were tested by kanamycin resistance, a PCR analy-
sis, and a Northern blot analysis.
Northern blot analysis of CCFMADS ex- pression in C. formosana
Young leaves were collected monthly from 2005 to 2006 for RNA extraction using the CTAB method by Chang et al. (1993).
Northern blotting was performed by the fol- lowing process. The extracted RNA was separated by 1.5% agarose gel electropho- resis under denaturing conditions. The gel was soaked in phosphate buffer (50 mM NaH2PO4, 50 mM NaOH, and 5 mM EDTA, pH 6.5) for 1 h, and then transferred into 20x SSPE for 30 min. The RNA was blotted onto positively charged nylon membranes (Roche Basel, Switzerland) and then a UV cross-linking process was performed. For probe hybridization, nylon membranes with RNA were soaked in hybridization solu- tion (0.2x SSPE, 0.5% laurylsarcosine, 1%
sodium dodecylsulfate (SDS), 1% blocking reagent (Roche)) at 65℃ for 3 h. The prim- ers 5’-CGGGATCCATGGGCCGTGGGAA GATTGAG-3’ and 5’-CGAGCTCGTCAGC CAAGCTGAAGCGTTGTTTGC-3’ were used to generate the DIG-labeled CCFMADS probe. The DIG-probe was generated by fol- lowing the DIG Northern Starter Kit and CDP-Star chemiluminescent substrate (ready- to-use) ((Roche).
RESULTS
Analysis of the sequence of full-length cDNA
A DNA fragment of 669 bp was cloned from the flower bud of C. formosana and named CCFMADS, which encoded 222 ami- no acids with a predicted molecular weight of 25.62 kDa. A GenBank BLAST search analysis of the CCFMADS protein sequence shared high homology with MADS-box pro-
teins of several gymnosperm species (83~99%
identity and 92~100% similarity) (Table 1). It shared the greatest similarity with the MADS- box transcription factor of Pin. resinosa.
Thus, we predicted that this putative protein is a MADS-box transcription factor, because it contained a conserved MADS-box domain
from residues Gly2 to His60 and the K-domain from Gln91 to Arg153 (Fig. 1).
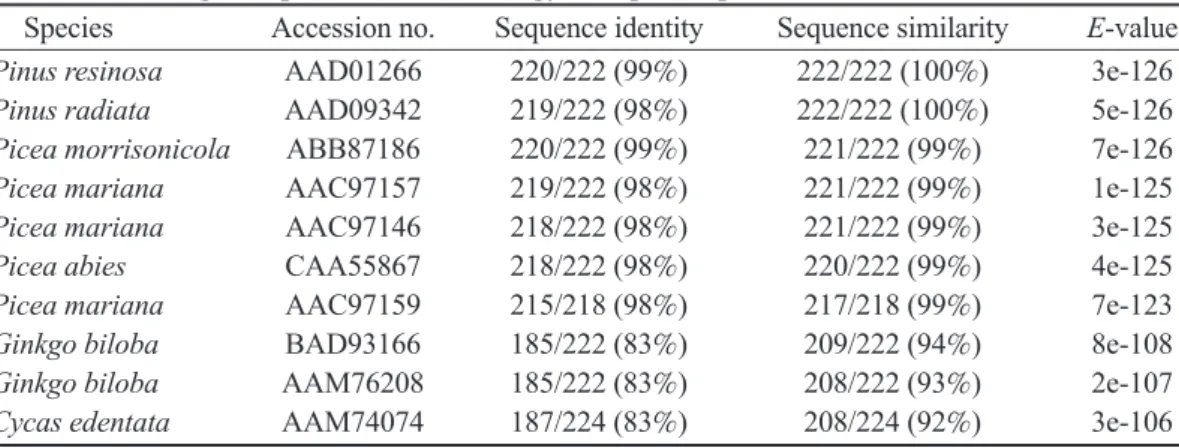
The conserved domain analysis of the CCFMADS MADS-box domain using a CDD (conserved domain database) at the NCBI showed the same result with other MADS-box genes that contain a MADS-box and K domain. We found Table 1. Comparison of the deduced, full-length amino acid sequence of the CCFMADS gene with the full-length sequences from other gymnosperm species.
