Association between fluconazole susceptibility and
genetic relatedness among Candida tropicalis
isolates in Taiwan
Jang-Shiun Wang,
1,2Shu-Ying Li,
3Yun-Liang Yang,
4Hsiao-Hui Chou
3and Hsiu-Jung Lo
2Correspondence Hsiu-Jung Lo hjlo@nhri.org.tw
1Graduate Institute of Life Sciences, National Defense Medical Center, Taipei, Taiwan,
Republic of China
2Division of Clinical Research, National Health Research Institutes, 35 Keyan Road, Zhunan Town,
Miaoli County 350, Taiwan, Republic of China
3Division of Laboratory Research and Development, Center for Disease Control, Taipei, Taiwan,
Republic of China
4Department of Biological Science and Technology, National Chiao Tung University, Hsinchu,
Taiwan, Republic of China
Received 6 April 2006 Accepted 2 January 2007
Among the 162 Candida tropicalis isolates collected in the Taiwan Surveillance of Antimicrobial Resistance of Yeasts in 1999, 23 (14.2 %) had fluconazole MICs ¢64 mg l”1, and thus fulfilled the definition of resistance. Random amplified polymorphic DNA assay showed that all 23 fluconazole-resistance C. tropicalis isolates collected from different hospitals around Taiwan were closely related. Two distinct pulsotypes associated with fluconazole susceptibility were identified when these 23 resistant isolates, along with 13 susceptible ones, were analysed by PFGE.
INTRODUCTION
Although Candida albicans is the most frequently isolated
yeast pathogen causing morbidity in seriously
immuno-compromised hosts (Cheng et al., 2004; Pfaller et al., 2000;
Yang et al., 2004), there has been a shift toward the more
treatment-resistant non-albicans Candida species (Sanglard
& Odds, 2002; Walsh et al., 2004; Yang et al., 2004). We
initiated a nationwide surveillance, the Taiwan Surveillance
of Antimicrobial Resistance of Yeasts (TSARY), in 1999 to
investigate the distribution and drug susceptibility of
Candida species (Yang et al., 2003, 2004). A total of 53
among the 632 isolates collected in the TSARY (in 1999) had
fluconazole MICs ¢64 mg l
21. Thus, they were considered
as resistant isolates (Clinical and Laboratory Standards
Institute, 1997). Among them, 23 were Candida tropicalis,
collected from 10 of the 22 participating hospitals.
Clinically, the increase in the rate of fluconazole resistance
in C. tropicalis is of considerable importance since this is
one of the most commonly isolated non-albicans Candida
species (Cheng et al., 2004; Pfaller et al., 2000; Yang et al.,
2004, 2005). Information regarding the genetic background
of the resistant and susceptible isolates may provide further
insight into the distribution pattern and origin of the
resistance. In this study, we hoped to determine whether
these resistant isolates of C. tropicalis are genetically related
using two methods, random amplified polymorphic DNA
(RAPD) assay and PFGE analysis.
METHODS
Organisms and media. Yeast isolates were collected from 22 hospitals participating in the TSARY in 1999 (Lo et al., 2001). These hospitals were located around the island and covered all four geographical regions: the North, Middle, South and East districts. Each hospital was asked to submit up to 10 C. albicans and 40 non-albicans Candida species between 15 April and 15 June 1999. Duplicate isolates from the same patients were excluded. All isolates were stored frozen at 270uC in bead-containing Microbank cryovials (Pro-Lab Diagnostics). In total, the susceptibilities to fluconazole of 162 C. tropicalis isolates were determined. Then, the genetic relatedness of all 23 fluconazole-resistant isolates, which were collected from 10 hospitals, along with 13 susceptible ones, were determined. These 10 hospitals included four in the North district, three in the South district, two in the East district and one in the Middle district of Taiwan. The number of resistant isolates from each hospital ranged from one to five. RAPD. RAPD assays were performed according to a modified protocol specifically for C. tropicalis (Roilides et al., 2003). Amplification reactions (50 ml) were performed with the RP02 (59-GCG ATC CCC A-39) primer and in accordance with the manufacturer’s protocol, except that 5 units Taq DNA polymerase (New England Biolabs) and 1 ml genomic DNA were added to the reaction. PCR was performed as follows: 94uC for 5 min; 36 uC for 5 min and 72uC for 5 min for 4 cycles; 94 uC for 1 min, 36 uC for 1 min and 72uC for 2 min for 30 cycles; and a 10 min extension period Abbreviations: RAPD, random amplified polymorphic DNA; TSARY,
Taiwan Surveillance of Antimicrobial Resistance of Yeasts.