Species Accession no. Sequence identity Sequence similarity E-value Pinus resinosa AAD01266 220/222 (99%) 222/222 (100%) 3e-126 Pinus radiata AAD09342 219/222 (98%) 222/222 (100%) 5e-126 Picea morrisonicola ABB87186 220/222 (99%) 221/222 (99%) 7e-126 Picea mariana AAC97157 219/222 (98%) 221/222 (99%) 1e-125 Picea mariana AAC97146 218/222 (98%) 221/222 (99%) 3e-125 Picea abies CAA55867 218/222 (98%) 220/222 (99%) 4e-125 Picea mariana AAC97159 215/218 (98%) 217/218 (99%) 7e-123 Ginkgo biloba BAD93166 185/222 (83%) 209/222 (94%) 8e-108 Ginkgo biloba AAM76208 185/222 (83%) 208/222 (93%) 2e-107 Cycas edentata AAM74074 187/224 (83%) 208/224 (92%) 3e-106
Fig. 1. Sequence of the CCFMADS gene and its amino acid sequence. The highly conserved region in gray is the MADS-box. The K-domain is underlined. The star indicates a stop codon of CCFMADS.
that the CCFMADS had some protein structures:
an α-helix in the MADS-box domain (Asn16 to Leu38), a pair of ß-sheets (Val42 to Phe48 and Leu54 to Ala58), and an α-helix in the intervening do- main, that immediately followed the MADS-box domain. The MADS-box domain contained char- acteristic functional sites such as a DNA-binding site, a dimerization interface formed as the protein function is activated, and phosphorylation (Fig. 2).
In addition, 2 highly conserved motifs, AG motif I and AG motif II, were identified in the C-terminal of the CCFMADS protein sequence (Fig. 3).
There are hydrophobic and polar residues at these 2 motifs. These highly conserved regions were defined as a synapomorphy of the AG-like gene subfamily in seed plants (Kramer et al. 2004).
Phylogenetic analysis of CCFMADS In order to understand the relationship
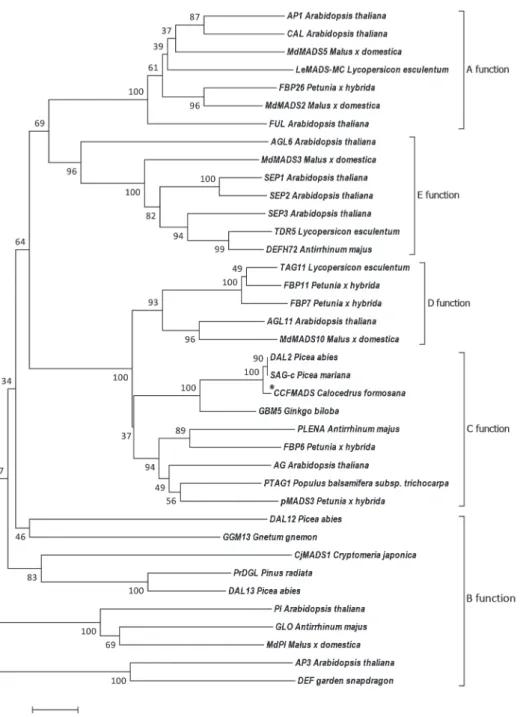
of the CCFMADS gene to the other different functional MADS-box genes, we selected 37 MADS-box genes (Fukui et al. 2001, Vandenbussche et al. 2003, Kim et al. 2005) including A, B, C, D, and E function genes to construct a phylogenetic tree of CCFMADS (Fig. 4). The phylogeny showed that differ- ent functions were clustered into separate groups, and the CCFMADS gene clustered with C-function MADS-box proteins. Two MADS-box proteins from Pic. mariana and Pic. abies, respectively named SAG-c and DAL2, showed 99% similarity with CCF- MADS. Data indicated that the GBM5 pro- tein was also closely related to CCFMADS, by showing 93% similarity with CCFMADS.
Thus, we classied CCFMADS as a C-function MADS-box gene that belonged to the AGA- MOUS family.
Fig. 2. Putative function site of the MADS domain in the CCFMADS. The position marked # is a putative function site.
Expression patterns of CCFMADS during the reproductive cycle of C. formosana
We collected and observed samples from April 2005 to March 2006. In the beginning of differentiation, vegetative buds became hook- like (Fig. 5A, B); this is a key feature of the conversion from vegetative buds to reproduc- tive buds. These reproductive buds can further develop into male or female cones. Through December 2005, 9 months after we first began our the observations, we could tell which were male cones (with pollen) or female cones (with pollination drops) (Fig. 5C-F).
A Northern blot analysis with a gene- specific CCFMADS probe was carried out to determine monthly expression patterns of CCFMADS in C. formosana. The Northern blot data showed that CCFMADS was only expressed in April and May (Fig. 6).
Phenotypes of transgenic Arabidopsis Twelve CCFMADS transgenic Arabidopsis plants (sense: S6, S8, S11, S15, S18, and S20;
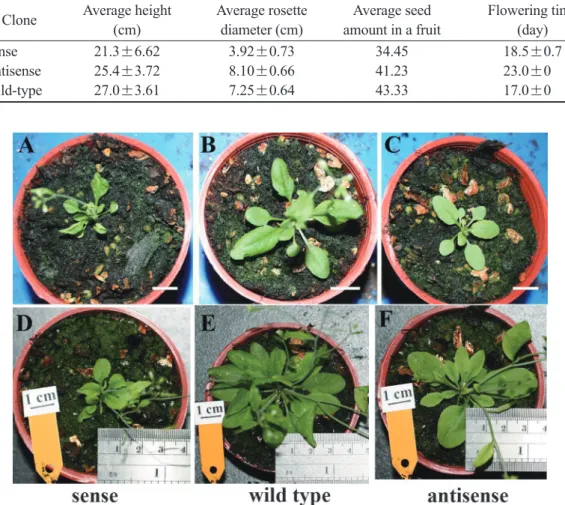
antisense: A2, A3, A5, A7, A11, and A13) were randomly selected for further investigation. At the moment the first fruit was mature and open, all heights of sense transgenic plants were re- duced with an average height of 21.3 cm, and the average rosette diameter was smaller (3.92 cm) than these of the antisense and wild-types
(Table 2). Sense transgenic plants leaves were smaller and curled or folded upwards or inwards from the margins (Figs. 7A, 8A, D).
The phenotypes of sense transgenic plants differed from those of wild-type plants: sepals and fruits were smaller than those of wild-type plants. In addition, 1 sense transgenic plant had abnormal phenotypes, such as petal-like sta- mens, abnormal flowers and fruits, carpel-like sepals, unseparated stems, and a sterile plant with poly-flowers, and poly-stems (Fig. 9).
Results showed no clearly different phe- notypes between wild-type and antisense trans- genic plants (Fig. 7B, C, 8B, C, E, F). At the moment the first fruit was mature and open, the average height of antisense transgenic plants was 25.4 cm, and the average rosette diameter was 8.1 cm (Table 2). The growth of antisense transgenic plants was better than that of sense transgenic plants. Comparing sense and anti- sense transgenic plants, the average time that the first flower appeared in antisense transgenic plants was 5~6 days later than in wild-type and sense plants (Table 2).
DISCUSSION
We isolated a MADS-box gene (CCF- MADS) from C. formosana. Its deduced protein sequence had a classic structure of Fig. 3. Alignment of the C-terminal region of the CCFMADS gene and other gymnosperm plant genes’ amino acid sequences. There is an AG motif I in the CCFMADS gene at Phe194 to Asp209, and an AG motif II at Phe211 to Gly222. Accession numbers of the sequences: DAL2 (CAA55867); CyAG (AAM74074); GBM5 (AAM76208); and GGM3 (CAB44449).
Fig. 4. Phylogenetic tree of MADS-box genes from different function types of the MADS box gene, CCFMADS (in the dotted box). The tree shown is a Neighbor-joining (NJ) method tree after calculating bootstrap with values 1000 repeats. The number on every divergence shows the confidence level. The scale shows the nucleotide substitutions per site.
MADS-box transcription factors: a K-domain
and highly conserved motifs (AG motifs I and II) at the C-terminus. AG motifs I and II exist in the C and D-function gene families,
Fig. 5. Photos of Calocedrus formosana. A, Vegetative bud converts to reproductive bud in April and May. (The picture was collected in May.) B, Comparison of vegetative buds and reproductive buds (right are vegetative buds, left are reproductive buds). C, Maturation of reproductive buds differs for male cones and female cones. D, Immature male cone; E, Mature male cone; F, female cone with pollination drops. Scale bars: A and B;1cm; C; 0.5cm;
D-F; 0.05 mm.