Journal of Medical Microbiology (2007), 56, 650–653 DOI 10.1099/jmm.0.46664-0
at 72uC. The RAPD reactions (10 ml) were analysed on a 2 % agarose gel containing 0.56 TBE plus ethidium bromide (0.4 mg l21). PFGE. PFGE analysis was performed as described in our previous report (Chen et al., 2005). The plug slices were placed in 200 ml buffer 3 solution (100 mM NaCl, 50 mM Tris/HCl, 10 mM MgCl2, 1 mM
DTT) (New England BioLabs) and incubated for 1 h at 50uC. Then they were transferred to 200 ml buffer 3 solution containing 4 units BssHII and incubated at 50 uC overnight. Electrophoreses were performed with a Biometra Rotaphor at pulse time 6–50 s, angle 120u, 180 V in 0.8 % agarose gel with 0.56 TBE for 36 h. After the electrophoresis, the gel was stained in ethidium bromide solution (0.4 mg l21) for 15 min and destained in distilled water.
Dendrogram analysis was performed with Bionumerics software, version 3.0 (Applied Maths). The similarity values of the fingerprints were based on the presence or absence of bands between each profile pair compared. The band inclusion window was adjusted by the size reference markers. The position tolerance was set at 1 % and optimization was set at 0 %. The Dice coefficient was used to analyse the similarities (SAB) of the band
patterns. UPGMA was used for the cluster analysis.
RESULTS AND DISCUSSION
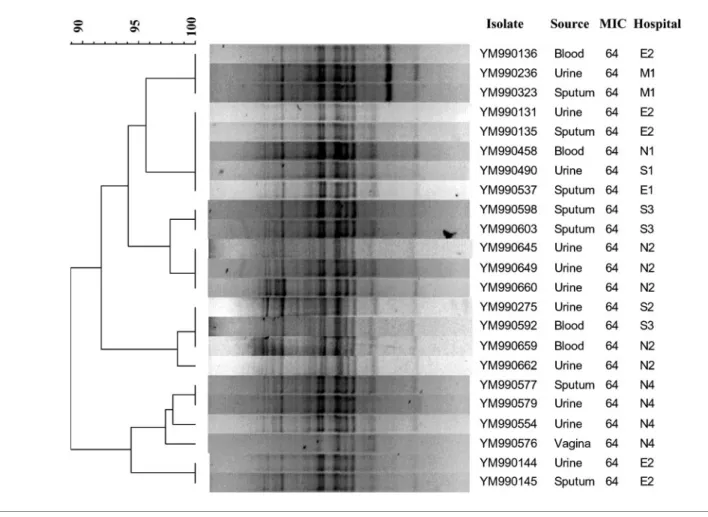
First of all, the genetic relatedness of all 23 resistant isolates
was analysed by RAPD assay as described by Roilides et al.
(2003). Interestingly, all the resistant isolates were closely
related (Fig. 1). There are two possible explanations for this
result. One is that all C. tropicalis isolates are genetically
related due to the intrinsic stability of their genomes. The
other is that all resistant ones were related due to clonal
spreading. To distinguish between these two possibilities,
we performed PFGE analysis as described previously (Chen
et al., 2005) to determine the genetic relatedness of the 23
resistant isolates along with 13 susceptible ones.
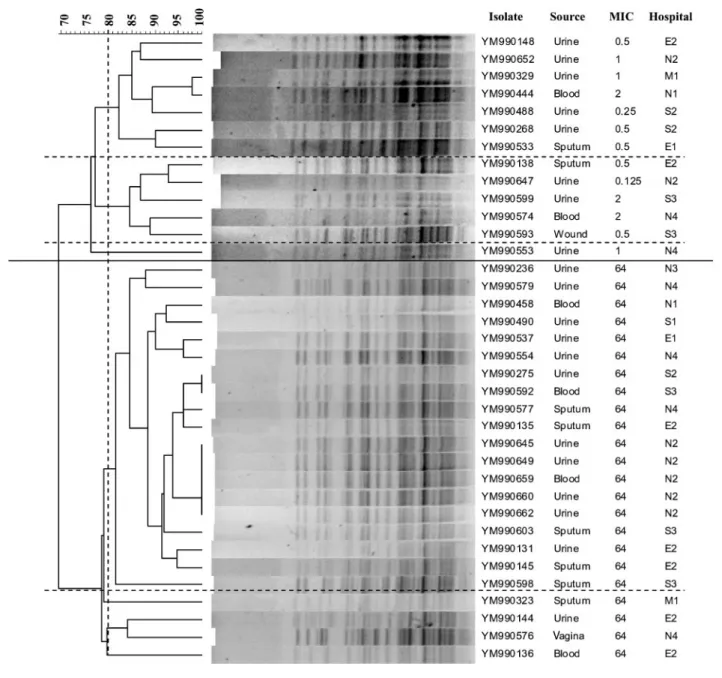
The result of the PFGE analysis is shown in Fig. 2. All of the
36 tested isolates were grouped into one of two pulsotypes,
which were independent of sources and hospitals, but were
closely associated with the fluconazole susceptibility. In
fact, all the 13 susceptible isolates tested belonged to one
pulsotype, while all the 23 resistant ones belonged to the
other. Among the 13 susceptible isolates, two subgroups
with .80 % relatedness were identified, one with 7 and the
other with 5 isolates. Among the 23 resistant ones, a major
subgroup consisting of 19 (82.6 %) isolates with .80 %
relatedness was also identified.