Fig. 6. Northern blot analysis of CCFMADS expression of Calocedrus formosana (Samples were tips of leaves) in different months. From lane 1 to lane 12 represents January to December.
Fig. 7. Phenotypes of CCFMADS transgenic Arabidopsis of about 2-weeks old. Scale bar is 1 cm.
Table 2. Growth of CCFMADS transgenic plants. Observed time was the time that the first fruit was broadcast, when the average plant height and diameter of leaf spread were measured. Sense clones: S6, S8, S11, S15, S18, S20. Antisense clone: A2, A3, A5, A7, A11 and A13.
Clone Average height Average rosette Average seed Flowering time (cm) diameter (cm) amount in a fruit (day)
Sense 21.3±6.62 3.92±0.73 34.45 18.5±0.7
Antisense 25.4±3.72 8.10±0.66 41.23 23.0±0
Wild-type 27.0±3.61 7.25±0.64 43.33 17.0±0
Fig. 8. Sense and antisense CCFMADS transgenic Arabidopsis. A-C, time of the first flower appearance; D-F, photographed at the time of collecting seeds.
belonging to the AGAMOUS family or AGL (AG-like) gene family (Kramer et al. 2004).
This MIKC-type domain structure exists universally in the MADS-box gene of vascu- lar plants (Winter et al. 1999; Santelli et al.
2000). The highly conserved protein struc- tures of the MADS-box domain may play the role for forming the higher-order complex formation. The higher-order complex of MADS domain proteins can provide struc- tural mechanisms for increasing functional specificity (Immink et al. 2010).
CCFMADS, DAL2 and SAG-c formed a clade which was a sister group to GBM5 of G.
biloba in the phylogenetic trees, such as the separation of Ginkgophyta and Pinophyta in morphology. Sequences of C and D-function genes are very homologous, but the functions differ (Colombo et al. 1995). For example, D-function genes are recognized for control- ling ovule identity, whereas the C-function genes lead to the formation of carpels (Co- lombo et al. 1995). The phylogenetic tree showed that gymnosperms and angiosperms
were separated in B and C-function clades. To the best of our knowledge, CCFMADS was the first C-function MADS-box gene cloned from C. formosana. For a clear understanding of MADS-box genes of C. formosana, further research is required in the future.
Most MADS-box genes are recognized as transcriptional regulators. Besides playing an important role in controlling floral organ formation, MADS-box genes have many kinds of functions such as regulating of the floral meristem, flowering times, and transi- tions of vegetative growth to reproductive growth (Ferrándiz et al. 2000, Hartmann et al.
2000, Samach et al. 2000, Carlsbecker et al.
2004, Ruokolainen et al. 2010).
Reproductive cycles of species in the Cupressaceae family are very complicated and dissimilar. We could find only 1 research paper on reproductive cycles of C. formosana (Chung and Kuo 2005). It reported that the appearance in the vegetative buds transfer to hook-like in May. It is a key feature of conversion from vegetative growth to reproductive growth. Af- ter that, reproductive buds continue to differen-
tiate to seed-cones or pollen-cones.
During April and May, the samples we collected were in the stage of conversion from vegetative to reproductive buds. The result of Northern blotting showed that the CCFMADS gene was expressed in April and May. Thus, the CCFMADS gene may play a role in the transition from vegetative growth to repro- ductive growth.
Sense and antisense transgenic CCF- MADS Arabidopsis plants were performed to evaluate putative functions of the CCFMADS gene. The phenotypes of sense CCFMADS transgenic plants in this study were very simi- lar to DAL2 transgenic Arabidopsis which had a reduced stem length and leaves that were small and curled or folded (Tandre et al. 1998). Thus, the CCFMADS gene might belong to the C-function MADS-box family and had the same ancestor that AGAMOUS had in the evolutional process. In observa- tions of the growth height of transgenic Arabodipsis, we found that all sense trans- genic plants were smaller than antisense and wild-type plants. We thought that curled or
Fig. 9. Abnormal shape of a CCFMADS sense transgene plant. A, Wild-type; B and C, abnormal flower and fruits; petal-like stamen; D and E, carpel-like sepal; F, unseparated stem of sense transgenic plant; G, sterile plant with polyflowers, and polystems. Scale bars:
A: 1 mm; B and C: 0.5 mm; D: 0.5 mm; F: 0.5 cm.
folded leaves caused by ectopic expressions of the CCFMADS gene may cause photosyn- thesis or other physiological abnormalities, eventually causing the plants to grow weakly.
The average seed amount of sense transgenic plants was fewer than those of the wild-type and antisense transgenic Arabidopsis plants.
Since the CCFMADS gene is very close to the AGAMOUS gene family, which controls the formation of stamens and carpels, the over- expression of the CCFMADS gene had an abnormal influence on floral organ formation, including the seed quantity and fruit size.
The CCFMADS amino acid sequence had 60% identity to SHP2 (accession no.:
AAU82057). The CCFMADS nucleotide se- quence showed 71% identity to Arabidopsis SHP2, which is one of the MADS-box genes expressed in carpels and ovules. The align- ment of the CCFMADS and SHP2 genes revealed high identity in the MADS domain with many functional sites. The CCFMADS sense transgenic plants revealed similar phe- notypes the SHP2 transgenic Arabidopsis which had curled or folded leaves, sepal- converted carpels, and earlier flowering (Pin- yopich et al. 2003).
The times of the first flowering were not very different from wild-type (17 d) and sense CCFMADS (18.5 d) Arabidopsis, but the times when antisense CCFMADS Arabidopsis initiated flowering were later than those of the wild-type Arabidopsis. Further research is needed to validate if the antisense CCFMADS gene inhibited normal gene expressions such as SHP2 which caused a flowering time de- lay. The MADS-box transcription factor plays key roles in various biological developmental processes. The formation of floral organs very complicatedly involves many genes.
For example, a MADS-box gene, EgAG2 ( of the AGAMOUS family), has 67% identity with the CCFMADS gene, cloned from Elaeis
guineensis, and it was suggested to be a C or D-function (or C/D mixed) gene that showed progressively increasing amounts during male and female inflorescence development and spatial expression. The expression of EgAG2 was also similar to expressions of the SHP1 and SHP2 genes in Ara. thaliana. However, EgAG2 transgenic Arabidopsis plants showed no phenotypic alterations compared to wild- type plants (Adam et al. 2007). Many previ- ous reports indicated that the C or D-function genes can interact with 2 or more closely related genes to control reproductive organ identity (Adam et al. 2007). In this report, the expression of the CCFMADS gene and trans- genic plants may provide useful information for understanding the MADS-box family of C.
formosana.
CONCLUSIONS
In this research, we cloned and charac- terized a MADS-box gene, CCFMADS, from C. formosana. The phylogenetic tree and sim- ilarity analysis showed that this gene could be grouped into Cfunction lineages belonging to the AGAMOUS family. In an analysis of transgenic plants, we found that vegetative growth was affected in sense and antisense transgenic lines compared to wild-type Ara.
thaliana. The CCFMADS gene was only ex- pressed at the time that vegetative buds con- verted to reproductive buds. This infers that the CCFMADS gene may play an important role in the conversion from the vegetative to the reproductive stage. In woody plants, most studies of MADS-box genes in gymno- sperms focused on Gnetum, Pinus, and Picea (Theißen et al. 2000). Characterization of MADS-box genes in Calocedus should help us understand the evolution of MADS-box genes and their roles in reproductive develop- ment. However, further studies are required
in order to understand the mechanism of MADS-box genes in the floral development of C. formosana.
LITERATURE CITED
Adam H, Jouannic S, Orieux Y, Morcillo F, Richaud F, Duval Y, Tregear JW. 2007.
Functional characterization of MADS box genes involved in the determination of oil palm flower structure. J Exp Bot 58:1245-59.
Brent R, Ptashne M. 1985. A bacterial repres- sor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene.
Nature 314:198.
Chang SJ, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Mol Bio Rep 11:113-6.
Carlsbecker A, Tandre K, Johanson U, Englund M, Engström P. 2004. The MADS- box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J 40:546-57.
Causier B, Schwarz-Sommer Z, Davies B.
2010. Floral organ identity: 20 years of ABCs.
Semin Cell Develop Biol 21:73-9.
Colombo L, Franken J, Koetje E, van Went J, Dons HJ, Angenent GC, van Tunen AJ.