Molecular epidemiological surveillance of Candida species
isolated from an intensive care unit has been performed
Fig. 1. Dendrogram of the 23 fluconazole-resistant C. tropicalis isolates. Cluster analysis was based on the patterns of the RAPD assay. MIC refers to fluconazole. An MIC of 64 indicates the resistant isolates with MIC ¢64 mg l”1. E, East district; M, Middle district; N, North district; S, South district of Taiwan.
Closely related drug-resistant C. tropicalis
using RAPD analysis (Ergon & Gulay, 2005). There were 20
patterns among the 38 C. albicans isolates tested, suggesting
that the source of C. albicans was mostly endogenous,
consistent with other reports (Chen et al., 2001; Li et al.,
2006). In contrast, there were only 3 genotypes among a set
of 15 C. tropicalis isolates (Ergon & Gulay, 2005).
Further-more, through multilocus sequence typing analysis on C.
tropicalis, a set of clustered flucytosine-resistant isolates has
been reported recently (Tavanti et al., 2005). Interestingly, we
have also found relatively few genotypes among the 36
isolates tested by PFGE analysis. In fact, they can be divided
into only two groups as showed in Fig. 2. Whether this
phenomenon is due to clonal spreading or genome stability
needs further investigation. Though resistant isolates were
related, only one group consisted of more than two isolates
exhibiting .90 % relatedness. Therefore, if those resistant
isolates were clonal, microevolution over time has diluted
their genetic closeness. Alternatively, there was selective
pressure that edited out other genetic patterns.
According to the guidelines of the CLSI (Clinical and
Labo-ratory Standards Institute, 1997), the MICs to fluconazole
Fig. 2. Dendrogram of the 23 resistant and 13 susceptible C. tropicalis isolates. Cluster analysis of the 36 isolates was based on the patterns of BssHII restriction endonuclease analysis of genomic DNA. The long solid horizontal line separates the two major pulsotypes. The long dotted horizontal lines show the boundaries between subpulsotypes with .80 % relatedness. MIC refers to fluconazole. An MIC of 64 indicates the resistant isolates with MICs ¢64 mg l”1. E, East district; M, Middle district; N,
North district; S, South district of Taiwan. J.-S. Wang and others
are defined as the MICs of drugs capable of reducing the
turbidity of cells by .50 % after incubation at 35
uC for 48 h.
Isolates with MIC ¢64 mg l
21are considered to be
fluconazole resistant. Among the phenomena associated
with resistance, ‘trailing’ describes the reduced but persistent
growth that some isolates exhibit at drug concentrations
above the MIC in broth dilution tests with azole antifungal
agents, such as fluconazole (Lee et al., 2004). Trailing may
interfere with the observation of resistance levels in vivo. The
number of isolates exhibiting trailing with a particular MIC
at 48 h is approximately fourfold higher than at 24 h.
Therefore, the trailing growth can make an isolate that
appears susceptible (MIC ,64 mg l
21) after 24 h of
incubation to appear resistant (MIC ¢64 mg l
21) at 48 h
(Arthington-Skaggs et al., 2002). Thus, we would also like to
determine whether the 23 fluconazole-resistant isolates
exhibit trailing. Of the 23 isolates with MICs ¢64 mg l
21at 48 h, only 5, including YM990236, YM990275,
YM-990576, YM990579 and YM990649, exhibited MICs ¢64
mg l
21at 24 h. Nevertheless, the data obtained from in vitro
susceptibility testing are not always correlated with in vivo
outcome. Whether these 23 isolates with MICs ¢64 mg l
21will cause treatment failure needs further investigation.
The most important finding from this study was that the
fluconazole-resistant C. tropicalis isolates appeared to be
genetically related. Potentially, this will allow the
develop-ment of easy and even rapid identification methods for
clinical fluconazole-resistant or reduced-susceptibility C.
tropicalis using the genotyping information.