1995. The petunia MADS box gene FBP11 de- termines ovule identity. Plant Cell 7:1859-68.
Chung JD, Kuo SR. 2005. Reproductive cycles of Calocedrus formosana. Taiwan J For Sci 20:315-29.
De Bodt S, Raes J, Van de Peer Y, Theißen G.
2003. And then there were many: MADS goes genomic. Trends Plant Sci 8:475-83.
Ferrándiz C, Gu Q, Martienssen R, Yanof- sky MF. 2000. Redundant regulation of meri- stem identity and plant architecture by FRUIT- FULL, APETALA1 and CAULIFLOWER.
Development 127:725-34.
Florin R. 1956. Nomenclatural notes on gen- era of living gymnosperms. Taxon 5:188-92.
Fukui M, Futamura N, Mukai Y, Wang Y, Nagao A, Shinohara K. 2001. Ancestral MADS-box genes in Sugi, Cryptomeria japon- ica D.Don (Taxodiaceae), homologous to the B function genes in angiosperms. Plant Cell Physiol 42:566-75.
Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. 2000. Mo- lecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21:351-60.
Immink RG, Kaufmann K, Angenent GC.
2010. The ‘ABC’ of MADS domain protein behavior and interactions. Semin Cell Develop Biol 21:87-93.
Irish VF. 2003. The evolution of floral homeo- tic gene function. BioEssays 25:637-46.
Kim S, Koh J, Ma H, Hu Y, Endress PK, Hauser BA, et al. 2005. Sequence and expres- sion studies of A-, B-, and E-function MADS- box homologues in Eupomatia (Eupomatia- cea): support for the bracteates origin of the calyptra. Plant Sci 166:185-98.
Kramer EM, Jaramillo MA, Di Stilio VS.
2004. Patterns of gene duplication and func- tional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166:1011-23.
Ng M, Yanofsky MF. 2001. Function and evo- lution of the plant MADS-Box gene family.
Nat Rev Genet 2:186-95.
Nilsson L, Carlsbecker A, Sundås-Larsson A, Vahala T. 2007. APETALA2 like genes from Picea abies show functional similari- ties to their Arabidopsis homologues. Planta 225:589-602.
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. 2000. B and C floral organ identity functions require SEPALLATA MADS- box genes. Nature 405:200-3.
Pinyopich A, Ditta GS, Savidge B, Lijegren SJ, Baumann E, Wisman E, Yanofsky MF.
2003. Assessing the redundancy of MADS-box
genes during carpel and ovule development.
Nature 424:85-8.
Ruokolainen S, Ng YP, Broholm SK, Albert VA, Elomaa P, Teeri TH. 2010. Characteriza- tion of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproduc- tive transition. BMC Plant Biol10:128.
Rutledge R, Regan S, Nicolas O, Fobert P, Côté C, Bosnich W,et al. 1998. Characteriza- tion of an AGAMOUS homologue from the conifer black spruce (Picea mariana) that produces floral homeotic conversions when expressed in Arabidopsis. Plant J 15:625-34.
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CON- STANS target genes in reproductive develop- ment of Arabidopsis. Science 288:1613-6.
Santelli E, Richmond TJ. 2000. Crystal struc- ture of MEF2A core bound to DNA at 1.5 Å resolution. J Mol Biol 297:437-49.
Skipper M, Johansen LB, Pedersen KB, Frederiksen S, Johansen BB. 2006. Cloning and transcription analysis of an AGAMOUS and SEEDSTICK ortholog in the orchid Dendrobium thyrsiflorum (Reichb. f.). Gene 366:266-74.
Tandre K, Albert VA, Sundås V, Engström P.
1995. Conifer homologues to genes that con- trol floral development in angiosperms. Plant Mol Biol 27:69-78.
Tandre K, Svenson M, Svensson ME, Eng- ström P. 1998. Conservation of gene structure and activity in the regulation of reproductive organ development of conifers and angio- sperms. Plant J 15:615-23.
Theißen G. 2001. Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4:75-85.
Theißen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, et al. 2000. A short his- tory of MADS-box genes in plants. Plant Mol Biol 45:115-49.
Vandenbussche M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, et al. 2003. To- ward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPAL- LATA-like MADS box genes in petunia. Plant Cell 15:2680-93.
Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theißen G. 1999. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants.
Proc Natl Acad Sci USA 96:7342-7.