ACKNOWLEDGEMENTS
We would like to thank Pfizer for supplying fluconazole. We thank TSARY participating hospitals for providing clinical isolates and information regarding those isolates. We would like to express additional gratitude to the 10 hospitals providing resistant isolates. They are: Chang Gung Memorial Hospital, Buddhist Tzu-Chi General Hospital, Kaohsiung Medical University Chung-Ho Memorial Hospital, Hsin-Chu Hospital, Department of Health, the Executive Yuan, Linkou, Tainan Municipal Hospital, Hsu Foundation, Lo-Tung Poh Ai Hospital, Mackay Memorial Hospital Taitung Branch, Taipei Municipal Zen Ai Hospital, Veterans General Hospital – Kaohsiung, Veterans General Hospital – Taichung. This work was supported in parts by grants DOH94-DC-1102 from Center for Disease Control, Republic of China, NSC-94-2320-B-400-001 and NSC-94-2320-B-009-001 from the National Science Council, and CL-94-PP-05 from the National Health Research Institutes.
REFERENCES
Arthington-Skaggs, B. A., Lee-Yang, W., Ciblak, M. A., Frade, J. P., Brandt, M. E., Hajjeh, R. A., Harrison, L. H., Sofair, A. N. & Warnock, D. W. (2002).Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob Agents Chemother 46, 2477–2481.
Chen, Y. C., Chang, S. C., Tai, H. M., Hsueh, P. R. & Luh, K. T. (2001). Molecular epidemiology of Candida colonizing critically ill patients in intensive care units. J Formos Med Assoc 100, 791–797.
Chen, K. W., Lo, H. J., Lin, Y. H. & Li, S. Y. (2005).Comparison of four molecular typing methods to assess genetic relatedness of Candida albicans clinical isolates in Taiwan. J Med Microbiol 54, 249–258. Cheng, M. F., Yu, K. W., Tang, R. B., Fan, Y. H., Yang, Y. L., Hsieh, K. S., Ho, M. & Lo, H. J. (2004).Distribution and antifungal susceptibility of Candida species causing candidemia from 1996 to 1999. Diagn Microbiol Infect Dis 48, 33–37.
Clinical and Laboratory Standards Institute (1997). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, approved standard M27-A. Wayne, PA: Clinical and Laboratory Standards Institute.
Ergon, M. C. & Gulay, Z. (2005).Molecular epidemiology of Candida species isolated from urine at an intensive care unit. Mycoses 48, 126–131.
Lee, M. K., Williams, L. E., Warnock, D. W. & Arthington-Skaggs, B. A. (2004).Drug resistance genes and trailing growth in Candida albicans isolates. J Antimicrob Chemother 53, 217–224.
Li, S. Y., Yang, Y. L., Chen, K. W., Cheng, H. H., Chiou, C. S., Wang, T. H., Lauderdale, T. L., Hung, C. C. & Lo, H. J. (2006).Molecular epidemiology of long-term colonization of Candida albicans strains from HIV-infected patients. Epidemiol Infect 134, 265–269. Lo, H.-J., Ho, Y.-A. & Ho, M. (2001). Factors accounting for misidentification of Candida species. J Microbiol Immunol Infect 34, 171–177.
Pfaller, M. A., Jones, R. N., Doern, G. V., Sader, H. S., Messer, S. A., Houston, A., Coffman, S. & Hollis, R. J. (2000). Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997–1998. Antimicrob Agents Chemother 44, 747–751.
Roilides, E., Farmaki, E., Evdoridou, J., Francesconi, A., Kasai, M., Filioti, J., Tsivitanidou, M., Sofianou, D., Kremenopoulos, G. & Walsh, T. J. (2003). Candida tropicalis in a neonatal intensive care unit: epidemiologic and molecular analysis of an outbreak of infection with an uncommon neonatal pathogen. J Clin Microbiol 41, 735–741. Sanglard, D. & Odds, F. C. (2002).Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis 2, 73–85.
Tavanti, A., Davidson, A. D., Johnson, E. M., Maiden, M. C., Shaw, D. J., Gow, N. A. & Odds, F. C. (2005). Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Microbiol 43, 5593–5600.
Walsh, T. J., Groll, A., Hiemenz, J., Fleming, R., Roilides, E. & Anaissie, E. (2004). Infections due to emerging and uncommon medically important fungal pathogens. Clin Microbiol Infect 10 (Suppl. 1), 48–66.
Yang, Y. L., Cheng, H. H., Ho, Y. A., Hsiao, C. F. & Lo, H. J. (2003). Fluconazole resistance rate of Candida species from different regions and hospital types in Taiwan. J Microbiol Immunol Infect 36, 187–191.
Yang, Y. L., Ho, Y. A., Cheng, H. H., Ho, M. & Lo, H. J. (2004). Susceptibilities of Candida species to amphotericin B and fluconazole: the emergence of fluconazole resistance in Candida tropicalis. Infect Control Hosp Epidemiol 25, 60–64.
Yang, Y. L., Ho, Y. A., Cheng, H. H. & Lo, H. J. (2005).Distribution and susceptibility to amphotericin B and fluconazole of Candida spp. isolated from Taiwan. Epidemiol Infect 133, 325–330.
Closely related drug-resistant C. tropicalis